Abstract
Ligand-gated ion channels are important in sensory and synaptic transduction. The PKD1L3–PKD2L1 channel complex is a sour taste receptor candidate that is activated by acids. Here, we report that the proton-activated PKD1L3–PKD2L1 ion channels have the unique ability to be activated after the removal of an acid stimulus. We refer to this property as the off-response (previously described as a delayed response). Electrophysiological analyses show that acid-induced responses are observed only after the removal of an acid solution at less than pH 3.0. A small increase in pH is sufficient for PKD1L3–PKD2L1 channel activation, after exposure to an acid at pH 2.5. These results indicate that this channel is a new type of ion channel—designated as an ‘off-channel'—which is activated during stimulus application but not gated open until the removal of the stimulus. The off-response property of PKD1L3–PKD2L1 channels might explain the physiological phenomena occurring during sour taste sensation.
Keywords: acid sensation, off-response, PKD1L3, PKD2L1
Introduction
Ligand-gated ion channels have diverse roles in processing internal and external information such as sensory and synaptic transductions. For example, several proton-activated ion channels are proposed to mediate sour taste sensations, which are used by animals to evaluate the acidity of food, a crucial cue that prevents animals from consuming spoilt food or harmful solutions (Ugawa et al, 1998; Stevens et al, 2001; Lin et al, 2004; Richter et al, 2004). Recently, PKD1L3 and PKD2L1 were reported to be expressed in mouse taste cells (Huang et al, 2006; Ishimaru et al, 2006) and to form an acid-activated ion channel (Ishimaru et al, 2006). These proteins are coexpressed in a subset of taste receptor cells found in specific taste areas. The cells expressing them are distinct from taste cells expressing TRPM5 (transient receptor potential melastatin 5), a molecule involved in bitter, sweet or umami (taste of l-amino acids) taste sensation (Zhang et al, 2003). PKD1L3 and PKD2L1 physically interact to form a functional receptor, and their coexpression is necessary for their efficient translocation to the cell surface. Electrophysiological analysis has shown that these proteins form ion channels that are activated by various acids, including citric acid and HCl (Ishimaru et al, 2006). Furthermore, diphtheria toxin (DTA)-mediated ablation of PKD2L1-expressing taste cells is reported to cause a complete loss of gustatory nerve response to sour stimuli in vivo (Huang et al, 2006). As PKD1L3 and PKD2L1 coexpression is essential for their functional cell-surface expression in mammalian cells (Ishimaru et al, 2006), the PKD1L3–PKD2L1 channel complex has been proposed to form a sour taste receptor in mammalian cells.
Previously, we reported that PKD1L3–PKD2L1 channels are activated by transient acid exposure. However, this activation was slightly delayed and was followed by rapid desensitization (Ishimaru et al, 2006). Here, we show that PKD1L3–PKD2L1 channel activation is not a delayed response, but rather an off-response. This suggests that this channel is gated open only after the removal of an acid stimulus, although initial acid exposure is essential. Both Ca2+ imaging and whole-cell patch-clamp recordings show that acid-induced responses occurred only after the removal of an acid stimulus at a pH of less than 3.0. Furthermore, distinct single-channel openings were observed after the removal of the acid solution in the outside-out but not in the inside-out configuration. A small pH increase seems to be sufficient to open the PKD1L3–PKD2L1 channel after exposure to an acid solution at pH 2.5. These results indicate that the PKD1L3–PKD2L1 channel is a new type of ion channel, designated as an off-channel.
