Abstract
A novel mutation, repro5, was isolated in a forward genetic screen for infertility mutations induced by ENU mutagenesis. Homozygous mutant mice were phenotypically normal but were infertile. Oocytes from mutant females appeared normal, but were severely maturation-defective in that they had reduced ability to progress to metaphase II (MII), and those reaching MII were unable to progress beyond the two pronuclei stage following in vitro fertilization (IVF). Mutant males exhibited defective spermiogenesis, resulting in oligoasthenoteratospermia. Genetic mapping, positional cloning, and complementation studies with a disruption allele led to the identification of a mutation in Brwd1 (Bromodomain and WD repeat domain containing 1) as the causative lesion. Bromodomain-containing proteins typically interact with regions of chromatin containing histones hyperacetylated at lysine residues, a characteristic of chromatin in early spermiogenesis before eventual replacement of histones by the protamines. Previous data indicated that Brwd1 is broadly expressed, encoding a putative transcriptional regulator that is believed to act on chromatin through interactions with the Brg1-dependent SWI/SNF chromatin-remodeling pathway. Brwd1 represents one of a small number of genes whose elimination disrupts gametogenesis in both sexes after the major events of meiotic prophase I have been completed.
Keywords: spermiogenesis, bromodomain, chromatin remodeling, infertility, mouse
INTRODUCTION
Approximately 15% of all couples experience infertility, regardless of race or ethnicity (Matzuk and Lamb, 2002). Still, the molecular mechanisms underlying commonly observed gamete defects, such as oligospermia and teratospermia, are largely unknown. The genetic causes of human infertility are poorly defined due to difficulties such as identifying families segregating discrete infertility alleles (Gianotten et al., 2004). Cases of infertility can have complex and disparate molecular etiologies, further confounding genetic analysis.
Most of the progress in understanding the genetics of human infertility has come from analysis of gene-targeted mutations in laboratory mice. This type of “reverse genetic” approach – the selection of genes to mutate based on an assumed role in a biological process – is inherently biased upon pre-existing knowledge leading to selection of a gene for further functional analysis. Nevertheless, these mouse models have provided a foundation for identification of genetic causes of human infertility. Candidate gene sequencing of infertile patients has led to the identification of mutations or sequence variants in the recombination gene DMC1 (Mandon-Pepin et al., 2002) and the synaptonemal complex gene SYCP3 (Miyamoto et al., 2003). However, whether these variants are actually responsible for infertility in these patients is unknown. Overall, progress in identifying genetic causes of human infertility has been limited.
Clearly, one major factor in the slow progress in our understanding of the genetics of human infertility is related to our as-yet incomplete knowledge of the genes required for all aspects of gamete production. Despite progress in defining mouse infertility genes via targeted mutagenesis, most genes required for gametogenesis remain to be identified and/or modeled. It has been estimated that at least 2,000 genes may be required for normal spermatogenesis (Hargreave, 2000). Forward genetic screens address those genes that are not previously implicated in a particular biological process. In previous work, we conducted the first genome-wide forward genetic screens for infertility mutants in mice (Ward et al., 2003), and this led to a large-scale “ReproGenomics” program that has generated scores of new infertility models by ethylnitrosourea (ENU) germline mutagenesis (Handel et al., 2006).
One mutation identified in the ReproGenomics screen, repro5, is recessive and causes both male and female infertility. Most of the mutations that cause infertility in both sexes disrupt fundamental processes in meiosis I (Matzuk and Lamb, 2002). repro5 is unique in that males are infertile due to spermiogenesis defects, and females are infertile due to defect(s) in oocyte maturation after meiotic prophase. Therefore, it seemed likely that the underlying molecular defect would reveal a novel mechanism in common between spermiogenesis and oocyte maturation.
Here, we report the positional cloning of repro5 and identification of a point mutation in the gene Brwd1, formerly known as Wdr9. BRWD1 is a member of a novel protein family that has both WD40 repeat domains (seven) and dual bromodomains. WD40 repeats are present in diverse eukaryotic proteins such as G proteins, phosphatases and transcription regulators (Smith et al., 1999). These ~40 amino acid repeats form structures that interact with other proteins or ligands. The bromodomain is an ~110 aa motif found in organisms ranging from yeast to mammals (Haynes et al., 1992). Bromodomain-containing proteins are involved in diverse cellular functions including transcriptional activation, transcriptional silencing, chromatin remodeling, mRNA splicing, and DNA replication.
