Abstract
ERp44 mediates thiol-dependent retention in the early secretory pathway, forming mixed disulphides with substrate proteins through its conserved CRFS motif. Here, we present its crystal structure at a resolution of 2.6 Å. Three thioredoxin domains—a, b and b′—are arranged in a clover-like structure. A flexible carboxy-terminal tail turns back to the b′ and a domains, shielding a hydrophobic pocket in domain b′ and a hydrophobic patch around the CRFS motif in domain a. Mutational and functional studies indicate that the C-terminal tail gates the CRFS area and the adjacent hydrophobic pocket, dynamically regulating protein quality control.
Introduction
The endoplasmic reticulum (ER) contains many chaperones and enzymes, providing a unique environment for oxidative folding, other post-translational modifications and the quality control of secretory proteins (Sitia & Braakman, 2003). Oxidative protein folding proceeds through protein-driven disulphide interchange relays, with protein disulphide isomerase (PDI) having a crucial role (Ellgaard & Ruddock, 2005). ERp44 is a member of the PDI family and is composed of an amino-terminal thioredoxin (Trx) domain a, endowed with a CRFS motif, followed by two redox inactive Trx-like domains, b and b′, and contains a carboxy-terminal ER retrieval signal. Consistent with its role in folding and transport in the secretory pathway (Anelli et al, 2002, 2003, 2007), ERp44 is induced during ER stress and abundantly expressed in secretory tissue cells (www.hpr.se).
The existence of numerous PDI members in the early secretory apparatus with various numbers, redox potential and arrangements of Trx-like domains suggests subtle specificities in their functions (Maattanen et al, 2006). Having a CRFS motif, ERp44 cannot be an efficient oxidoreductase. However, the absence of the second cysteine allows longer-lived mixed disulphides to be formed, a characteristic compatible with its role in quality control. ERp44 contributes to retain Ero1α and Ero1β intracellularly, the two rate-limiting factors in PDI oxidation (Mezghrani et al, 2001), by forming mixed disulphides through Cys 29 in its CRFS motif (Otsu et al, 2006). Similarly, ERp44 forms transient disulphides with immunoglobulin M subunits (Anelli et al, 2002, 2003, 2007), adiponectin (Wang et al, 2007) or formylglycine-generating enzyme (FGE; Mariappan et al, 2008), regulating their transport. In addition to its crucial role in thiol-mediated retention and quality control, ERp44 interacts with inositol 1,4,5-trisphosphate receptor type I, inhibiting its Ca2+ channel activity (Higo et al, 2005). These crucial regulatory roles prompted us to investigate the structure of ERp44. So far, among eukaryotic enzymes catalysing oxidative folding, full-length three-dimensional structures are available for yeast PDI (yPDI; Tian et al, 2006), and isolated domains of human PDI (hPDI; Gruber et al, 2006) and ERp57 (Kozlov et al, 2006). Here, we describe the crystal structure of human ERp44 (ERp44) at 2.6 Å, the first complete structure of a mammalian PDI family member. Biochemical and functional analyses of wild type and mutants suggest a regulatory role for the C-terminal tail (C-tail) in regulating substrate binding and release.