Results
To gain further insight into the mechanisms by which PKD1L3–PKD2L1 channels are activated, we used Ca2+ imaging and patch-clamp techniques (Fig 1) to measure acid-induced responses in PKD1L3–PKD2L1-expressing cells. First, we examined whether delayed PKD1L3–PKD2L1 channel activation could be observed by Ca2+ imaging. The intracellular Ca2+ concentration ([Ca2+]i) was not observed to increase before or during acid exposure (Fig 1A–C). However, a robust [Ca2+]i increase was detected after the removal of the acid. This [Ca2+]i increase was absent in cells that were transfected with vectors, PKD1L3 complementary DNA (cDNA) alone or PKD2L1 cDNA alone (supplementary Fig 1A online). This unique response of PKD1L3–PKD2L1 channels to acid stimuli was also noted in the patch-clamp recordings (Fig 1D,E). First, we confirmed that no current responses, except those mediated by endogenously expressed acid-sensing ion channels (ASICs), were induced in HEK293T (human embryonic kidney) cells transfected with vectors, PKD1L3 cDNA alone or PKD2L1 cDNA alone (supplementary Fig 1B online). As our previous studies showed that citric acid is a potent activator of PKD1L3 and PKD2L1, we treated PKD1L3–PKD2L1-expressing cells with 25 mM citric acid at pH 2.8 for 1–15 s. Surprisingly, acid-induced currents were observed only when the acidic solutions were replaced with a solution at pH 7.4, regardless of the duration of treatment (Fig 1D). With a short duration of acid treatment, repeated activation was noted without significant desensitization (Fig 1D, left). This was probably due to small current sizes (supplementary Fig 2 online). Large currents were occasionally induced by short-term acid exposure for 1–2 s (supplementary Fig 3 online), whereas a 5 s exposure seemed to be sufficient to cause maximum activation (data not shown). This acid-induced current activation was also detected at the single-channel level in the outside-out (Fig 1E), but not the inside-out configuration (Fig 1F), after acid removal (pH 2.5). The single-channel openings seemed to be flickering. The unitary conductance for Na+ was calculated as approximately 144 pS at negative potentials, a value similar to that reported for homomeric PKD2L1 channels in oocytes (137 pS; Chen et al, 1999). These findings strongly suggest that PKD1L3–PKD2L1 channels can be gated open by pH changes in the extracellular solution, although initial acid exposure is essential. We performed a third experiment to confirm this further. After removing the initial acid stimulus, an extracellular solution of pH 7.4 was applied. After PKD1L3–PKD2L1 channel activation, the extracellular solution was replaced with an acid solution. This treatment resulted in a sudden inhibition of the currents followed by re-activation after neutralization (Fig 1G). These results indicate that PKD1L3–PKD2L1 channels have an off-response property, whereby they are gated open by removal of an acid stimulus. On the basis of this unique property, we concluded that the PKD1L3–PKD2L1 channel is a new type of ion channel, designated as an ‘off-channel', whereas traditional ligand-gated channels can be considered to be ‘on-channels' that are activated during stimulus application (supplementary Fig 4 online).
Figure 1.
Off-response property of the PKD1L3–PKD2L1 channel. (A–C) Human embryonic kidney (HEK)293T cells expressing PKD1L3–PKD2L1 were exposed to an acid stimulus (25 mM citric acid; pH 2.8) for 15 s. (A) Changes in [Ca2+]i after acid exposure, as indicated by the fura-2 ratio with pseudocolour expression. (B,C) Profiles of the [Ca2+]i changes recorded for (B) selected individual cells and (C) average changes recorded among experiments (n=12). Red and black bars indicate acid and ionomycin exposure, respectively. (D–F) Patch-clamp analysis of HEK293T cells expressing PKD1L3–PKD2L1. (D) Whole-cell currents induced after removal of the acid stimulus after initial exposure for various durations. HEK293T cells were exposed to 25 mM citric acid (pH 2.8; red bars) at −60 mV for various durations (1–5 and 15 s). No off-response was induced on treatment with 25 mM citric acid neutralized to pH 7.4 (data not shown). (E) Single-channel currents induced after the removal of the acid stimulus in the outside-out configuration. Profiles recorded (a) during or (b) after acid exposure (red bar) are enlarged in the lower panels. A patch membrane was exposed to an acid (pH 2.5) at −60 mV for 5 s; the basal currents were reduced after acid exposure. (F) A representative current in the inside-out configuration. A patch membrane from PKD1L3–PKD21-expressing cells was exposed to an acid (pH 2.8) for 5 s (red bar) at +60 mV. (G) Inhibition of acid-induced whole-cell currents after acid re-exposure. HEK293T cells expressing PKD1L3–PKD2L1 channels were exposed to an acid (pH 2.5) at −60 mV for 10 s. On induction of a high-magnitude inward current after acid removal, an acid was re-applied for 8 s. Red bars indicate acid exposure.