The studies presented herein paint an enigmatic picture of a shared genetic defect leading to disparate gamete phenotypes in male vs female repro5 mice. Identification of Brwd1 as the causal gene enables us to speculate that the common molecular basis for defects in spermiogenesis and egg maturation may be misregulated gene expression or aberrant chromatin remodeling at a crucial time after completion of the major events of meiotic prophase I.
MATERIALS AND METHODS
Mutagenesis and genetic mapping
Male C57BL/6J (B6) mice were mutagenized with ethylnitrosourea (ENU) as described (Wilson et al., 2005), then bred to C3HeB/FeJ (C3H) females to create “G1” male founders of pedigrees. These males were backcrossed to C3H females, and G2 daughters were backcrossed to the G1 male to yield G3 offspring that were tested for infertility (Lessard et al., 2004). DNA was isolated from infertile G3 animals in the repro5 pedigree, and a genome scan was performed with microsatellite markers polymorphic between C3H and B6.
After linkage to distal Chr 16 was obtained, and additional breeding was performed as described in RESULTS, some recombinants in the critical region had to be evaluated by progeny testing (because the recombinant chromosome was in trans to a wild-type chromosome). This was done by crossing to a known heterozygote. Because the critical region had few polymorphisms between B6 and C3H, several recombinants were crossed to strain 129SvImJ (“129”) to exploit several known B6 vs 129 SNPs in the region. Critical for this study were SNPs rs4221076 and rs4221077, located 26bp apart. The following primers were designed to amplify across both SNPs.
Kcnj6F:TAGTGTGGACTGCCTGTCTC and Kcnj6R: AGCTTTCCTGCCTTTAAAAC. The genotype of recombinants at SNP loci was evaluated using a Transgenomic WAVE denaturing HPLC machine.
Gene trap mice
The AW0417 ES cell line was obtained from the Sanger Institute Gene Trap Resource (SIGTR). Chimeras were generated by microinjection of the ES cells into C57BL/6J blastocysts, using standard procedures.
Histology
Gonads were fixed in Bouin’s overnight, paraffin-embedded, and stained with either hemotoxylin and eosin (H&E) or periodic Acid Schiff (PAS). For transmission electron microscopy, testes were fixed overnight at 4°C in 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.2). After a wash in cacodylate buffer and post fixation in 1% osmium tetroxide in cacodylate buffer, the preparations were then dehydrated and embedded in Embed-812 resin (Electron Microscopy Sciences, Hatfield, PA). Sections were stained with 2% uranyl acetate and Reynolds lead citrate and evaluated using a JEOL1230 transmission electron microscope. The classification of spermatid step was based on Russell et al (Russell et al., 1990).
Sperm analysis and meiotic chromosome spreads
Epididymides and vasa deferentia were removed immediately postmortem, minced, suspended in one milliliter of phosphate buffered saline and left at 37 degrees for 60 minutes in 5% CO2 to allow sperm to swim out. Motility was qualitatively assessed at this point. Aliquots were removed for quantification on a hemacytometer. For photography under phase contrast, sperm were placed at 4° for >1 hour to reduce or eliminate motility, then aliquots were placed on microscope slides under a cover glass. Preparation and immunolabelling of surface spread spermatocyte chromosomes were done as described (Reinholdt et al., 2004). Chromosome constitution of spermatocytes was evaluated from air-dried chromosome preparations (Evans et al., 1964). Briefly, spermatocytes from repro5 homozygous and heterozygous littermates were collected by centrifugation and washed in 2.2% sodium citrate. The air-dried chromosome preparations were stained with Gurr Giemsa (Invitrogen). Metaphase I and metaphase II chromosomes were imaged at 400X using a Hamamatsu C5810 cool-chilled camera (Photonic System, Bridgewater, NJ), and adjusted in Adobe Photoshop.
RT-PCR Analysis
Total RNA was collected from testes and ovaries using RNeasy mini kits (Qiagen). RNA was quantified using a NanoDrop apparatus. RT-PCR was performed using random hexamer primers on 1 µg of total RNA with SuperScript III, according to the manufacturer’s instructions. PCR was then performed using 3 µL of cDNA in a 30 µL total reaction volume. Primer sequences are available upon request.
