Abstract
Polyglutamylation is a post-translational modification in which glutamate side chains of variable lengths are formed on the modified protein. It is evolutionarily conserved from protists to mammals and its most prominent substrate is tubulin, the microtubule (MT) building block. Various polyglutamylation states of MTs can be distinguished within a single cell and they are also characteristic of specific cell types or organelles. Polyglutamylation has been proposed to be involved in the functional adaptation of MTs, as it occurs within the carboxy-terminal tubulin tails that participate directly in the binding of many structural and motor MT-associated proteins. The discovery of a new family of enzymes that catalyse this modification has brought new insight into the mechanism of polyglutamylation and now allows for direct functional studies of the role of tubulin polyglutamylation. Moreover, the recent identification of new substrates of polyglutamylation indicates that this post-translational modification could be a potential regulator of diverse cellular processes.
Keywords: microtubules, polyglutamylase, post-translational modification, TTLL, tubulin
Introduction
The enzymatic addition or removal of chemical cues—referred to as post-translational modification—provides powerful mechanisms to regulate protein function rapidly and reversibly. Polyglutamylation is a modification that consists of the generation of polyglutamate side chains at specific sites on a target protein (Fig 1). It was initially discovered on the proteins that build microtubules (MTs), the α-tubulins and β-tubulins (Edde et al, 1990; Alexander et al, 1991; Redeker et al, 1992; Rüdiger et al, 1992). The length of the added side chains can vary from 1 to about 20 glutamyl units, which allows diverse signals to be encoded on a single modification site. The selectivity for α- or β-tubulin, as well as the presence of several modification sites within a single tubulin molecule, increase further the complexity of the signals that polyglutamylation can generate on MTs. Understanding the impact of the information encoded by polyglutamylation on the regulation of cellular function is therefore especially challenging (Sidebar A). Here, we review the current knowledge of polyglutamylation, from its discovery on tubulin to the recent identification of the enzymes that catalyse it.
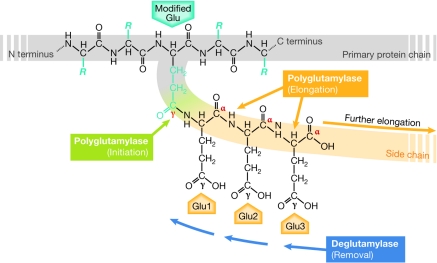
Figure 1.
Mechanism of polyglutamylation. Polyglutamylation takes place on a glutamate residue within the primary sequence of the modified protein (modified Glu). The first reaction, which is initiation (green), generates an amide bond between the γ-carboxyl group of the modified glutamate and the amino group of the first glutamyl unit (Glu1). Subsequent elongation reactions (orange) link the α-carboxyl group of the preceding glutamyl unit with the amino group of the next unit (Glu2, Glu3 and so on). Deglutamylase activity can shorten the side chain. C terminus, carboxy terminus; N terminus, amino terminus.
The discovery of tubulin polyglutamylation
MTs can be dedicated to many distinct functions, even within the same cell. The generation of functionally diverse MTs results from the molecular heterogeneity of tubulin. Besides the fact that MTs can be diversified by the incorporation of different α- and β-tubulin isotypes (Luduena, 1998), it was appreciated at an early stage that post-translational modifications of α- and β-tubulin can rapidly and locally adapt MTs to specific functions. Several groups searched for such modifications by analysing the particularly high number of tubulin variants observed in the brain (Field & Lee, 1988; Wolff et al, 1982). The team of Bernard Eddé and Philippe Denoulet found that a glutamate residue located within the carboxy-terminal tail of α-tubulin is modified by the attachment of polyglutamate side chains of variable lengths (Edde et al, 1990). The first glutamate is added by the formation of a covalent bond between its amino group and the γ-carboxyl group of the glutamate that is modified, in a reaction called initiation. Subsequent elongation reactions are responsible for the formation of glutamate side chains. Although the α- and γ-carboxyl groups of the glutamyl units of the side chains are both available for elongation, the new glutamates are linked mostly to the α-carboxyl groups of the preceding glutamyl unit (Fig 1; Redeker et al, 1991; Wolff et al, 1994).