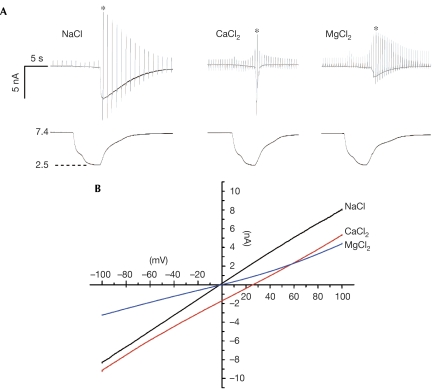
The current–voltage (I–V) relationship of the PKD1L3–PKD2L1 channel current during off-response showed the reversal potential to be −2.9±0.7 mV (n=9) in a bath solution containing 140 mM NaCl. This suggested that PKD1L3–PKD2L1 channels showed similar permeabilities to Na+ and K+ (Fig 2B). This is consistent with the I–V relationship during the delayed response in our previous report (Ishimaru et al, 2006). The ion permeability of the channel was independent of the induced current size (data not shown). In the bath solution containing 110 mM CaCl2 or MgCl2, the reversal potential was shifted to 26.5±1.1 mV (n=7) or –2.2±1.5 mV (n=8), respectively, indicating that the PKD1L3–PKD2L1 channel showed higher permeabilities to Ca2+ and Mg2+ than to Na+ and K+ (PNa:PK:PCa:PMg=1:1.1:11.0:1.8). Furthermore, desensitization was markedly accelerated in the bath solution containing Ca2+ (Fig 2A). The τ of desensitization in 110 mM CaCl2 was 0.08±0.06 s (n=7), whereas it was similar to that in the standard bath solution (10.0±6.3 s; n=11) in 140 mM NaCl (10.4±3.4 s; n=7). The desensitization was also slightly accelerated in 110 mM MgCl2 (1.9±0.4 s; n=6). The current induced during acid stimulation showed a positive shift of the reversal potential in the bath solution containing 140 mM NaCl, suggesting that Na+-selective channels were activated, a current property consistent with that of ASICs (supplementary Fig 5 online).
Figure 2.
Ion selectivity of PKD1L3–PKD2L1 channels. (A) Representative activation profiles in the bath solution containing 140 mM NaCl, 110 mM CaCl2 or 110 mM MgCl2. Voltage ramp pulses (from −100 to +100 mV in 100 ms) were repeatedly applied at 1 s intervals for NaCl or 500 ms intervals for CaCl2 and MgCl2. Traces at the bottom indicate the pH of bath solution. (B) Current–voltage (I–V) relationship in the bath solution containing 140 mM NaCl, 110 mM CaCl2 or 110 mM MgCl2. Traces indicated by asterisks in (A) are presented. The acidic and neutral solution compositions are described in the supplementary information online.