Oocyte maturation and fertilization in vitro
Adult females were injected with 5 IU equine chorionic gonadotropin and ovaries removed 44 hours later. Oocyte-cumulus cells complexes were isolated and matured for 15 hours in vitro as described previously (O'Brien et al., 2003) except the medium for maturation was supplemented with 5% fetal bovine serum. After removal of cumulus cells with hyaluronidase, matured oocytes were carefully inspected using a stereomicroscope for the presence or absence of a polar body. Those exhibiting a polar body were presumed to have progressed to MII. MII oocytes were then fertilized in vitro using capacitated B6SJLF1 cauda epididymal sperm and inseminated eggs were cultured as described previously (O'Brien et al., 2003).
RESULTS
The ENU-induced repro5 mutation causes infertility in both sexes
The repro5 mutation was identified in a whole-genome ENU mutagenesis screen designed to uncover mouse infertility mutants (Lessard et al., 2004). In brief, mutant pedigrees were founded by male offspring (G1 generation) of ENU-treated C57BL/6J (“B6”) males who were outcrossed to C3HeB/FeJ (“C3H”) females. G1 founders were mated to C3H females to produce several G2 daughters, each of which was backcrossed to the G1 to produce G3 animals for fertility testing. The pedigree segregating repro5 contained G2 females that, upon mating to the G1 founder, produced infertile male and female offspring (although 1 genetically mutant female gave birth to a single offspring) that were otherwise phenotypically normal. Subsequent analyses (enabled by direct molecular genotyping, see below) revealed that repro5 segregates as a simple recessive Mendelian trait, in which homozygotes were produced from intercrosses of heterozygotes at a frequency (21.5%) slightly, but insignificantly less that 25% (84/390; χ2 = 2.5; P=0.114).
repro5 affects the oocyte-to-embryo transition
To elucidate the basis for female infertility in repro5/repro5 mice, we first examined ovarian histology. repro5 mutant ovaries were grossly normal compared to controls. They contained follicles and oocytes at various stages of development, and corpora lutea were present (data not shown). To address whether repro5 affects processes required for the maintenance of pregnancy, wild-type C57BL/6J embryos were transferred into control and repro5/repro5 pseudopregnant females. Both sustained full-term pregnancies (data not shown).
Fully-grown germinal vesicle (GV) stage mutant oocytes (isolated 44 hours following stimulation with eCG; see Materials and Methods) were morphologically indistinguishable from WT controls. To assess the developmental capacity of oocytes produced by infertile mutant females, in vitro maturation and insemination was performed. Included in the analysis is a disruption allele of repro5, Brwd1Gt, which is described below. Only 44% of mutant oocytes progressed to MII (vs 94% of controls), and nearly all inseminated mutant MII oocytes developed only to the two pronuclear stage and did not cleave (Table 1; Fig. 1). No inseminated mutant oocytes developed to the blastocyst stage in vitro (compared to 36% of controls). Taken together, these results show that mutation of Brwd1 does not overtly affect the morphology of follicular development or fully-grown oocytes, however, oocyte competence to complete the oocyte-to-embryo transition is dramatically impaired.
TABLE 1. In vitro oocyte maturation and embryo development after IVF.
“Gt” refers to the gene trap allele, Brwd1AW0417. All data are from cumulus-surrounded oocytes. Subtotals from the 6 mutant animals and 4 non-mutant animals are presented. The +/+ animals were littermates of repro5 and Gt mutants. Only MII eggs were inseminated. The number of eggs at the pronuclear and 2-cell stage was determined; 24 hours after IVF; the percentage is based on the number of MII eggs. The number of blastocysts was determined after culture of 2-cell or pronuclear stage embryos for an additional four days; the percentage is based on the number of MII eggs.