Results And Discussion
Overall structure of ERp44
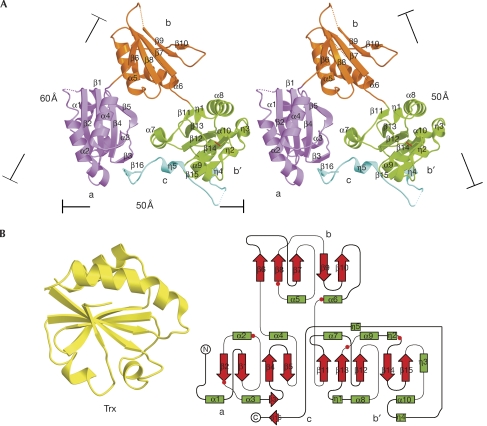
ERp44 was crystallized into space group P3121. The crystal structure of a selenomethioninyl derivative was solved and refined to 2.6 Å with 87% residues traced clearly into electron density maps (supplementary Table S1 online). The overall structure resembles a clover composed of three domains—a, b and b′—and a C-tail, which bridges domains b′ and a, resulting in a compact appearance (Fig 1A). Domains a, b and b′ all share the Trx fold (βαβαβαββα) with some distinctive variations (Fig 1B). Domain a (residues 1–112) lacks the first β strand and comprises a four-stranded central β-sheet (β1, β2, β4 and β5) flanked by two α-helices (α1 and α3, α2 and α4) on each side, and one additional β-strand (β3) between α3 and β4. Domain b (residues 113–215) has two segments (residues 119–130 and 170–177) at the corresponding loci of the two canonical α-helices with poor electron density for modelling. Finally, b′ (residues 216–325) has three additional 310 helices (η1, η2 and η3). The C-tail (residues 326–377) is flexible and mainly composed of random coils in addition to one short strand β16 (residues 369–370), which is antiparallel to β3 in domain a, and two short 310 helices: η4 (residues 326–328) and η5 (residues 358–360) filling the cavity lined by helices α7 and α9 in domain b′ (Fig 1A).
Figure 1.
Overall structure of human ERp44. (A) Stereo ribbon view of the structure of ERp44. The dotted lines denotes the regions (residues 51–53, 119–130, 170–177 and 332–350) that are not resolved. The dimensions of the clover-like molecule are indicated. (B) Secondary structure topology of Trx (left) and ERp44 (right). The latter was made using TopDraw (Bond, 2003): green box, helix; red arrow, β sheet; and red dot, cysteine residue. ERp44, human ERp44; Trx, thioredoxin.
ERp44 contains six cysteine residues (29, 63, 160, 212, 272 and 289) with Cys 272 and Cys 289 forming a disulphide bond in domain b′ (Fig 1A,B). The short distance (4.3 Å) between Cys 160 and Cys 212 in domain b suggests a strong potential to form a covalent linkage, which was found in the structure of an ERp44 crystal soaked in 1 mM glutathione/0.2 mM glutathione disulphide (data not shown). This disulphide bond is likely to be present in vivo, which is consistent with our previous observations (Anelli et al, 2002, 2003).
The CRFS motif in domain a
The a domains of ERp44 and yPDI can be superimposed with a root mean square deviation (r.m.s.d.) of 1.1 Å for 103 structurally homologous Cα atoms (Fig 2A). The domains differ in the active site motif: CRFS (residues 29–32) in ERp44 instead of the canonical CXXC in yPDI. In ERp44 the Cys 29 sulphydryl is within hydrogen-bonding distance to the side-chain hydroxyl of Ser 32, in the equivalent position of Cys 64 in yPDI, and the hydroxyl of Thr 369 in the C-tail (Fig 2B). The side chain of Arg 30 extends at the surface and shows high flexibility in the crystal structure. Phe 31 occupies the equivalent position of His 63 in yPDI and forms one part of a hydrophobic patch around the CRFS motif, together with Met 34, Pro 37, Ile 38, Val 45 and Val 100. The other part of the hydrophobic patch is formed by the highly conserved Trp 28, together with Ala 70, Ile 75, Tyr 78, Pro 79, Leu 81 and Tyr 94 on the other side of the CRFS motif. The latter is similar to the hydrophobic patch surrounding the CGHC motif in yPDI but is shielded by the C-tail (Fig 2C). The resulting contiguous hydrophobic patch around the CRFS could be involved in client protein binding, as in PDI (Tian et al, 2006).
Figure 2.
Features of the CRFS motif in domain a. (A) Structural superposition of the a domains from ERp44 (pink) and yPDI (cyan) with a close-up view of the CRFS region (right). (B) Stereo view of the CRFS motif region with the straight 2FoFc omit map (contoured at 1.5σ). Hydrogen-bonding interactions are depicted as a black dotted line with distance indicated. (C) The hydrophobic patch (yellow) around the CRFS motif (red) and the C-tail (orange ribbon). ERp44, human ERp44; yPDI, yeast protein disulphide isomerase.