Soon after the discovery of polyglutamylation on α-tubulin, it was shown that this modification also targets the C-terminal tail regions of β-tubulin (Alexander et al, 1991; Redeker et al, 1992; Rüdiger et al, 1992). Moreover, indirect observations pointed to the existence of an enzymatic activity that removes the polyglutamylation from tubulin, and therefore implied that polyglutamylation could be a reversible process (Audebert et al, 1993; Regnard et al, 1999). The balance of modifying and demodifying enzymes could provide a flexible control mechanism for polyglutamylation, making the modification a highly modular regulatory event.
Tubulin polyglutamylation discriminates MT species
Detailed studies of the distribution of tubulin polyglutamylation were made possible by the development of the first monoclonal antibody specific to the modification, glutamylated tubulin clone number 335 (GT335). This antibody was raised against a peptide that mimics the C-terminal tail of α-tubulin modified with a side chain of two glutamyl units. GT335 detects tubulin glutamylation irrespective of the length of the side chains, and it labels all of the glutamylated forms of α- and β-tubulin on western blots after two-dimensional gel electrophoresis. In immunofluorescence, GT335 strongly labels all of the MTs in neurons (Wolff et al, 1992). In most other cell types, cytoplasmic MTs have relatively low polyglutamylation levels; only centrioles, which are specialized MT structures at the cores of centrosomes, are strongly polyglutamylated in all cell types (Bobinnec et al, 1998a)—an observation that led to an unexpected career for GT335 as a centriole marker. In addition, a temporary and locally restricted appearance of polyglutamylation was found in cell division, during which a notable increase in β-tubulin modification occurs on the MTs of the mitotic spindle and the midbody (Bobinnec et al, 1998a; Regnard et al, 1999). Another MT structure with high levels of polyglutamylation is the axoneme, which is the central structure of cilia and flagella. The presence of the modification in mammalian spermatozoa (Fouquet et al, 1994), as well as in ciliated and flagellated unicellular organisms (Bre et al, 1994; Weber et al, 1997), further indicated that polyglutamylation is evolutionarily conserved from protists to mammals.
Thus GT335 helped to identify the hotspots of polyglutamylation as the MTs in neurons, axonemes and centrioles, as well as cell-cycle-related MT structures such as the mitotic spindle and the midbody (Fig 2A). More-detailed analysis of the modification in these different MT species using mass spectrometry and two-dimensional gel electrophoresis showed that the length of the glutamyl side chains and the distribution of the modification between α- and β-tubulin are highly diverse (Fig 2B). The possibility of modulating these two parameters allows polyglutamylation to generate a large range of modification patterns on MTs, which could contribute to their adaptation to various functions.
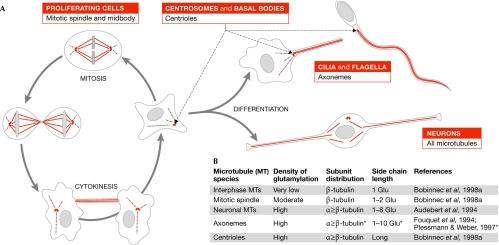
Figure 2.
The occurrence of tubulin polyglutamylation in mammalian cells. (A) Schematic representation of microtubule (MT) species that are modified by polyglutamylation. In proliferating cells, cytoplasmic MTs are polyglutamylated at low levels in interphase. During cell division, polyglutamylation is locally increased on the mitotic spindle and the midbody. Polyglutamylation is also enriched in neurons, as well as on specialized MT structures such as centrioles and axonemes. (B) Parameters of polyglutamylation patterns observed on different MT species in mammals. Values can differ in more distant species. Asterisks indicate that conflicting results about the final length of the side chains have been obtained, probably owing to the methods applied.