| Genotype | Total | GV | MI | MII | Pronuclei | 2-cell | Blasts |
|---|---|---|---|---|---|---|---|
| Gt/Gt | 18 | 2 | 9 | 7 | 7 | 0 | 0 |
| repro5/repro5 | 24 | 2 | 17 | 5 | 5 | 0 | 0 |
| repro5/repro5 | 54 | 0 | 27 | 27 | 16 | 4 | 0 |
| repro5/repro5 | 9 | 1 | 7 | 1 | 0 | 0 | |
| repro5/repro5 | 10 | 1 | 4 | 5 | 1 | 0 | |
| repro5/repro5 | 15 | 0 | 9 | 6 | 2 | 0 | |
| Totals | 116 | 6 (5%) | 73 (63%) | 51 (44%) | 31 (61%) | 4 (6%) | 0 |
| Gt/+ | 34 | 0 | 0 | 34 | 0 | 34 | 14 |
| +/+ | 14 | 2 | 0 | 12 | 0 | 10 | 7 |
| +/+ | 19 | 1 | 1 | 17 | 0 | 11 | 4 |
| +/+ | 20 | 0 | 0 | 20 | 0 | 11 | 5 |
| Totals | 87 | 3 (3%) | 1 (1%) | 83 (94%) | 0 | 66 (80%) | 30 (36) |
FIG. 1. Effect of the repro5 mutation on oocyte morphology and zygotic development.
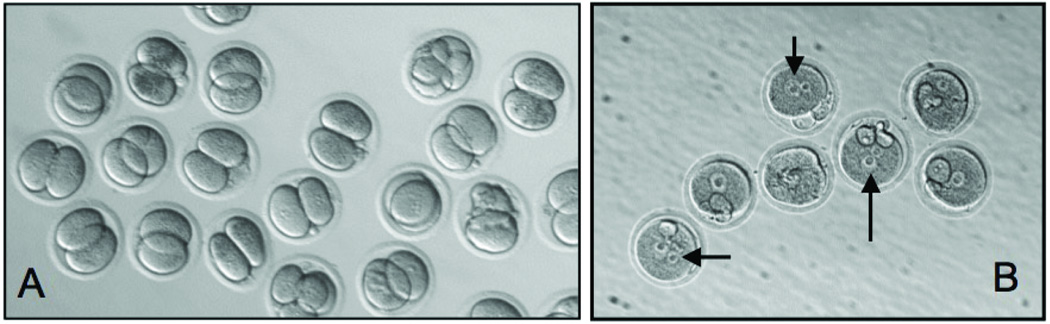
Panels A (WT) and B (mutant) are micrographs taken 24 hours after insemination and show that the WT oocytes cleaved normally while the mutant eggs all arrested at the two pronuclear stage. The mutant eggs failed to undergo further development. Arrows point to unfused pronuclei.
repro5/repro5 males are oligoasthenoteratospermic and exhibit abnormal spermiogenesis
repro5/repro5 males were invariably infertile. To explore the cause, we assessed the sperm count and quality by microscopy, histological analysis of testes, and IVF. Mutant mice had extremely reduced epididymal sperm concentrations (Table 2). In one litter, two mutants (# 6&7) had sperm concentrations ~20 fold lower than 2 heterozygous (#1&2) littermates (5×104 vs 1.05×106/ml). The vast majority of epididymal sperm recovered from mutants were non-motile or poorly motile. Furthermore, the majority of mutant sperm were dead as determined by viability staining with Eosin-Nigrosin (not shown) (Bjorndahl et al., 2003). Epididymal sperm from four G3 mutants failed to fertilize WT eggs in IVF experiments, as judged by the virtual absence of oocyte development to the 2-cell stage (N=1/118 oocytes; but because a concurrent no-sperm control also produced two 2-cell embryos, it is assumed that these cleaved eggs had undergone parthenogenetic activation), compared to heterozygote controls, which exhibited a fertilization rate of 86% (77/89 oocytes). In G3 animals, >90% of the mutant sperm were morphologically abnormal, particularly with respect to head shape, in contrast to <10% in wild type littermates (Table 2). Whereas normal mouse sperm have a hook-shaped head (Fig. 2A), mutant sperm had a variety of abnormal shapes (Fig. 2B). Additionally, mutant sperm tails were often defective. Most notable was a degenerated or “ragged” appearance of the midpiece, the structure largely comprised of a mitochondrial sheath (Fig. 2B). Because mutant males have low sperm counts, morphologically abnormal sperm, and poorly motile sperm, they can be classified by oligoasthenoteratozoospermia.
Table 2. Sperm counts of mice with various allele combinations.