Similar to other Trx family members (Tian et al, 2006), residue Pro 79 in the cis conformation neighbouring the CRFS motif (Fig 2A) could be important for maintaining the substrate binding ability and reactivity of the active site. Much attention has been paid to buried salt bridges that are formed by Glu 55/Gln 87 in yPDI (Tian et al, 2006), Asp 26/Lys 57 in Trx (Carvalho et al, 2006) and Glu 24/Lys 58 in DsbA (Jacobi et al, 1997), for their possible role in regulating the redox state of the N-terminal cysteine in CXXC active sites. However, the corresponding residues in ERp44 (Asn 23 and Arg 60) do not form a salt bridge (Fig 2A); therefore, the special residue environment around the CRFS motif might specifically modulate the binding of physiological partner proteins.
A hydrophobic pocket in domain b′ shielded by the C-tail
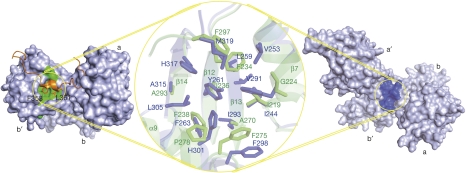
In both hPDI and yPDI, the b′ domain is thought to be a primary substrate binding site (Klappa et al, 1998; Tian et al, 2006). A similar hydrophobic pocket in ERp44 is shown by superposition of the b′ domains of ERp44 and yPDI with an r.m.s.d. of 1.8 Å against 95 aligned Cα atoms (Fig 3). In ERp44, this pocket is formed by three strands (β12, β13 and β14) flanked by two helices (α7 and α9) with a diameter of approximately 16 Å, a depth of about 5 Å and a solvent-accessible surface area of about 200 Å2. The upper portion of this pocket is composed of Ile 219, Gly 224, Phe 234, Ala 270, Phe 275, Pro 278 and Phe 297, and the lower part is composed of Ile 236 and Phe 238, with Ala 293 at the bottom (Fig 3). Sequence alignment based on the structures of ERp44 and yPDI indicated high similarity of the residues composing the hydrophobic pocket (supplementary Fig S1 online). The hydrophobic pocket in domain b′ and the adjacent hydrophobic patch in domain a (supplementary Fig S2 online) might act as a docking site for ERp44 substrates. However, this region is partly covered by the C-tail extending from domain b′ to domain a (Fig 3). The η5 helix of the C-tail settles onto the hydrophobic pocket similar to a lid, with Phe 358 and Leu 361 providing additional anchors (Fig 3), which might reduce the accessibility to client proteins. The importance of hydrophobic interactions is supported by the observation that non-covalent interactions between ERp44 and FGE can mediate ER retention (Mariappan et al, 2008). Hence, the hydrophobic surface in domain b′ of ERp44 shows strong structural analogies to the binding site of yPDI, but its accessibility is limited by the observed conformation of the C-tail.
Figure 3.
Hydrophobic pockets in the b′ domains of human ERp44 and yeast protein disulphide isomerase. The pockets are indicated by circles, with the surface in green for ERp44 (left) and dark blue for yPDI (right). The C-terminal tail of ERp44 is indicated by the orange ribbon. The middle shows a close-up superposition view of the hydrophobic pockets from ERp44 (green) and yPDI (dark blue) using stick models to represent the residues that compose the hydrophobic pockets. ERp44, human ERp44; yPDI, yeast protein disulphide isomerase.
Removal of the C-tail increases ERp44 activity
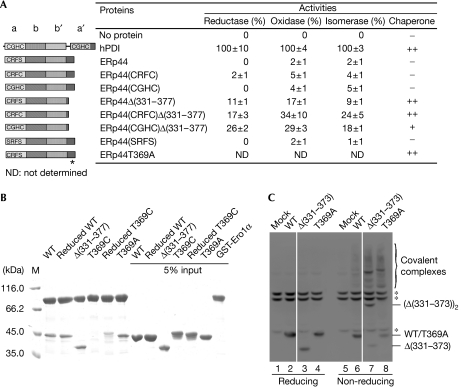
To analyse whether the C-tail could have a regulatory role in substrate binding, we analysed a panel of mutants. As shown in Fig 4A, wild-type ERp44 showed little reductase, oxidase or isomerase activities. Replacing Ser 32 by a cysteine or transplanting CGHC (the PDI active motif) for CRFS did not significantly restore enzyme activities. Compared with the full-length counterparts, however, the ERp44Δ(331–377) deletion mutant showed a significant increase (about fivefold) in enzyme activities, which is consistent with a regulatory role of the tail.