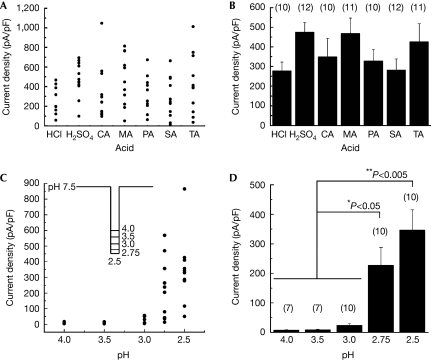
The acid-induced PKD1L3–PKD2L1 channel activation did not show any acid specificity (Fig 3A,B). The average current densities observed after treatment with various acids, including two strong and five weak acids, were similar. We did not use acetic acid because the HEK293T cells showed a nonspecific response both during and after its application (data not shown). Then, we examined the pH dependency of PKD1L3–PKD2L1 channel activation. It was difficult to normalize the acid-induced currents by activation of the PKD1L3–PKD2L1 channel because this channel showed rapid and irreversible desensitization depending on the induced current size (supplementary Fig 2 online). When a large current was induced by the first acid stimulus, little or no current was induced by the second acid stimulus (supplementary Fig 2 online), suggesting the involvement of activity-dependent mechanisms in the process. Therefore, we compared the current densities in a series of experiments performed on different cells. PKD1L3–PKD2L1-expressing cells were exposed to acids at various pH values (2.5, 2.75, 3.0, 3.5 and 4.0) towards neutralization for 5 s each (Fig 3C, inset). No off-response currents were observed after acid exposure at pH 4.0 or 3.5 (Fig 3C,D), although acid stimulation in this pH range evoked small transient currents, indicating ASIC activation (data not shown). However, off-response currents were induced by acids at a pH of less than 3.0 and their magnitudes increased as the pH decreased.
Figure 3.
Acid specificity and threshold pH for PKD1L3–PKD2L1 channel activation. (A) Current-density distribution on application of various acids, including HCl, H2SO4, citric acid (CA), malic acid (MA), phosphoric acid (PA), succinic acid (SA) and tartaric acid (TA). The pH of all the acids, except SA (pH 2.6), was adjusted to 2.5. The acidic and neutral solution compositions are described in the supplementary information online. Cells expressing PKD1L3–PKD2L1 channels were exposed to acid solutions at pH 2.5 for 5 s. (B) Average current densities induced by various acid solutions. Error bars and the values in parentheses indicate the s.e.m. and number of cells, respectively. (C) Current-density distribution induced by acids at various pH values. Cells expressing PKD1L3–PKD2L1 channels were exposed to acids at various pH values (2.5, 2.75, 3.0, 3.5 or 4.0) for 5 s each (inset). (D) Average current densities induced by the acid solutions at various pH values. Error bars and values in parentheses indicate the s.e.m. and number of cells, respectively.
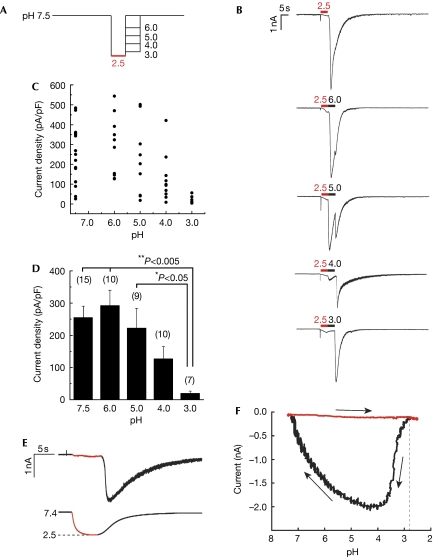
Subsequently, we examined the extent of pH increase from 2.5, the pH value required to open PKD1L3–PKD2L1 channels. PKD1L3–PKD2L1-expressing cells were first exposed to an acidic solution at pH 2.5 for 5 s and then to increasing pH values of 3.0, 4.0, 5.0, 6.0 or 7.5 for 5 s each (Fig 4A). After the initial acid stimulus (pH 2.5), exposure at pH 3.0 induced little off-responses, whereas treatment with solutions between pH 4.0 and 7.5 induced large off-response currents (Fig 4B). The induced PKD1L3–PKD2L1 current magnitude increased with the pH and plateaued at a pH value higher than 5.0 (Fig 4B–D). Finally, to determine the pH threshold for activation, we generated pH-response profiles (Fig 4E,F). On pH reduction to 2.5, off-currents were induced at pH 2.7±0.1 (n=8). No significant difference was observed among the strong and weak acids (data not shown). These results indicate that a slight increase in pH is sufficient to open PKD1L3–PKD2L1 channels after acid exposure at pH 2.5.