The allelotypes are abbreviated as in the text.
| # Genotype | Sperm/ml* | % abnormal Sperm^ |
|---|---|---|
| 1 repro5/+ | 1.3×106 | 6% |
| 2 repro5/+ | 8.0×105 | 6% |
| 3 Gt/+ | 1.0×106 | 4% |
| 4 +/+ | 1.3×106 | 8% |
| 5 Gt/+ | 9.0×105 | 11% |
| Avg | 1.0×106 | 7% |
| 6 repro5/repro5 | 5.0×104 | 98% |
| 7 repro5/repro5 | 5.0×104 | 97% |
| Avg | 5.0×104 | 97.5% |
| 8 repro5/Gt | 2.0×105 | 65% |
| 9 repro5/Gt | 1.7×105 | 96% |
| Avg | 1.85×105 | 81% |
| 10(Gt/Gt | 3.3×105 | 81% |
| 1 Gt/Gt | 1.9×105 | 86% |
| 1;Gt/Gt | 4.5×105 | 95% |
| Avg | 3.2×105 | 87.3% |
Sperm concentration was determined by mincing 1 epididymis into 2 ml of PBS, and counting after 1 hour.
The percentage of abnormal sperm was determined by counting 100 sperm from each mouse. The following groups of animals were littermates: (1,2,6,7); (3,11,12); (4,5,8,9). Animals 10 was sired by the same parents as 11 & 12. All males were 2–4 months of age.
FIG. 2. Abnormal sperm development in repro5 mutants.
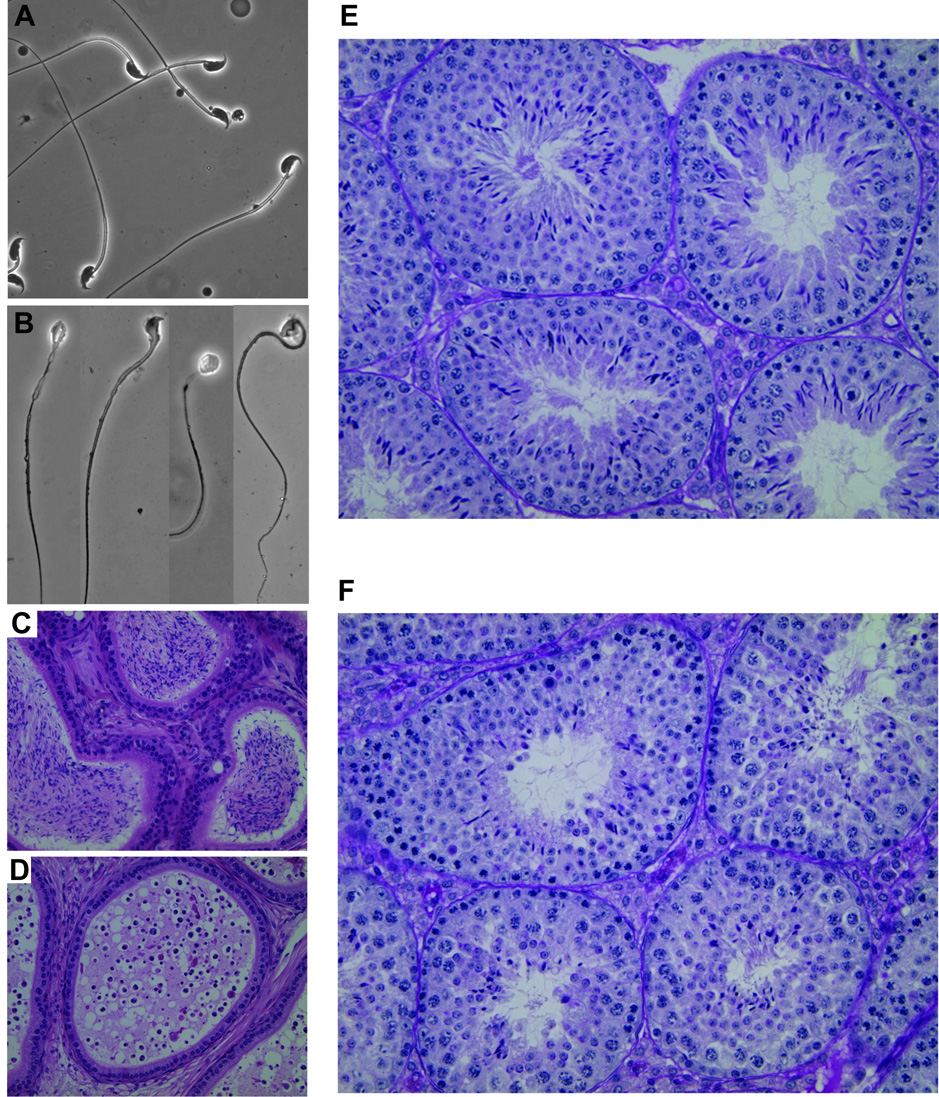
(A) Wild-type sperm. (B) Sperm from a repro5/repro5 littermate. The phase-contrast images (~60X) of mutant sperm were taken individually, due to low concentration. All have abnormal head shape, and some have abnormal midpieces with a ragged appearance. (C) Wild-type section of an epididymis, with sperm-filled lumens. (D) Section of a repro5/repro5 epididymis. Note the prevalent round cells. (E) PAS-stained section of wild-type testis showing several seminiferous tubules containing elongated spermatids with typical hook-shaped heads. (F) PAS-stained mutant testis section showing multiple seminiferous tubules containing spermatids with abnormally shaped heads. The wild-type and mutant animals were littermates.