Figure 4.
The C-terminal tail of human ERp44 restricts substrate binding in vitro and in vivo. (A) Schematic representation of the wild-type and mutant ERp44 proteins with the CRFS motif mutated and/or with the C-terminal (residues 331–377) truncated or mutated is shown on the left, and their enzyme and chaperone activities on the right. Data of enzyme activities are expressed as mean±s.d. (n=3), and inhibition of rhodanese aggregation as no activity (–), weak activity (+) and strong activity (++). The asterisk indicates the T369A mutation site. (B) Pull-down assay of ERp44 proteins. Glutathiol Sepharose 4 fast-flow beads (GE Healthcare, Buckinghamshire, UK) were incubated at 25°C and pH 7.5 for 1 h with 10−6 M GST-Ero1α and various ERp44 proteins, as indicated, and then analysed by SDS–12% polyacrylamide gel electrophoresis (SDS–12% PAGE) under reducing conditions after being washed five times. (C) The tail deletion (ERp44Δ(331–373)) and replacement (ERp44T369A) mutants accumulate intracellularly mostly as mixed disulphides with client proteins. Lysates from HeLa cells transiently transfected with the indicated vectors were prepared 48 h after transfection and resolved by SDS–12% PAGE under reducing (lanes 1–4) or non-reducing (lanes 5–8) conditions, blotted and decorated with monoclonal HA antibodies. The asterisks indicate anti-HA reactive background bands, present also in mock-transfected cells. White lines separate lanes 2 and 3, and 6 and 7, which were juxtaposed from the same gel. ERp44, human ERp44; HA, haemagglutinin epitope tag; M, marker; WT, wild-type ERp44; T369C, ERp44T369C; T369A, ERp44T369A; Δ(331–373), ERp44Δ(331–373); Δ(331–377), ERp44Δ(331–377).
Next, we compared ERp44 and ERp44Δ(331–377) for their ability to suppress the aggregation of denatured rhodanese during refolding on dilution. ERp44 showed no chaperone activity; by contrast, ERp44Δ(331–377) suppressed aggregation as effectively as hPDI (Fig 4A; supplementary Fig S3 online). Both the rate and extent of rhodanese aggregation decreased with the increase in the concentrations of ERp44Δ(331–377) (supplementary Fig S4 online). Also, ERp44Δ(331–377) bound to Ero1α in vitro more efficiently than ERp44 (Fig 4B). The deletion mutant also bound to Ero1α and Ig-μ chains more efficiently when expressed in HeLa cells (supplementary Fig S4 online). However, it showed similar stability, detergent solubility and circular dichroism patterns (supplementary Fig S5 online), and bound to the ER–Golgi intermediate compartment marker 53 with a similar efficiency (supplementary Fig S4 online), excluding gross structural alterations.
Further evidence for a regulatory role of the C-tail originates from the effects of two point mutations, T369C and T369A. ERp44T369C showed an impaired binding to Ero1α in vitro; pretreatment with dithiothreitol partly restored binding, suggesting that an intramolecular bond is formed between Cys 29 and Cys 369 that locks the tail and impedes substrate binding. By contrast, ERp44T369A, unable to form a hydrogen bond with Cys 29, bound to Ero1α more efficiently, which is in line with an increased accessibility of the CRFS motif and the hydrophobic surfaces (Fig 4B). In addition, T369A efficiently prevented rhodanese aggregation (Fig 4A; supplementary Fig S3A online). Thus, shielding of client protein binding by the C-tail occurs in solution and is not an artefact of crystallization.