The potential roles of tubulin polyglutamylation
Polyglutamylation occurs within the C-terminal tail domain of tubulins, which are the binding sites for many MT-associated proteins (MAPs) and molecular motors. The presence of a highly modular post-translational modification at these sites indicated that it could regulate MT–MAP interactions (Fig 3). Support for this hypothesis was obtained by blot-overlay assays of brain tubulin that had been resolved in two-dimensional electrophoresis. In these experiments, the affinity of MAP1B, MAP2, tau and neuronal kinesin increased progressively for tubulins bearing 1–3 glutamyl units, and then decreased sharply for longer chains. By contrast, MAP1A maintained an affinity for tubulin with long side chains (Bonnet et al, 2001; Boucher et al, 1994; Larcher et al, 1996). These results are not conclusive because the tubulin molecule is denatured under the experimental conditions that were used. Nonetheless, they imply that polyglutamylation could regulate interactions between MTs and MAPs, and that this regulation might be based on the presence or absence of polyglutamylation, and also be fine-tuned by the length of the glutamyl side chains. Moreover, polyglutamylation could regulate not only the binding but also the activity of some MAPs and motors. For example, it has been shown that the processivity of kinesins is mediated by electrostatic interactions between a basic neck-linker region of the motor and the C-terminal tail of the tubulins. Reinforcing or reducing these interactions by artificially adding positive or negative charges to the kinesin neck region respectively increased or decreased the processivity of the motor (Thorn et al, 2000). Therefore, polyglutamylation might, by increasing the negative charges on the C-terminal tubulin tails, act as a regulator of motor function.
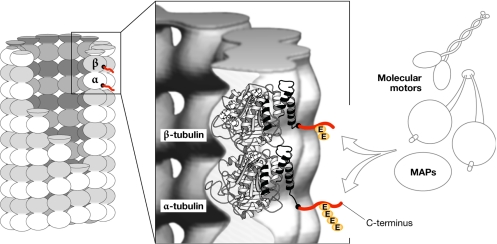
Figure 3.
Localization of polyglutamylation sites on microtubules. Schematic representation of the arrangement of α–β-tubulin dimer within the microtubule (MT) lattice. The positions of the acidic carboxy-terminal tails of both α- and β-tubulin, which are not resolved in the crystal structure, are indicated in red. Located on the surface of MTs, the tails are the main interaction sites for MT-associated proteins (MAPs) and motors. Polyglutamylation, which occurs on the C-terminal tails, might regulate these interactions. The central panel has been modified with permission from Amos, 2000.
Initial functional studies on tubulin polyglutamylation were performed by masking the modification with the glutamylation-specific antibody GT335 in vivo. Incubation of permeabilized spermatozoa with GT335 reduced the amplitude of flagellar beating, suggesting that polyglutamylation is important for the interactions of axonemal MTs with ciliary dyneins that generate the flagellar beat (Gagnon et al, 1996). In a similar experiment, the injection of GT335 into HeLa cells caused the disruption of centrioles and the dispersion of the pericentriolar material. This indicated that the high polyglutamylation levels on centrioles might be important for the integrity of these MT-based structures and, consequently, for centrosome function (Bobinnec et al, 1998b).
Another approach used to investigate the function of polyglutamylation was the systematic mutation of all known modification sites on either α- or β-tubulin in Tetrahymena thermophila. Mutations of the modification sites on α-tubulin had no effect, whereas those on β-tubulin were lethal (Xia et al, 2000). The phenotypes of the β-tubulin mutants included cytokinesis defects, as well as ciliary paralysis resulting from structural defects in axonemes (Thazhath et al, 2004, 2002). However, these phenotypes might also be related to the suppression of another post-translational modification of tubulin, polyglycylation (Redeker et al, 1994), which occurs at similar sites. Therefore, the results show that the modification of β-tubulin is essential for Tetrahymena, although it is not possible to specify whether the observed phenotypes are related to the loss of polyglutamylation or polyglycylation. Nevertheless, this genetic approach has provided strong evidence of the importance of tubulin polymodifications.
Although all of these experiments imply that polyglutamylation has an important role in diverse MT-related functions, they do not provide unambiguous evidence for this hypothesis. More-conclusive functional results might be obtained by directly modulating the polyglutamylation levels of tubulin, which has stimulated the search for the enzymes that catalyse polyglutamylation.