Both the decrease in postmeiotic sperm number and abnormal morphology were evident in epididymal and testicular histology. In comparison to control littermates (Fig. 2C), epididymides of mutant mice were depleted of sperm; they contained mostly round cells of unknown nature (Fig. 2D). Unlike control seminiferous tubule sections, where uniform cohorts of adluminal elongating spermatids exhibiting typical head and acrosome morphologies (revealed by PAS staining; Fig. 2E), there were decreased numbers of elongating spermatids in repro5/repro5 testes, and both spermatid heads and acrosomes were malformed (Fig. 2F). Transmission electron microscopy revealed that the initial stages of spermiogenesis in repro5 mutant testes appeared normal. Compared to heterozygous littermates, the number and morphology of round spermatids in repro5 mutant were not different (Fig. 3A,B), and no abnormalities were detected prior to step 6 of spermiogenesis. However, the numbers of spermatids of later steps were reduced, and the morphology of these spermatids was aberrant. Malformed spermatid heads were evident (Fig. 3C–F), exhibiting defective chromatin condensation, and failure of normal acrosome mophogenesis. In spite of these postmeiotic abnormalities, germ cell development before spermiogenesis appeared normal.
FIG. 3. Spermiogenic abnormalities in repro5 mutants.
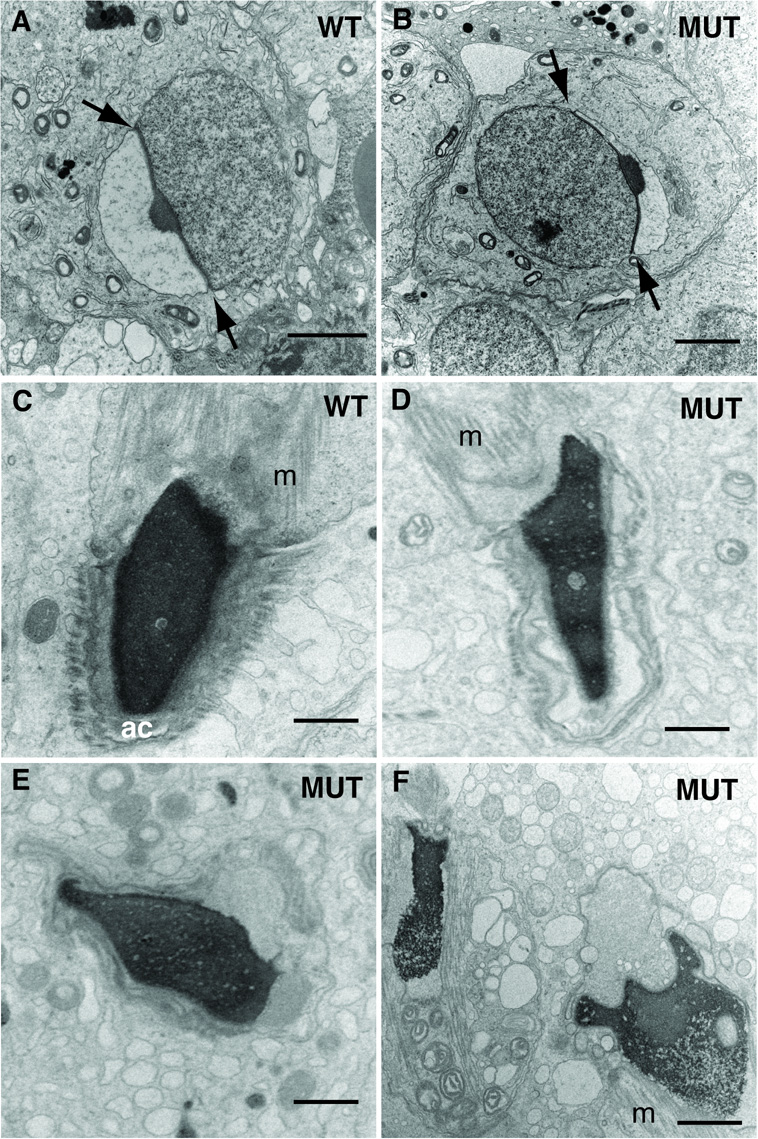
Transmission electron microscopy revealed aspects of spermatid head and acrosome abnormalities after step 6 of spermiogenesis. (A) Heterozygous (“WT”) step 6 spermatid, and (B) repro5/repro5 (“MUT”) spermatid. In both, the angle subtended by the acrosome (between arrows) extends to about 100°. Bar (A−B) = 2 µm. (C) The heads of spermatids in heterozygous repro5 spermatids reveal evenly distributed condensed chromatin in the nucleus, and normal morphology of the acrosome (“ac”) and manchette (“m”). (D–F) show typical irregular shapes of spermatid heads in repro5/repro5 mutant males. These images reveal observed defects and irregularities in chromatin condensation and abnormalities of acrosome differentiation. Bar (C−F) = 1 µm.