Figure 4.
pH dependency of PKD1L3–PKD2L1 channel activation after acid exposure. (A) Cells expressing PKD1L3–PKD2L1 channels were exposed to an acid at pH 2.5 for 5 s and then to solutions at various pH values (3.0, 4.0, 5.0, 6.0 or 7.5) for 5 s each toward neutralization (pH 7.5). (B) Representative whole-cell current profiles obtained by the protocol described in (A). Red and black bars represent treatment with the acid at pH 2.5 and with solutions at various pH values, respectively. (C) Distribution and (D) average of the current densities induced by treatment with the second solutions at various pH values. Error bars and values in parentheses indicate the s.e.m. and number of cells, respectively. (E) A representative citric acid-induced current (upper) with corresponding pH (bottom). Red parts indicate a current during the acid application. (F) A representative pH-response profile made from traces in (E). The red part indicates acid application shown in (E). Arrows indicate time course of current–pH relationship.
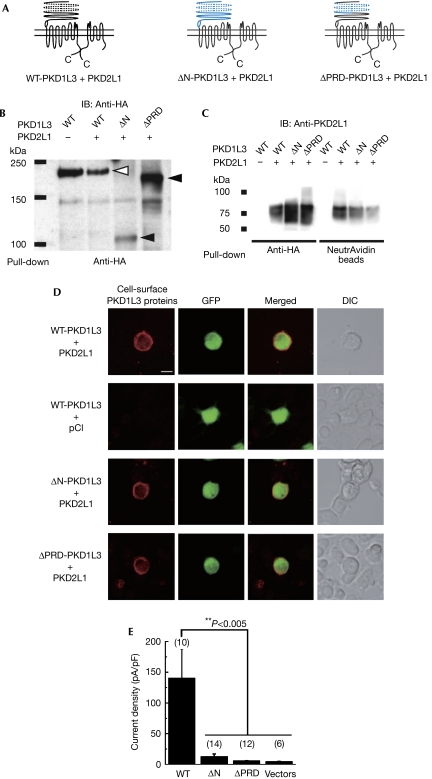
We performed a deletion analysis of the PKD1L3 protein to obtain insights into the structural basis of PKD1L3–PKD2L1 channel activation (Fig 5A). We focused on the extracellular amino-terminal region of the PKD1L3 protein for the following reasons: (i) the results of the patch-clamp analysis strongly suggested that extracellular acidification can affect the activity of PKD1L3–PKD2L1 channels; (ii) the PKD1L3 protein has a large extracellular N-terminal region, containing a proline–serine (Pro–Ser)-rich domain (179–606 aa) and a PKD homology domain (1,007–1,056 aa). We generated a deletion of the entire extracellular N-terminal region (ΔN), as well as a partial deletion of this region that removed the Pro–Ser-rich domain (ΔPRD). The expression of these deletion mutants was confirmed by immunoblotting (Fig 5B). Deletion of the entire extracellular N-terminal region of the PKD1L3 protein resulted in a significant decrease in its expression. Interactions between ΔPRD or ΔN and wild-type PKD2L1, as well as the surface expression of the deletion mutants, were detected by immunoblotting and immunostaining (Fig 5C,D). The surface expression of PKD2L1 was confirmed by a biotinylation assay (Fig 5C). ΔN or ΔPRD expression resulted in a significant reduction in channel activity, as measured by average current densities after acid treatment (Fig 5E). These results indicate that the Pro–Ser-rich extracellular N-terminal domain of PKD1L3 might be important in the correct response to acid stimulus. However, the possibility remains that deletion of the extracellular N-terminal region of the PKD1L3 protein might disrupt the PKD1L3–PKD2L1 channel function.