Polyglutamylating enzymes
Since the discovery of polyglutamylation, much effort has been dedicated to the identification of the modifying enzymes; however, the road to the final discovery has been a bumpy one. By using mouse brain as starting material, Catherine Regnard and Bernard Eddé set up a biochemical procedure to enrich the polyglutamylating activity present in this tissue, which allowed for a detailed analysis of the enzymatic properties of neuronal polyglutamylase (Regnard et al, 1998). Using a similar approach on extracts from proliferating cells (Regnard et al, 1999), they showed for the first time the existence of different types of glutamylating enzymes. Notably, the activities of neuronal and HeLa cell polyglutamylases vary in their specificity towards α- or β-tubulin, and in the length of the side chains that they generate.
As attempts to purify polyglutamylase further using classical biochemical methods resulted in a loss of enzymatic activity, monoclonal antibodies that specifically immunoprecipitate the enzyme were developed and used in a large-scale immunopurification. Several proteins were co-purified together with the neuronal polyglutamylase activity (Regnard et al, 2003). The first protein to be identified, polyglutamylase subunit 1 (PGs1), corresponded to the product of the GTRGEO22 gene, a mutation of which had been implicated in male sterility and neuronal defects in mice (Campbell et al, 2002). Subsequent analysis of the GTRGEO22 mutant mouse revealed disturbed MT-based neuronal traffic and a notable loss of polyglutamylation on α-tubulin (Ikegami et al, 2007), showing that PGs1 is important for tubulin polyglutamylation. Although PGs1 is not directly involved in the catalysis of polyglutamylation, its localization to the main sites of polyglutamylation—neurons, axonemes and centrioles—indicates that it acts as a localization subunit of a bigger polyglutamylase complex (Regnard et al, 2003).
Indeed, PGs1 was shown to form a multiprotein complex with four additional subunits (Janke et al, 2005), among which only one, PGs3, has obvious homologies to a known protein, tubulin tyrosine ligase (TTL; Ersfeld et al, 1993). This similarity to an amino-acid ligase of tubulin indicated that PGs3, which is now called TTL-like protein 1 (TTLL1), is a strong candidate for the catalytic subunit of the neuronal polyglutamylase. However, no enzymatic activity was detected after the production of recombinant TTLL1 in different expression systems. The subsequent investigation of additional members of the TTLL family in T. thermophila led to the discovery of the first polyglutamylase with autonomous enzymatic activity, Ttll6Ap (Janke et al, 2005). This finding established that tubulin polyglutamylases are members of the TTLL protein family.
The systematic study of the mammalian TTLL family revealed the existence of 13 TTLL proteins that are characterized by the presence of the highly homologous core TTL domain. Within this group, six autonomously active polyglutamylases where identified. They all share with TTLL1 an additional homology motif, the extended TTL domain. This suggests that the extended TTL domain is a general characteristic of polyglutamylases and could help to predict the enzymatic activities of other TTLLs (van Dijk et al, 2007). The subsequent enzymatic characterization of the six newly discovered TTLL polyglutamylases showed that each enzyme has a specific substrate and reaction preference (Fig 4), suggesting that the particular polyglutamylation patterns that are found on different MT subtypes (Fig 2) are directly determined by the specificities of the enzymes involved in their generation (van Dijk et al, 2007). How the activity of the enzymes is coordinated to generate locally restricted and well-defined polyglutamylation patterns on MTs remains to be established.
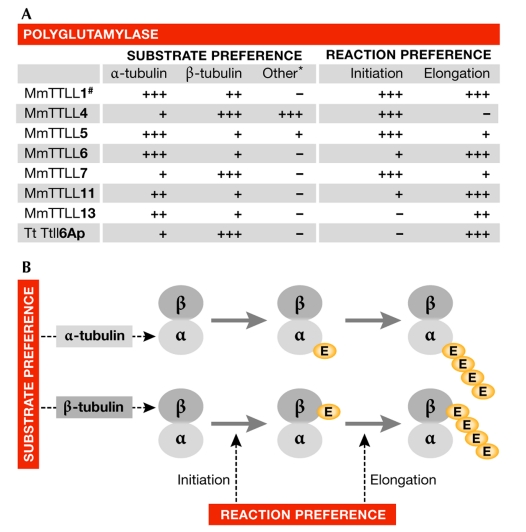
Figure 4.
