Abstract
The short genes encoding transfer RNA (tRNA) molecules are highly conserved in both sequence and structure, reflecting the central role of tRNA in protein biosynthesis. The frequent occurrence of fragmented intron-containing tRNAs that require processing to form contiguous molecules is therefore surprising. Recent discoveries of permuted and split tRNA genes have added to the apparent creativity of nature regarding the organization of these fragmented genes. Here, we provide an overview of the various types of fragmented tRNA genes and examine the hypothesis that the integration of mobile genetic elements—including viruses and plasmids—established such genes in pieces.
Keywords: genome integration, intron, mobile elements, splicing, tRNA
Introduction
Central to the process of protein biosynthesis are transfer RNAs (tRNAs), which translate individual codons of a messenger RNA (mRNA) into the corresponding amino acid, and thereby form the link between the genetic code and protein sequence. To fulfil this role, a subset of tRNA species exists for each of the canonical 20 amino acids, which is recognized and aminoacylated specifically by its cognate aminoacyl-tRNA synthetase (reviewed by Ibba & Söll, 2000). The resulting aminoacyl-tRNAs are then delivered to the translating ribosome where they are required to fit into its tRNA-binding sites (reviewed by Ramakrishnan, 2002). Therefore, all canonical tRNAs show a conserved cloverleaf-shaped secondary structure that folds into an L-shaped tertiary structure. The central function of tRNAs in these crucial cellular processes accounts for their highly conserved sequence and structure, as well as their restricted length, which averages 76 nucleotides (nt; Marck & Grosjean, 2002).
Here, we focus on the remarkably diverse arrangements of tRNA genes that are not directly transcribed into a standard contiguous tRNA. Instead, in these instances, the initial transcript is a disrupted precursor that is subsequently enzymatically processed to form mature tRNA. Given the small size of tRNA, it is remarkable to see the diverse ways in which these genes are disrupted. Here we give a synopsis of these fragmented tRNA genes and describe how the cell assembles the functional tRNAs. Finally, we present our view on the evolution of such ‘tRNA genes in pieces'.
Introns
The most common and best-described disruption of tRNA genes is the presence of an intron, which was initially found to be inserted exclusively 1 nt after the anticodon-adjacent position 37 of eukaryotic tRNAs (Goodman et al, 1977; Valenzuela et al, 1978). This intron is removed by the eukaryal splicing endonuclease that recognizes the body (mature domain) of a tRNA molecule and measures the fixed distance to the splice sites (Reyes & Abelson, 1988). Subsequently, an RNA ligase joins the resultant tRNA halves and a 2′-phosphotransferase removes the 2′-phosphate from the splice junction.
By contrast, bacterial tRNAs are found to contain a different type of intron, the group I intron ribozyme, in their anticodon loops. This is able to self-splice either without the help of protein cofactors (Adams et al, 2004; Reinhold-Hurek & Shub, 1992) or with the help of splicing factors, including mitochondrial tyrosyl-tRNA synthetase (Paukstelis et al, 2008).
Finally, the analysis of archaeal genomes allowed the identification and investigation of several more varied introns inserted at numerous positions within the tRNA molecule (reviewed by Marck & Grosjean, 2003). It became clear that in certain members of the Archaea, introns are also located in the anticodon stem, the acceptor stem, the D-loop, the T-loop and the variable region. Common to all archaeal tRNA introns, a structural motif is formed at the intron–exon junction that is recognized and cleaved by the archaeal splicing endonuclease. This motif is known as the bulge–helix–bulge motif (BHB) and is characterized by a central conserved 4 base-pair (bp) helix flanked by two 3-nt bulges with an approximate twofold symmetry (Fig 1A,B; Diener & Moore, 1998; Xue et al, 2006). The eukaryal splicing endonuclease naturally excises introns based on their distance to the mature tRNA domain; however, it has also been shown to recognize and cleave BHB motifs both in vitro and in vivo (Di Segni et al, 2005; Fabbri et al, 1998). The activity of archaeal splicing endonucleases relies solely on the presence of these BHB motifs in a so-called ‘mature-domain-independent fashion'. The nature of the archaeal RNA ligase is still unknown.