Aside from those depleting the germ cell pool, nearly all mouse mutations causing infertility in both sexes produce defects in meiosis. Meiotic defects in males typically leads to early arrest of spermatogenesis in Prophase I, characterized by massive germ cell loss and smaller testis size. The repro5 male phenotype is clearly different, and the normal histological morphology of spermatogonia and spermatocytes suggests that the defects are post-meiotic rather than meiotic. Nonetheless, there are cases, like the PL/J strain of mice, in which germ cells progress through meiosis despite a high load of aneuploidy, resulting in elevated numbers of abnormal sperm and lower fertility (Pyle and Handel, 2003). Therefore, to determine more definitively if the repro5 allele causes subtle meiotic defects, chromosomes were visualized after surface-spreading of chromatin and immunolabelling with antibodies against two proteins: SYCP3 (a component of the axial element of the synaptonemal complex) and phosphorylated histone variant H2AX (γH2AX), which is present at the sites of double strand breaks (DSBs), the sex body during pachynema, and asynapsed regions of pachytene chromosomes. Together, these antibodies allow assessment of chromosome pairing, status of DSB repair, and meiotic nuclear morphology.
In normal pachytene spermatocytes, 19 fully synapsed autosomes plus the terminally synapsed sex chromosomes are present (Fig. 4A). As shown in Fig. 4B, repro5 spermatocytes showed normal pairing of chromosomes, and a typical sex body (as indicated by intense staining with anti-γH2AX). From these observations, we conclude that there is no gross perturbation to either chromosome synapsis or DSB processing in repro5/repro5 spermatocytes. Additionally, spermatocytes at metaphase I and metaphase II were examined from both repro5 homozygous and heterozygous males; these displayed normal number and configuration of chromosomes (Fig. 4C–F) further confirming lack of obvious meiotic abnormalities.
FIG. 4. Normal meiotic chromosome pairing in repro5 spermatocytes.