Substrate and reaction preferences of polyglutamylases. (A) Substrate and reaction preferences of known polyglutamylases. Table adapted from van Dijk et al, 2007. *For details of other substrates, see van Dijk et al, 2008. #Enzymatic characteristics deduced from measurements with enriched brain polyglutamylase activity (Janke et al, 2005). (B) Preferences of polyglutamylases determine the distribution of the modification between α- and β-tubulin, and the length of the side chain. Mm, Mus musculus; Tt, Tetrahymena thermophila; TTLL, tubulin tyrosine ligase-like.
Perspectives
The identification of enzymes that catalyse polyglutamylation, and the discovery of their predetermined substrate and reaction preferences, now allow for more-systematic functional studies. The depletion of the β-tubulin-preferring polyglutamylase TTLL7 in the neuron-like cell line PC12 showed that the outgrowth of neurites depends on tubulin polyglutamylation (Ikegami et al, 2006). The importance of polyglutamylation for ciliary assembly and function became apparent when the glutamylating enzyme TTLL6 was depleted in zebrafish, which caused aberrations in the formation, stability and function of a subset of cilia (Pathak et al, 2007). Moreover, a complete inhibition of ciliary beating was obtained by the overexpression of Ttll6Ap in T. thermophila, which induced a hyper-polyglutamylation of ciliary MTs (Janke et al, 2005). The newly discovered polyglutamylases have therefore provided the first evidence of the importance of polyglutamylation in MT functions. However, a potential problem for the studies of tubulin polyglutamylation could be the functional redundancy of some of these glutamylating enzymes. The identification of enzymes that remove the modification—the deglutamylases—could further advance the functional studies by providing additional experimental tools for interfering with polyglutamylation.
Polyglutamylation is generally recognized as a modification of tubulin, but it is not restricted to tubulin. Two prominent non-tubulin substrates, nucleosome-assembly protein (NAP)1 and NAP2, were identified 10 years after the initial discovery of tubulin polyglutamylation (Regnard et al, 2000). Notably, NAPs were recently shown to share another post-translational modification with tubulin, polyglycylation. Although tubulin glycylases remain to be identified, TTLL10, which is a member of the TTLL family that does not belong to the polyglutamylase TTLL subgroup, was shown to catalyse NAP glycylation (Ikegami et al, 2008). It is therefore likely that tubulin glycylases are also members of the TTLL protein family. In addition, a proteomic approach identified more than100 proteins that bind specifically to the glutamylation-specific antibody GT335. A detailed analysis of these proteins led to the discovery of new substrates of polyglutamylation, including several chromatin-associated proteins (van Dijk et al, 2008). These new substrates are modified by specific TTLL polyglutamylases, although it is not known how polyglutamylation affects their biological function. Nevertheless, the discovery of a broad range of substrates shows that polyglutamylation is a general post-translational protein modification and should be considered as a potential regulator of diverse cellular functions.
Sidebar A | In need of answers.
Polyglutamylation is a reversible post-translational modification; however, the identity of the deglutamylating enzymes is still unknown.
In mammals, 13 tubulin tyrosine ligase-like (TTLL) proteins have been identified, nine of which are polyglutamylases, and one was recently shown to be a glycylase. Are the remaining TTLL family members also amino-acid ligases?
We still do not know much about the functions of polyglutamylation on microtubules, or what impact the diversity of the polyglutamylation pattern could have on the regulation of these functions.
We do not know which effectors are regulated by polyglutamylation.
Several non-tubulin proteins have been shown to be polyglutamylated, but how does this modification affect the functions of these proteins?
From left: Krzysztof Rogowski, Juliette van Dijk & Carsten Janke
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Universities Montpellier 2 and 1, the Association de la Recherche contre le Cancer (ARC) awards CR504/7817 and 3140 (to C.J.), the French National Research Agency (ANR) award JC05_42022, the European Molecular Biology Organization (EMBO) Young Investigator Program (YIP; to C.J.), the EMBO long-term fellowship ALTF 546-2006 (to K.R.) and the Ligue contre le Cancer fellowship JG/VP-5454. We especially thank B. Eddé (Université Paris 6, France) for groundbreaking work in the field, and for his mentoring and instructive discussions. The authors are grateful to B. Lacroix, J. Miro, M. Bosch-Grau (Centre de Recherches de Biochimie Macromoléculaire), M. Bettencourt-Dias (Instituto Gulbenkian de Ciência, Oeiras, Portugal) and M. Magiera (Institute of Molecular Genetics of Montpellier, Montpellier, France) for critical reading of the manuscript. Owing to the space restrictions, we could cite only a selection of the literature relevant to this review; we apologize to all the authors of valuable publications that have not been cited.