To confirm the in vitro data, we expressed the mutants in human cell lines. Removal of the C-tail or replacing Thr 369 by alanine increased significantly the fraction of ERp44 that forms mixed disulphides with endogenous proteins in HeLa cells (Fig 4C), as well as in human embryonic kidney (HEK)293 or human hepatocellular liver carcinoma (HepG2) cells (data not shown). These data also suggest that in living cells the flexible tail dynamically hampers the reactivity of ERp44 with its endogenous client proteins (supplementary Fig S2 online), perhaps restricting its specificity. In this respect, an interesting analogy is found in yeast Ero1, the flexible loop of which regulates electron fluxes (Sevier & Kaiser, 2007). It will be of interest to identify the mechanisms that control the ERp44 tail movements.
Concluding remarks
Although ERp44 interacts with substrates through mixed disulphide bonds (Anelli et al, 2002, 2003; Otsu et al, 2006), hydrophobic interactions are important in aligning Cys 29 to target cysteines in the client proteins. Our findings suggest a role of the ERp44 C-tail in regulating substrate binding and release during protein quality control in the early secretory apparatus.
Methods
Crystallization. The complementary DNA encoding ERp44 (accession code CAC87611) without signal sequence (Anelli et al, 2002) was subcloned into pGEX-6P-1 (Amersham, Buckinghamshire, UK) at BamHI and XhoI sites (also for the template of all the mutations (Fig 4A)) and expressed in BL21 (DE3) plysS (Novagen, San Diego, CA, USA). After removing the glutathione S-transferase (GST) tag, the purified protein was methylated using borane-dimethylamine complex (Aldrich, St Louis, MO, USA; Liu et al, 2005) and crystallized in 20 mM Tris–HCl (pH 7.5), 1.2 M disodium succinate and 0.1% N-dodecyl-β-D-maltopyranoside at 290 K by using hanging-drop vapour diffusion. Methylation does not affect binding of ERp44 to Ero1α (data not shown). Selenomethioninyl derivatives were prepared as described previously (Doublie, 1997).
Structural determination. Crystals were soaked in 3.3 M sodium formate with 0.5 M disodium succinate for 2 h for dehydration and flash-frozen in liquid nitrogen. One single-wavelength anomalous diffraction data of selenomethioninyl derivate crystal was collected at 100 K (λ=0.9790 Å) to 2.6 Å at the beamline BL5A of Photon Factory, Japan. Data processing is described in the supplementary information online. All figures for surfaces, ribbons, balls and sticks were generated with Pymol (http://pymol.sourseforge.net) and/or BobScript 2.6b (Esnouf, 1997).
Activity determination. Thiol protein reductase activity was assayed as described previously (Ke et al, 2006) and isomerase and oxidase activities were assayed according to Wilkinson et al (2005) at 25°C. The relative activity was calculated as (A−A0)/(A1−A0) × 100%, where A is the activity of ERp44 protein, A1 the activity of recombinant hPDI (Li et al, 2006) and A0 the activity of blank in the absence of protein. Chaperone activity was determined as described previously (Song & Wang, 1995).
Analysis of mixed disulphides between ERp44 and endogenous client proteins. HeLa, HEK293 and HepG2 cell lines were obtained from ATCC (Manassas, VA, USA), transfected and analysed as described previously (Anelli et al, 2002, 2003).