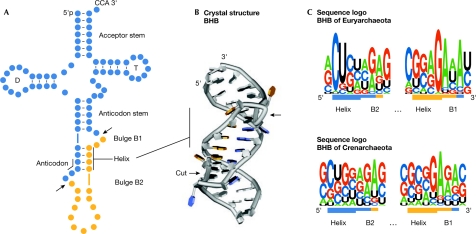
Figure 1.
Characteristics of the archaeal bulge–helix–bulge splicing motif. The colouring scheme in all three panels indicates the transfer RNA (tRNA; blue) and the intron (orange). Splice sites are indicated by arrows. (A) Schematic secondary structure of an intron-containing pre-tRNA. The conserved features of a tRNA are indicated, D and T indicate the D-loop and the T-loop respectively. (B) Crystal structure of a bulge–helix–bulge (BHB) motif taken from the complex structure with splicing endonuclease from Archaeoglobus fulgidus (Xue et al, 2006). (C) Sequence logo representation of 44 euryarchaeal (top) and 137 crenarchaeal (bottom) BHB motifs (extracted from Marck & Grosjean, 2003; Sugahara et al, 2007).
The extent of conservation of both sequence and structure of the BHB motif varies among the archaeal kingdoms. The members of the Euryarchaeota show a relatively conserved BHB motif, which is located mostly in the anticodon loop and is recognized by homomeric splicing endonucleases (Li et al, 1998; Lykke-Andersen & Garrett, 1997). By contrast, the members of the Crenarchaeota show more relaxed BHB motifs, which are also inserted in non-anticodon tRNA regions that are processed by a heterotetrameric splicing endonuclease (Calvin et al, 2005; Randau et al, 2005a; Tocchini-Valentini et al, 2005; Yoshinari et al, 2005). In these organisms, the structural splicing motif can sometimes be reduced to the central 4-bp helix flanked by only one 3-nt bulge and a loop of varying length. Sequence conservation of the BHB motif can be examined by a sequence logo representation of an alignment of archaeal BHB sequences in which the height of each letter stack represents the extent of conservation (Fig 1C; Crooks et al, 2004). The initial 2 nt of the euryarchaeal BHB helix are most often C and U; the conserved nucleotides interact with ribosomal RNA and precede the anticodon of the tRNA. The bulge containing two intron-derived nucleotides (B1 in Fig 1) shows a greater conservation with predominant guanosine and adenosine residues at the helix–bulge B1 junction (Fig 1C). This adenosine was shown to be the most important recognition element by the archaeal splicing endonuclease and is positioned at a sharp bend in a conserved cross-subunit arginine–nucleotide–arginine stack (Xue et al, 2006). Nevertheless, the overall conservation of the BHB sequence seems to be lower than originally thought (Kleman-Leyer et al, 1997). It should be noted that BHB motifs were also found at archaeal introns of both ribosomal RNA (rRNA) and mRNA (Kjems & Garrett, 1988; Tang et al, 2002; Watanabe et al, 2002).
Recent computational studies predicted the simultaneous presence of two introns in certain crenarchaeal tRNAs and even three introns in tRNAPro of Thermofilum pendens Hrk 5 (Sugahara et al, 2007). In this specific case, the pre-tRNA folds into a conformation that allows the splicing of a third intron only after the splicing of the first two introns (Fig 2B). The average length of reported archaeal tRNA introns is approximately 20–25 nt, and the individual length varies between 11 and 175 nt. It is likely that such long introns—longer than the tRNA—have a functional role. This is exemplified by the 104-bp intron of Halobacterium volcanii tRNATrp, which is suggested to mediate the 2′-O-methylation in cis of 2 nt of tRNA (Clouet-d'Orval et al, 2005).
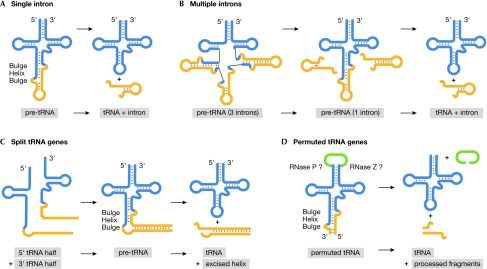
Figure 2.