Figure 5.
Deletion analysis of the amino-terminal region of the PKD1L3 protein. (A) Predicted structure of PKD1L3 deletion mutants. Either the entire N-terminal sequence (1,068 amino acids (aa)) or a part of it, including the proline–serine (Pro–Ser)-rich domain (499 aa; shown in blue) was deleted (ΔN and ΔPRD, respectively). The Pro/Ser-rich domain and the PKD homology domain are indicated as a dotted line and an oval, respectively. PKD2L1 contains six transmembrane domains with a putative pore-forming region. (B–D) Surface expression of the PKD1L3 deletion mutants and their interaction with PKD2L1. (B) Membrane proteins were prepared from human embryonic kidney (HEK)293T cells expressing haemagglutinin (HA)-tagged wild-type PKD1L3 (WT) and the deletion mutants (ΔN and ΔPRD) in the presence of PKD2L1. After immunoprecipitation with an HA antibody, the PKD1L3 proteins were detected using an HA antibody. The open and filled arrowheads indicate HA-tagged wild-type PKD1L3 and the deletion mutants, respectively. (C) Association between HA-tagged wild-type PKD1L3 (WT) or the deletion mutants (ΔN and ΔPRD) and the PKD2L1 protein was confirmed by a pull-down assay. The HA-tagged PKD1L3 proteins were co-immunoprecipitated with the PKD2L1 proteins. Furthermore, the cell-surface expression of PKD2L1 was confirmed by a cell-surface biotinylation. (D) The surface expressions of the PKD1L3 deletion mutants were confirmed by immunostaining by using a confocal microscope. HEK293T cells expressing HA-tagged wild-type PKD1L3 (WT-PKD1L3) and the deletion mutants (ΔN-PKD1L3 and ΔPRD-PKD1L3) in the presence of PKD2L1 were stained with HA antibodies under non-permeabilizing conditions (scale bar, 10 μm). HEK293T cells expressing wild-type PKD1L3 in the absence of PKD2L1 were stained under the same conditions as the negative control. (E) Average current densities induced by treatment with an acid solution at pH 2.5 for 5 s in HEK293T cells expressing wild-type PKD1L3 (WT) or the deletion mutants (ΔN and ΔPRD) in the presence of PKD2L1. The vectors control indicates the data obtained from cells transfected with pDisplay/pCI. Error bars and values in parentheses indicate the s.e.m. and number of cells, respectively. The details are described in the supplementary information online. DIC, differential interference contrast; GFP, green fluorescent protein.
Discussion
Here, we have shown that PKD1L3–PKD2L1 channels have a unique property, an off-response, whereby the channels are gated open only after removal of an acid stimulus, although initial acid exposure is essential. Furthermore, we have also clarified that the apparent delayed response observed in our previous report was actually not a delayed response, but rather an off-response. We performed a deletion analysis of PKD1L3 to obtain further insight into the structural basis of PKD1L3–PKD2L1 channel activation (Fig 5). As there was little current response in the deletion mutants, we could not clearly identify the pH-sensing domain in PKD1L3; however, the extracellular N-terminal region could be a potential candidate. Although the mechanism of PKD1L3–PKD2L1 channel activation remains to be explained, this channel could represent the first example of an off-channel, a new type of ion channel, gated open by the removal of an initial stimulus.
An off-response has often been observed to be induced by stimuli associated with sour taste sensation in mammals (DeSimone et al, 1995; Danilova et al, 2002; Lin et al, 2002). In rats, for example, additional off-responses have been reported to occur in the chorda tympani (CT) nerves and in some acid-responsive taste receptor cells (DeSimone et al, 1995; Lin et al, 2002) after the removal of an acid stimulus. A strong off-response to acidic stimuli has also been detected in both the whole CT and glossopharyngeal nerves, as well as single CT fibre recordings in a New-World monkey, the common marmoset (Danilova et al, 2002). The off-response property of PKD1L3–PKD2L1 channels provides the most feasible explanation for the off-response associated with the perception of sour taste in mammals.