Coordinates. The structure has been deposited with the Protein Data Bank (accession code 2R2J).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Z.Y. Lou for data collection. We greatly appreciate the helpful suggestions from members of our laboratories. This work was supported by grants from the Chinese Ministry of Science and Technology (2006CB910903, 2006CB806508 and 2006CB806506) and the China National Science Foundation (30620130109), and from Associaziona Italiana Ricerca sul Cancro, Fondazione Cariplo (Progetto NOBEL), Ministero dell'Istruzione, dell'Università e della Ricerca (CoFin) and Telethon Italy.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R (2002) ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J 21: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Alessio M, Bachi A, Bergamelli L, Bertoli G, Camerini S, Mezghrani A, Ruffato E, Simmen T, Sitia R (2003) Thiol-mediated protein retention in the endoplasmic reticulum: the role of ERp44. EMBO J 22: 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Ceppi S, Bergamelli L, Cortini M, Masciarelli S, Valetti C, Sitia R (2007) Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J 26: 4177–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS (2003) TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics 19: 311–312 [DOI] [PubMed] [Google Scholar]
- Carvalho AP, Fernandes PA, Ramos MJ (2006) Similarities and differences in the thioredoxin superfamily. Prog Biophys Mol Biol 91: 229–248 [DOI] [PubMed] [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. Method Enzymol 276: 523–530 [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW (2005) The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep 6: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15: 132–134, 112–113 [DOI] [PubMed] [Google Scholar]
- Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ (2006) Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci 31: 455–464 [DOI] [PubMed] [Google Scholar]
- Higo T, Hattori M, Nakamura T, Natsume T, Michikawa T, Mikoshiba K (2005) Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell 120: 85–98 [DOI] [PubMed] [Google Scholar]
- Jacobi A, Huber-Wunderlich M, Hennecke J, Glockshuber R (1997) Elimination of all charged residues in the vicinity of the active-site helix of the disulfide oxidoreductase DsbA. Influence of electrostatic interactions on stability and redox properties. J Biol Chem 272: 21692–21699 [DOI] [PubMed] [Google Scholar]
- Ke H, Zhang S, Li J, Howlett GJ, Wang CC (2006) Folding of Escherichia coli DsbC: characterization of a monomeric folding intermediate. Biochemistry 45: 15100–15110 [DOI] [PubMed] [Google Scholar]
- Klappa P, Ruddock LW, Darby NJ, Freedman RB (1998) The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J 17: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Maattanen P, Schrag JD, Pollock S, Cygler M, Nagar B, Thomas DY, Gehring K (2006) Crystal structure of the bb′ domains of the protein disulfide isomerase ERp57. Structure 14: 1331–1339 [DOI] [PubMed] [Google Scholar]
- Li SJ, Hong XG, Shi YY, Li H, Wang CC (2006) Annular arrangement and collaborative actions of four domains of protein-disulfide isomerase: a small angle X-ray scattering study in solution. J Biol Chem 281: 6581–6588 [DOI] [PubMed] [Google Scholar]
- Liu ZJ et al. (2005) Salvaging Pyrococcus furiosus protein targets at SECSG. J Struct Funct Genomics 6: 121–127 [DOI] [PubMed] [Google Scholar]
- Maattanen P, Kozlov G, Gehring K, Thomas DY (2006) ERp57 and PDI: multifunctional protein disulfide isomerases with similar domain architectures but differing substrate–partner associations. Biochem Cell Biol 84: 881–889 [DOI] [PubMed] [Google Scholar]
- Mariappan M, Radhakrishnan K, Dierks T, Schmidt B, von Figura K (2008) ERp44 mediates a thiol independent retention of formylglycine generating enzyme in the endoplasmic reticulum. J Biol Chem 283: 6375–6383 [DOI] [PubMed] [Google Scholar]
- Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R (2001) Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J 20: 6288–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu M, Bertoli G, Fagioli C, Guerini-Rocco E, Nerini-Molteni S, Ruffato E, Sitia R (2006) Dynamic retention of Ero1α and Ero1β in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid Redox Sign 8: 274–282 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA (2008) Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta 1783: 549–556 [DOI] [PubMed] [Google Scholar]
- Sitia R, Braakman I (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894 [DOI] [PubMed] [Google Scholar]
- Song JL, Wang CC (1995) Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. Eur J Biochem 231: 312–316 [DOI] [PubMed] [Google Scholar]
- Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H (2006) The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 124: 61–73 [DOI] [PubMed] [Google Scholar]
- Wang ZV, Schraw TD, Kim JY, Khan T, Rajala MW, Follenzi A, Scherer PE (2007) Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol Cell Biol 27: 3716–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B, Xiao R, Gilbert HF (2005) A structural disulfide of yeast protein-disulfide isomerase destabilizes the active site disulfide of the N-terminal thioredoxin domain. J Biol Chem 280: 11483–11487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information