Schematic overview of pre-transfer RNA processing of unusual transfer RNA gene products. (A) Processing of single introns in members of the Archaea. The conserved structural bulge–helix–bulge (BHB) motif is recognized by the splicing endonuclease. (B) Processing of up to three introns in Thermofilum pendens. Two BHB motif-containing elements are spliced out leading to the formation of pre-transfer RNA (pre-tRNA) with a third intron. (C) Split tRNAs in Nanoarchaeum equitans are assembled by long reverse-complementary sequences. The helix–tRNA junctions fold into the BHB motif, which are recognized by the splicing endonuclease. (D) Permuted tRNAs in Cyanidioschyzon merolae show a splicing motif on folding of the reversed tRNA sequences. The circular product is probably processed by RNase P and RNase Z to yield the mature 5′ and 3′ termini.
Split tRNA genes
A unique type of tRNA gene disruption was found in the genome of Nanoarchaeum equitans, which is the only known organism to contain tRNA half-genes as well as four intron-containing tRNA genes (Randau et al, 2005b). Both a 5′ tRNA half-gene and a 3′ tRNA half-gene exist for six tRNA genes, which are distributed widely throughout the genome (Randau et al, 2005c). One exception is that two 5′ tRNAGlu isoacceptor halves share the same 3′ tRNAGlu half, which raises the interesting questions of whether and how transcription is controlled to guarantee the production of these molecules in a 1:1 ratio (from 2:1 half-genes). Each split tRNA gene is preceded by its own promoter, so that the initial transcripts are tRNA half-molecules that need to find each other in the cell (Fig 2C). This is guaranteed by a 12–14 bp GC-rich sequence after the 5′ tRNA half that has a perfect reverse-complementary match to a sequence preceding its corresponding 3′-tRNA half. Once these sequences are annealed, a helix forms that is proposed to facilitate the folding of the tRNA body. The junctions of these helices and the tRNAs form the intron-characteristic BHB motifs that are then recognized and spliced by the N. equitans splicing endonuclease (Randau et al, 2005a). This enzyme shows the heteromeric subunit conformation found in members of the Crenarchaeota that seems to be optimized for the recognition of non-canonical introns. As the six split tRNAs, as well as the four intron-containing tRNAs, are processed by the same splicing machinery, a common origin of both phenomena seems likely.
N. equitans shows another unique feature in the organization of its tRNA genes: it is the only known organism that has been shown to circumvent the need for RNase P, which is the otherwise universal ribozyme required for the removal of 5′ leader sequences from pre-tRNA transcripts (Altman et al, 1989). In N. equitans, accurate promoter placement ensures that all tRNA genes begin transcription at the first nucleotide of the mature tRNA, resulting in leaderless tRNAs. Transcription initiation can only proceed with a purine; therefore, the three tRNAs that require a pyrimidine at position one contain an additional 5′ purine base for tRNA genes. The apparent loss of RNase P in this organism is also supported by the detection of mature 5′-triphosphorylated tRNA species in total RNA extracts (Randau et al, 2008).
Permuted tRNA genes
Recently, another example of an unusual tRNA gene disruption was found in the nuclear genome of the red alga Cyanidioschyzon merolae (Soma et al, 2007). In total, 11 tRNA genes were identified in which the 3′ region of the tRNA is located upstream of the 5′ half of the tRNA gene. The two regions are separated by an intervening sequence 7–74 bp in length. On folding of the tRNA, this intervening sequence connects the mature termini of the tRNA in a circular fashion (Fig 2D). It is proposed that the enzymes usually involved in processing the 5′ terminus (RNase P) and the 3′ terminus (for example, RNase Z) of a pre-tRNA molecule are able to precisely cut and remove this sequence from the circular intermediate. Therefore, the first step in the maturation of this tRNA is the removal of the 5′-leader and 3′-trailer sequences unusually located in the anticodon region of the tRNA (Fig 2D). BHB motifs were proposed to form at the junctions of these sequences and the tRNA body, which would then be recognized and cleaved by the C. merolae splicing endonuclease. Subsequently, an RNA ligase would circularize these tRNA precursors. This mechanism raises several interesting questions. First is how these tRNAs are transcribed, as eukaryotic tRNAs typically contain RNA polymerase III elements located within the tRNA gene. These elements are known as A box and B box, and overlap with conserved regions of the D-arm and T-arm portions of the tRNA gene. This promoter arrangement is not possible in permuted tRNA genes, in which the T-arm lies unusually upstream of the D-arm. To compensate for the lack of the standard internal promoter, an upstream sequence has been proposed to be a potential external promoter resembling the archaeal promoter organization. Second, as the occurrence of BHB-splicing motifs was previously thought to be an archaeal phenomenon, it will be of interest to determine to what extent the C. merolae splicing endonuclease resembles its archaeal counterpart, and to investigate how—and why—this peculiar algae adapted the described archaeal-like tRNA-processing elements.