References
- Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, MacDonald TL, Sundberg RJ, Rebhun LI, Frankfurter A (1991) Characterization of posttranslational modifications in neuron-specific class III β-tubulin by mass spectrometry. Proc Natl Acad Sci USA 88: 4685–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA (2000) Focusing-in on microtubules. Curr Opin Struct Biol 10: 236–241 [DOI] [PubMed] [Google Scholar]
- Audebert S, Desbruyeres E, Gruszczynski C, Koulakoff A, Gros F, Denoulet P, Edde B (1993) Reversible polyglutamylation of α- and β-tubulin and microtubule dynamics in mouse brain neurons. Mol Biol Cell 4: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, Edde B (1994) Developmental regulation of polyglutamylated α- and β-tubulin in mouse brain neurons. J Cell Sci 107: 2313–2322 [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Moudjou M, Fouquet JP, Desbruyeres E, Edde B, Bornens M (1998a) Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton 39: 223–232 [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M (1998b) Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol 143: 1575–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet C, Boucher D, Lazereg S, Pedrotti B, Islam K, Denoulet P, Larcher JC (2001) Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J Biol Chem 276: 12839–12848 [DOI] [PubMed] [Google Scholar]
- Boucher D, Larcher JC, Gros F, Denoulet P (1994) Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry 33: 12471–12477 [DOI] [PubMed] [Google Scholar]
- Bre MH, de Nechaud B, Wolff A, Fleury A (1994) Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335. Cell Motil Cytoskeleton 27: 337–349 [DOI] [PubMed] [Google Scholar]
- Campbell PK et al. (2002) Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice. Genetics 162: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P (1990) Posttranslational glutamylation of α-tubulin. Science 247: 83–85 [DOI] [PubMed] [Google Scholar]
- Ersfeld K, Wehland J, Plessmann U, Dodemont H, Gerke V, Weber K (1993) Characterization of the tubulin-tyrosine ligase. J Cell Biol 120: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field DJ, Lee JC (1988) Analysis of tubulin proteins and peptides in neuronal and non-neuronal tissues using immobilized pH gradients. Electrophoresis 9: 555–562 [DOI] [PubMed] [Google Scholar]
- Fouquet JP, Edde B, Kann ML, Wolff A, Desbruyeres E, Denoulet P (1994) Differential distribution of glutamylated tubulin during spermatogenesis in mammalian testis. Cell Motil Cytoskeleton 27: 49–58 [DOI] [PubMed] [Google Scholar]
- Gagnon C, White D, Cosson J, Huitorel P, Edde B, Desbruyeres E, Paturle-Lafanechere L, Multigner L, Job D, Cibert C (1996) The polyglutamylated lateral chain of α-tubulin plays a key role in flagellar motility. J Cell Sci 109: 1545–1553 [DOI] [PubMed] [Google Scholar]
- Ikegami K, Mukai M, Tsuchida J-I, Heier RL, Macgregor GR, Setou M (2006) TTLL7 is a mammalian β-tubulin polyglutamylase required for growth of MAP2-positive neurites. J Biol Chem 281: 30707–30716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K et al. (2007) Loss of α-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci USA 104: 3213–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Horigome D, Mukai M, Livnat I, Macgregor GR, Setou M (2008) TTLL10 is a protein polyglycylase that can modify nucleosome assembly protein 1. FEBS Lett 582: 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C et al. (2005) Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308: 1758–1762 [DOI] [PubMed] [Google Scholar]
- Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P (1996) Interaction of kinesin motor domains with α- and β-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J Biol Chem 271: 22117–22124 [DOI] [PubMed] [Google Scholar]
- Luduena RF (1998) Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol 178: 207–275 [DOI] [PubMed] [Google Scholar]