What is the physiological significance of the PKD1L3–PKD2L1 channel off-responses? The sour-stimulus-induced on-responses noted in the electrical recordings of taste receptor cells and nerve fibres contribute to sour taste sensation in mammals, whereas the off-response property of PKD1L3–PKD2L1 channels seems to be inconsistent with this phenomenon in vivo. This might be because sour taste sensation involves two different mechanisms. The first might occur through an on-response induced by a moderately low pH and involves other sour taste receptors, whereas the second mechanism might be related to an off-response caused by an extremely low pH. Both of these mechanisms might contribute to sour taste sensation in mammals. The off-response property of PKD1L3–PKD2L1 channels might be important in the prolonged sensation of sour taste induced by an extremely low pH.
Another possible explanation is that the off-response itself has a significant role in sour taste detection in vivo. The taste receptor cells located in the circumvallate and foliate papillae, in which PKD1L3 and PKD2L1 are coexpressed, contain salivary glands (von Ebner's glands) at the bottom of the cleft. As sour stimuli are known to enhance the secretion of saliva by salivary glands, such as the parotid glands (Hodson & Linden, 2006), acidic solutions seem to be instantaneously diluted by saliva in the mouth, resulting in an increase in pH. It is possible that the activation of PKD1L3–PKD2L1 channels might occur shortly after acid stimulation, as a slight increase in pH from the initial acidification at pH 2.5 is sufficient to open the channels.
It still remains to be solved whether the off-response of taste cells in vivo is mediated by the PKD1L3–PKD2L1 channel complex and how the off-response of the channel complex contributes to sour taste sensation. DTA-mediated ablation of PKD2L1-expressing taste cells clearly showed that these cells are essential for responses to sour stimuli in vivo (Huang et al, 2006). However, the DTA-mediated ablation of the taste cells should also ablate a coexpressed ‘on'-sour receptor. It is possible that our heterologous expression system using HEK293T cells might be lacking additional taste cell components that might confer different properties onto the PKD1L3–PKD2L1 channel complex. Generation and analyses of PKD1L3 and PKD2L1 gene knockout mice will help to define further the roles of PKD1L3 and PKD2L1 in vivo.
Methods
Ca2+ imaging and electrophysiological analysis. The following plasmid constructs have been reported previously: mouse PKD1L3 cDNA (AB290926) and mouse PKD2L1 cDNA (AB290927; Ishimaru et al, 2006). HEK293T cells were transfected by using Lipofectamine2000 for Ca2+ imaging and Lipofectamine for patch-clamp analysis. The pDsRed (BD Bioscience Clontech, Franklin Lakes, NJ, USA) and pGreen Lantern-1 (Life Technologies, Carlsbad, CA, USA) plasmids were used as expression markers for Ca2+ imaging and patch-clamp analysis, respectively. HEK293T cells were transfected and incubated for 30–36 h before the experiments, which were carried out at room temperature (23–25°C). Whole-cell patch-clamp recordings were sampled at 10 kHz and filtered at 5 kHz for analysis using an Axopatch 200B amplifier equipped with pCLAMP software (Molecular Devices, Sunnyvale, CA, USA) The standard bath solution that was used for fura-2-mediated Ca2+ imaging and for the whole-cell recordings contained 140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM HEPES and 10 mM glucose (pH 7.4, adjusted using NaOH). The pipette solution contained 140 mM KCl, 5 mM EGTA and 10 mM HEPES (pH 7.4, adjusted using KOH). In the experiments conducted for the outside-out patch configuration and in the whole-cell recordings that were performed in pH-dependent steps, we used bath solutions at a controlled pH. These solutions contained 140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 