Evolution of disrupted tRNA genes
The situations described above have led us to consider the evolution of the archaeal tRNA gene disruptions. Initially, one has to ask, what came first: did contiguous tRNA genes acquire introns and other gene disruptions over time, or did ancient fragmented tRNA genes evolve into connected versions?
There is substantial evidence that the modern tRNA gene originated from tRNA halves. The finding that a minihelix—that is, the acceptor stem plus T-arm—is an authentic substrate for several aminoacyl-tRNA synthetases provides what is possibly the best support for a hypothesis stating that this highly conserved part of the tRNA is older than the anticodon-arm–D-arm portion of the same molecule (Schimmel & Ribas de Pouplana, 1995; Weiner & Maizels, 1987; Widmann et al, 2005).
Nevertheless, it is questionable whether this origin of tRNA from two different hairpin portions is still reflected in modern fragmented tRNA genes. In this context, it is important to understand that these two coaxially stacked hairpins (minihelix and anticodon-arm–D-arm) do not comprise the exact regions of 5′ and 3′ half molecules separated in the anticodon loop. The tRNA halves created by canonical introns cannot fold to form the acceptor stem and the anticodon stem.
It has also been argued that the split tRNA genes of the supposedly early diverging N. equitans represent the ancient tRNA gene organization (Di Giulio, 2006). However, recent reinvestigation of the phylogenetic position of N. equitans placed this organism as a member of a fast-evolving euryarchaeal lineage with significant parasitic adaptation (Brochier et al, 2005; Das et al, 2006). Therefore, it seems likely that the split tRNA genes are derived from intron-containing genes that were separated in the process of genome rearrangement. This is in accordance with the observation that many conserved gene clusters are found separated in N. equitans, and that several split genes encoding conserved and usually contiguous proteins are present in its genome (Waters et al, 2003).
In our opinion, a scenario of late acquisition of introns by contiguous tRNA genes seems more plausible. Possibly the most convincing evidence for this derives from the identification of the highly variable intron-insertion sites in crenarchaeal tRNA genes. Here, the BHB-type introns do not simply disconnect two assumed ancient tRNA halves in the anticodon loop, but are inserted in different tRNA domains. For example, the introns in the acceptor stem of three tRNAs of the crenarchaeon Pyrobaculum aerophilum disconnect only the first 3 nt from the rest of the tRNA. Further evidence is provided by the simultaneous presence of two or three partly overlapping introns in tRNAs from P. aerophilum, T. pendens or Methanopyrus kandleri, which indicates a cumulative acquisition of these introns.
Integration at tRNA genes
What causes tRNA genes to maintain introns? We believe that to answer this question it is desirable to investigate the interactions of mobile genetic elements such as conjugative plasmids and viruses—both of which integrate into tRNA genes—with their hosts. Viruses are described as the most abundant biological entity on Earth, clearly outnumbering their hosts (Bergh et al, 1989). Although much less is known about archaeal viruses than their bacterial counterparts, their abundance is evident and many archaeal viruses have been isolated from hydrothermal environments, which are also home to the tRNA-intron-rich extremophiles discussed above (Rice et al, 2001). The viruses and virus-like particles with crenarchaeal hosts all show unusual and diverse morphologies, and are known to infect the genera Sulfolobus, Acidianus, Thermoproteus and Pyrobaculum. The remarkable range of these viruses is exemplified by electron-microscopic studies of the viral particles found in two hot springs at Yellowstone National Park in the USA (Rachel et al, 2002). Potential hosts include the archaeal genera Thermophilum, Thermoproteus, Pyrobaculum, Thermospheaera and Archaeoglobus, as well as members of the archaeal groups Desulfurococcales, Korarchaeota and Nanoarchaeota (Rachel et al, 2002).