- Pathak N, Obara T, Mangos S, Liu Y, Drummond IA (2007) The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell 18: 4353–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessmann U, Weber K (1997) Mammalian sperm tubulin: an exceptionally large number of variants based on several posttranslational modifications. J Protein Chem 16: 385–390 [DOI] [PubMed] [Google Scholar]
- Redeker V, Le Caer JP, Rossier J, Prome JC (1991) Structure of the polyglutamyl side chain posttranslationally added to α-tubulin. J Biol Chem 266: 23461–23466 [PubMed] [Google Scholar]
- Redeker V, Melki R, Prome D, Le Caer JP, Rossier J (1992) Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major α and β tubulin isotypes from pig brain are glutamylated. FEBS Lett 313: 185–192 [DOI] [PubMed] [Google Scholar]
- Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bre MH (1994) Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science 266: 1688–1691 [DOI] [PubMed] [Google Scholar]
- Regnard C, Audebert S, Desbruyeres, Denoulet P, Edde B (1998) Tubulin polyglutamylase: partial purification and enzymatic properties. Biochemistry 37: 8395–8404 [DOI] [PubMed] [Google Scholar]
- Regnard C, Desbruyeres E, Denoulet P, Edde B (1999) Tubulin polyglutamylase: isozymic variants and regulation during the cell cycle in HeLa cells. J Cell Sci 112: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Regnard C, Desbruyeres E, Huet JC, Beauvallet C, Pernollet JC, Edde B (2000) Polyglutamylation of nucleosome assembly proteins. J Biol Chem 275: 15969–15976 [DOI] [PubMed] [Google Scholar]
- Regnard C, Fesquet D, Janke C, Boucher D, Desbruyeres E, Koulakoff A, Insina C, Travo P, Edde B (2003) Characterisation of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J Cell Sci 116: 4181–4190 [DOI] [PubMed] [Google Scholar]
- Rüdiger M, Plessman U, Kloppel KD, Wehland J, Weber K (1992) Class II tubulin, the major brain β tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett 308: 101–105 [DOI] [PubMed] [Google Scholar]
- Thazhath R, Liu C, Gaertig J (2002) Polyglycylation domain of β-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat Cell Biol 4: 256–259 [DOI] [PubMed] [Google Scholar]
- Thazhath R, Jerka-Dziadosz M, Duan J, Wloga D, Gorovsky MA, Frankel J, Gaertig J (2004) Cell context-specific effects of the β-tubulin glycylation domain on assembly and size of microtubular organelles. Mol Biol Cell 15: 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD (2000) Engineering the processive run length of the kinesin motor. J Cell Biol 151: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C (2007) A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell 26: 437–448 [DOI] [PubMed] [Google Scholar]
- van Dijk J, Miro J, Strub J-M, Lacroix B, van Dorsselaer A, Edde B, Janke C (2008) Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem 283: 3915–3922 [DOI] [PubMed] [Google Scholar]
- Weber K, Schneider A, Westermann S, Muller N, Plessmann U (1997) Posttranslational modifications of α- and β-tubulin in Giardia lamblia, an ancient eukaryote. FEBS Lett 419: 87–91 [DOI] [PubMed] [Google Scholar]
- Wolff A, Denoulet P, Jeantet C (1982) High level of tubulin microheterogeneity in the mouse brain. Neurosci Lett 31: 323–328 [DOI] [PubMed] [Google Scholar]
- Wolff A, de Nechaud B, Chillet D, Mazarguil H, Desbruyeres E, Audebert S, Edde B, Gros F, Denoulet P (1992) Distribution of glutamylated α- and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol 59: 425–432 [PubMed] [Google Scholar]
- Wolff A, Houdayer M, Chillet D, de Nechaud B, Denoulet P (1994) Structure of the polyglutamyl chain of tubulin: occurrence of α and γ linkages between glutamyl units revealed by monoreactive polyclonal antibodies. Biol Cell 81: 11–16 [DOI] [PubMed] [Google Scholar]
- Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, Bre MH, Levilliers N, Gorovsky MA, Gaertig J (2000) Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J Cell Biol 149: 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]