5 mM HEPES, 5 mM MES, 5 mM citric acid and 5 mM glucose (pH adjusted using HCl or NaOH). The Perfusion Fast-Step system (SF-77B; Warner Instruments, Hamden, CT, USA) was used for the application of acidic solutions for short durations (1–4 s). The bath solution used for the inside-out configuration contained 140 mM KCl, 5 mM EGTA and 10 mM HEPES, in the presence (pH 2.8) or absence (pH 7.4) of 25 mM citric acid. The pipette solution contained 140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 5 mM HEPES, 5 mM MES, 5 mM citric acid and 5 mM glucose (pH 7.5, adjusted using NaOH). The pH of the bath solution was monitored as voltage using a pH electrode (AMANI-650 pH sensor, Warner Instruments) with a pH monitor buffer device (Inter Medical Co, Nagoya, Japan) and calculated after recordings. Statistical significance was assessed by performing pairwise multiple comparisons using the Holm's method.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Method
Acknowledgments
We thank Y. Kubo, Y. Okamura and Y. Ninomiya for their useful suggestions and the staff of Tominaga Laboratory. This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) to M.T., a National Institutes of Health grant DC05782 and DC 008967 to H.M., a Japan Society for the Promotion of Science fellowship to Y.I. and a Grant-in-Aid for Young Scientists (Start-up) 19880008 to Y.I. from the MEXT.
Footnotes
The authors declare that they have no conflict of interest.
References
- Chen XZ, Vassilev PM, Basora N, Peng JB, Nomura H, Segal Y, Brown EM, Reeders ST, Hediger MA, Zhou J (1999) Polycystin-L is a calcium-regulated cation channel permeable to calcium ions. Nature 401: 383–386 [DOI] [PubMed] [Google Scholar]
- Danilova V, Danilov Y, Roberts T, Tinti JM, Nofre C, Hellekant G (2002) Sense of taste in a New World monkey, the common marmoset: recordings from the chorda tympani and glossopharyngeal nerves. J Neurophysiol 88: 579–594 [DOI] [PubMed] [Google Scholar]
- DeSimone JA, Callaham EM, Heck GL (1995) Chorda tympani taste response of rat to hydrochloric acid subject to voltage-clamped lingual receptive field. Am J Physiol 268: C1295–C1300 [DOI] [PubMed] [Google Scholar]
- Hodson NA, Linden RW (2006) The effect of monosodium glutamate on parotid salivary flow in comparison to the response to representatives of the other four basic tastes. Physiol Behav 89: 711–717 [DOI] [PubMed] [Google Scholar]
- Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Trankner D, Ryba NJ, Zuker CS (2006) The cells and logic for mammalian sour taste detection. Nature 442: 934–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H (2006) Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci USA 103: 12569–12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Ogura T, Kinnamon SC (2002) Acid-activated cation currents in rat vallate taste receptor cells. J Neurophysiol 88: 133–141 [DOI] [PubMed] [Google Scholar]
- Lin W, Burks CA, Hansen DR, Kinnamon SC, Gilbertson TA (2004) Taste receptor cells express pH-sensitive leak K+ channels. J Neurophysiol 92: 2909–2919 [DOI] [PubMed] [Google Scholar]
- Richter TA, Dvoryanchikov GA, Chaudhari N, Roper SD (2004) Acid-sensitive two-pore domain potassium (K2P) channels in mouse taste buds. J Neurophysiol 92: 1928–1936 [DOI] [PubMed] [Google Scholar]
- Stevens DR, Seifert R, Bufe B, Muller F, Kremmer E, Gauss R, Meyerhof W, Kaupp UB, Lindemann B (2001) Hyperpolarization-activated channels HCN1 and HCN4 mediate responses to sour stimuli. Nature 413: 631–635 [DOI] [PubMed] [Google Scholar]
- Ugawa S, Minami Y, Guo W, Saishin Y, Takatsuji K, Yamamoto T, Tohyama M, Shimada S (1998) Receptor that leaves a sour taste in the mouth. Nature 395: 555–556 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ (2003) Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112: 293–301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Method