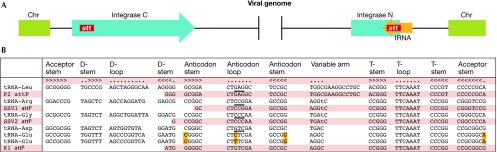
Many mobile genetic elements integrate into the archaeal chromosome by site-specific recombination occurring within a short region of perfect homology between the host chromosome (attB sequence) and the viral or plasmid DNA (attP sequence; Reiter et al, 1989). Their typical targets are tRNA genes, and this concept is considered to have been conserved during the evolution of integrating viruses of the Archaea, Bacteria and Eukaryota (Reiter et al, 1989; Winckler et al, 2005). The best-studied archaeal model for viral integration focuses on members of the family Fuselloviridae. Here, full-length copies of spindle-shaped virus (SSV) genomes are found integrated into the tRNA genes of their host. The viral-encoded SSV1 integrase acts as a site-specific endonuclease and ligase (She et al, 2004). On insertion into the host genome, the integrase gene is partitioned into an amino-terminal portion that overlaps with the reconstituted tRNA gene target and a carboxy-terminal portion with a direct repeat of the tRNA fragment-attachment site (Fig 3). The attB site of SSV1 is a 44-bp fragment of a tRNAArg gene (Reiter et al, 1989), SSV2 contains a 48-bp fragment identical to a portion of the host tRNAGly gene and SSV RH (isolated from the Ragged Hills area of Yellowstone National Park) contains a 59-bp attB site that is identical to a host tRNALeu gene (Wiedenheft et al, 2004). In most cases, the attB sites comprise the 3′ region of the tRNA with its highly conserved T-arm and the anticodon loop, which is the site of most introns (Fig 3B). One interesting example is the integration of the SSV K1 genome, which was shown to target the tRNAAsp gene and, surprisingly, also two tRNAGlu genes of Sulfolobus solfataricus (Fig 3B). The attB sites of tRNAAsp and the two tRNAGlu differ only in 4 nt, and the viral attP sequence comprises exactly the 49-nt of the 3′ terminal tRNAAsp. However, if integration into tRNAGlu occurs, the reconstituted tRNA gene acquires the viral attB site and is mutated (Fig 3B). This process introduces four mutations that alter the anticodon and the discriminator base of the tRNA, and switches its identity from tRNAGlu to tRNAAsp. As the attachment site is misinterpreted in the original paper (Wiedenheft et al, 2004), it has been adjusted to account for the correct orientation of the tRNA gene (Fig 3B). It would be interesting to investigate whether these chimeric tRNAs are functional in protein biosynthesis. This imprecise integration not only provides a fascinating insight into the evolutionary relationship of these two tRNA species but could also be an obvious reason why the host organism would attempt to prevent such viral integration.
Figure 3.
Integration of archaeal viruses into transfer RNA genes and attachment sites. (A) Schematic presentation of a spindle-shaped virus (SSV) genome integrated in its host chromosome (Chr). The amino-terminus of the partitioned integrase gene (integrase N) restores the 3′ terminal portion of the targeted tRNA gene (orange) with its attachment site (att). A direct repeat of this sequence is present in the carboxy-terminal integrase fragment (integrase C). (B) Four examples of attachment sites of SSV viruses and the corresponding transfer RNA (tRNA) sequences of the Sulfolobus host. The secondary structure of the tRNA is indicated as helices (‘<' and ‘>' symbols) and unpaired bases (dots). The integration of SSV K1 mutates potential tRNAGlu targets at four positions (yellow), which changes the identity of the tRNA species from Glu to Asp (modified from Wiedenheft et al, 2004).
Many diverse archaeal viruses have been discovered, and shown to be inserted into the 5′-distal and 3′-distal regions of tRNA genes (Krupovic & Bamford, 2008; Pagaling et al, 2007). Interestingly, there are also examples of viruses that contain tRNA genes, and even tRNA genes containing introns (Haring et al, 2005).
The highly conserved tRNA and rRNA genes are ideal integration targets, and, frequently, introns can be found in these genes. It can be easily argued that the presence of an intron in a tRNA gene is a valuable protection mechanism against the integration of viruses and other autonomous genetic elements. Similarly, split and permuted tRNA genes would be able to prevent such integration if the attB sites are disrupted. This scenario would explain the direct evolutionary pressure for maintaining disrupted tRNA genes instead of mutating them into a contiguous version. As the different attP sites are specific for the various individual mobile elements, only the directly targeted tRNA genes would benefit from the disruption of their corresponding attB sites. Following this line of thought, an organism would contain more introns either if it was a preferred target for an increased number of mobile elements or if the integrative elements were of particular harm to the organism. One example might be the extremely small and condensed genome of N. equitans; in this organism, the obvious need for genome reduction would benefit from a lack of integration.
A different perspective is that the formation of direct repeats of tRNA fragments on integration of viral particles might provide a mechanism for the generation of the more unusual tRNA gene disruptions discussed above. To understand the details of how permutations and split tRNAs might arise from insertion and excision events, it is desirable to investigate mobile genetic elements in organisms with a high tRNA-intron rate. Additional plasmids and viruses carrying integrase genes are no doubt awaiting discovery, and will aid our understanding of the full extent of their impact on genome evolution.

Dieter Söll (left) & Lennart Randau
Acknowledgments
We thank P. O'Donoghue, S. Palioura, L. Sherrer and J. Yuan for help and encouragement. This work was supported by grants from the National Institute of General Medical Sciences and the Department of Energy (to D.S.).
References
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA (2004) Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 [DOI] [PubMed] [Google Scholar]
- Altman S, Baer MF, Bartkiewicz M, Gold H, Guerrier-Takada C, Kirsebom LA, Lumelsky N, Peck K (1989) Catalysis by the RNA subunit of RNase P: a minireview. Gene 82: 63–64 [DOI] [PubMed] [Google Scholar]
- Bergh O, Borsheim KY, Bratbak G, Heldal M (1989) High abundance of viruses found in aquatic environments. Nature 340: 467–468 [DOI] [PubMed] [Google Scholar]
- Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P (2005) Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol 6: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvin K, Hall MD, Xu F, Xue S, Li H (2005) Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J Mol Biol 353: 952–960 [DOI] [PubMed] [Google Scholar]
- Clouet-d'Orval B, Gaspin C, Mougin A (2005) Two different mechanisms for tRNA ribose methylation in Archaea: a short survey. Biochimie 87: 889–895 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Paul S, Bag SK, Dutta C (2006) Analysis of Nanoarchaeum equitans genome and proteome composition: indications for hyperthermophilic and parasitic adaptation. BMC Genomics 7: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio M (2006) The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA). J Theor Biol 240: 343–352 [DOI] [PubMed] [Google Scholar]
- Di Segni G, Borghese L, Sebastiani S, Tocchini-Valentini GP (2005) A pre-tRNA carrying intron features typical of Archaea is spliced in yeast. RNA 11: 70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener JL, Moore PB (1998) Solution structure of a substrate for the archaeal pre-tRNA splicing endonucleases: the bulge–helix–bulge motif. Mol Cell 1: 883–894 [PubMed] [Google Scholar]
- Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola Negri E, Baldi MI, Attardi DG, Mattoccia E, Tocchini-Valentini GP (1998) Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280: 284–286 [DOI] [PubMed] [Google Scholar]
- Goodman HM, Olson MV, Hall BD (1977) Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci USA 74: 5453–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Vestergaard G, Brugger K, Rachel R, Garrett RA, Prangishvili D (2005) Structure and genome organization of AFV2, a novel archaeal lipothrixvirus with unusual terminal and core structures. J Bacteriol 187: 3855–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Söll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69: 617–650 [DOI] [PubMed] [Google Scholar]
- Kjems J, Garrett RA (1988) Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell 54: 693–703 [DOI] [PubMed] [Google Scholar]
- Kleman-Leyer K, Armbruster DW, Daniels CJ (1997) Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell 89: 839–847 [DOI] [PubMed] [Google Scholar]
- Krupovic M, Bamford DH (2008) Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology 25: 292–300 [DOI] [PubMed] [Google Scholar]
- Li H, Trotta CR, Abelson J (1998) Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280: 279–284 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Garrett RA (1997) RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J 16: 6290–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C, Grosjean H (2002) tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C, Grosjean H (2003) Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA 9: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagaling E, Haigh RD, Grant WD, Cowan DA, Jones BE, Ma Y, Ventosa A, Heaphy S (2007) Sequence analysis of an Archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics 8: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL (2008) Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature 451: 94–97 [DOI] [PubMed] [Google Scholar]
- Rachel R, Bettstetter M, Hedlund BP, Haring M, Kessler A, Stetter KO, Prangishvili D (2002) Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch Virol 147: 2419–2429 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V (2002) Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Randau L, Calvin K, Hall M, Yuan J, Podar M, Li H, Söll D (2005a) The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves non-canonical bulge-helix-bulge motifs of joined tRNA halves. Proc Natl Acad Sci USA 120: 17934–17939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, Münch R, Hohn MJ, Jahn D, Söll D (2005b) Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433: 537–541 [DOI] [PubMed] [Google Scholar]
- Randau L, Pearson M, Söll D (2005c) The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett 579: 2945–2947 [DOI] [PubMed] [Google Scholar]
- Randau L, Schröder I, Söll D (2008) Life without RNase P. Nature 453: 120–123 [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Shub DA (1992) Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357: 173–176 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Palm P, Yeats S (1989) Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res 17: 1907–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes VM, Abelson J (1988) Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55: 719–730 [DOI] [PubMed] [Google Scholar]
- Rice G, Stedman K, Snyder J, Wiedenheft B, Willits D, Brumfield S, McDermott T, Young MJ (2001) Viruses from extreme thermal environments. Proc Natl Acad Sci USA 98: 13341–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P, Ribas de Pouplana L (1995) Transfer RNA: from minihelix to genetic code. Cell 81: 983–986 [DOI] [PubMed] [Google Scholar]
- She Q, Shen B, Chen L (2004) Archaeal integrases and mechanisms of gene capture. Biochem Soc Trans 32: 222–226 [DOI] [PubMed] [Google Scholar]
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y (2007) Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318: 450–453 [DOI] [PubMed] [Google Scholar]
- Sugahara J, Yachie N, Arakawa K, Tomita M (2007) In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge–helix–bulge splicing motifs. RNA 13: 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B, Branlant C, Bachellerie JP, Brosius J, Huttenhofer A (2002) RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 30: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP (2005) Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc Natl Acad Sci USA 102: 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P, Venegas A, Weinberg F, Bishop R, Rutter WJ (1978) Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA. Proc Natl Acad Sci USA 75: 190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Yokobori S, Inaba T, Yamagishi A, Oshima T, Kawarabayasi Y, Kikuchi H, Kita K (2002) Introns in protein-coding genes in Archaea. FEBS Lett 510: 27–30 [DOI] [PubMed] [Google Scholar]
- Waters E et al. (2003) The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA 100: 12984–12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Maizels N (1987) tRNA-like structures tag the 3′ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci USA 84: 7383–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann J, Giulio MD, Yarus M, Knight R (2005) tRNA creation by hairpin duplication. J Mol Evol 61: 524–530 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M (2004) Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol 78: 1954–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler T, Szafranski K, Glockner G (2005) Transfer RNA gene-targeted integration: an adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome. Cytogenet Genome Res 110: 288–298 [DOI] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H (2006) RNA recognition and cleavage by a splicing endonuclease. Science 312: 906–910 [DOI] [PubMed] [Google Scholar]
- Yoshinari S, Fujita S, Masui R, Kuramitsu S, Yokobori S, Kita K, Watanabe Y (2005) Functional reconstitution of a crenarchaeal splicing endonuclease in vitro. Biochem Biophys Res Commun 334: 1254–1259 [DOI] [PubMed] [Google Scholar]