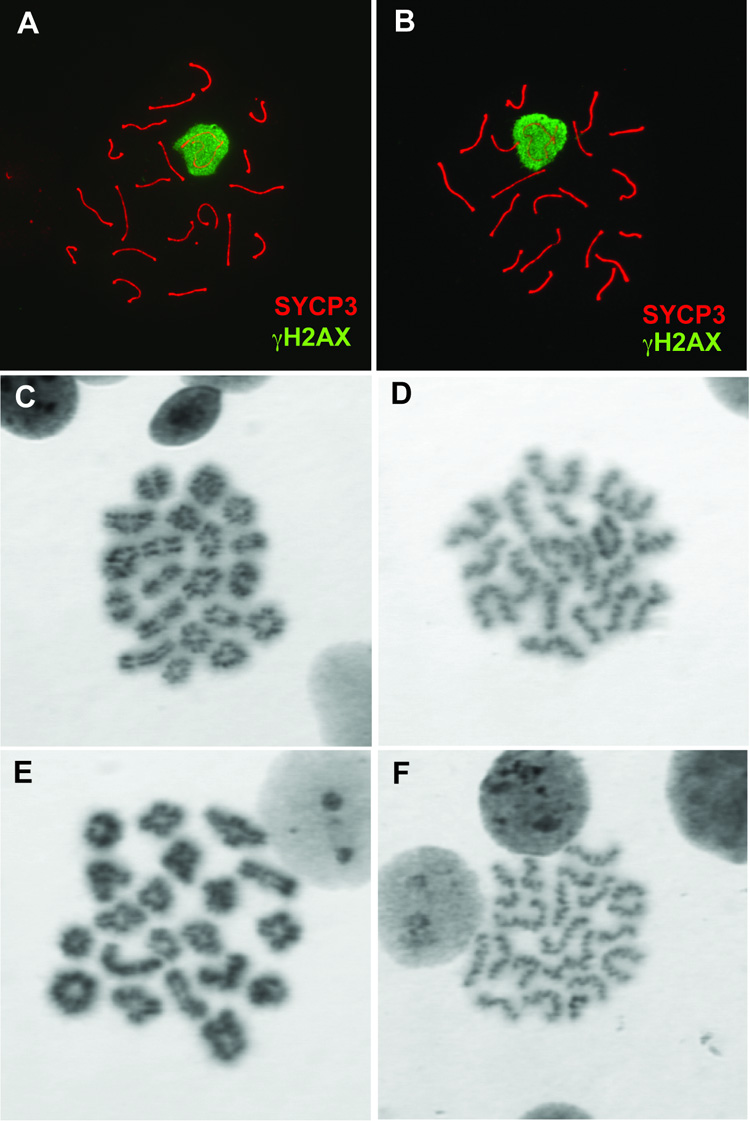
Surface-spread pachytene spermatocyte chromosomes from repro5 heterozygous (A) and homozygous (B) littermates co-immunostained with antibodies directed against SYCP3 (red) and γH2AX (green). In both samples, chromosomes are fully paired, and autosomes are devoid of γH2AX staining, which would otherwise indicate defects in synapsis or recombination. The transcriptionally silenced XY bodies are intensely stained with anti- γH2AX, as is typical for spermatocytes at this stage. In both heterozygous (C and D) and repro5/repro5 mutant (E and F) testes, 20 bivalents were observed in air-dried preparations of metaphase I (C and E) and metaphase II (D and F) spermatocytes.
Positional cloning of repro5 reveals a T to C transition that ablates a conserved splice site in Brwd1 and is responsible for the repro5 phenotype
To determine the location of the causative mutation for the observed gametogenic phenotypes, we took advantage of the mutagenesis breeding scheme to genetically map repro5. The mutation was induced in B6 males, then backcrossed twice to C3H to create the G1 and G2 animals, which were essentially at the F1 and N2 generation into C3H. We genotyped G3 offspring (G1 × G2) of proven G2 carriers with a genomewide panel (2–3 per chromosome) of polymorphic microsatellite markers, and identified regions that were homozygous for B6 in all sterile mice, but not in fertile mice. This analysis revealed linkage to a sub-region of distal chromosome 16, sharing conserved synteny with the human Down syndrome critical region. After this initial mapping, we continued backcrossing repro5 heterozygotes to C3H, and genotyping offspring for crossovers in the critical region. Recombinants were test-mated to known carriers to assess fertility of offspring inheriting the recombinant chromosome in trans to repro5 (characterized as such by having a B6 haplotype throughout the critical region). Analyses of 390 backcross animals narrowed the repro5 critical region to an interval between D16Mit86 and D16Mit71 (Fig. 5A), defined by 42 recombinants. These 42 were further analyzed with SNP markers within the region, leading to the identification of 5 recombinants between rs422107 and D16Mit71, an ~1.8 Mb region (according to the Mus musculus genome Build 36) within which repro5 must reside (Fig. 5A).
FIG. 5. Mapping and positional cloning of the repro5 mutation.
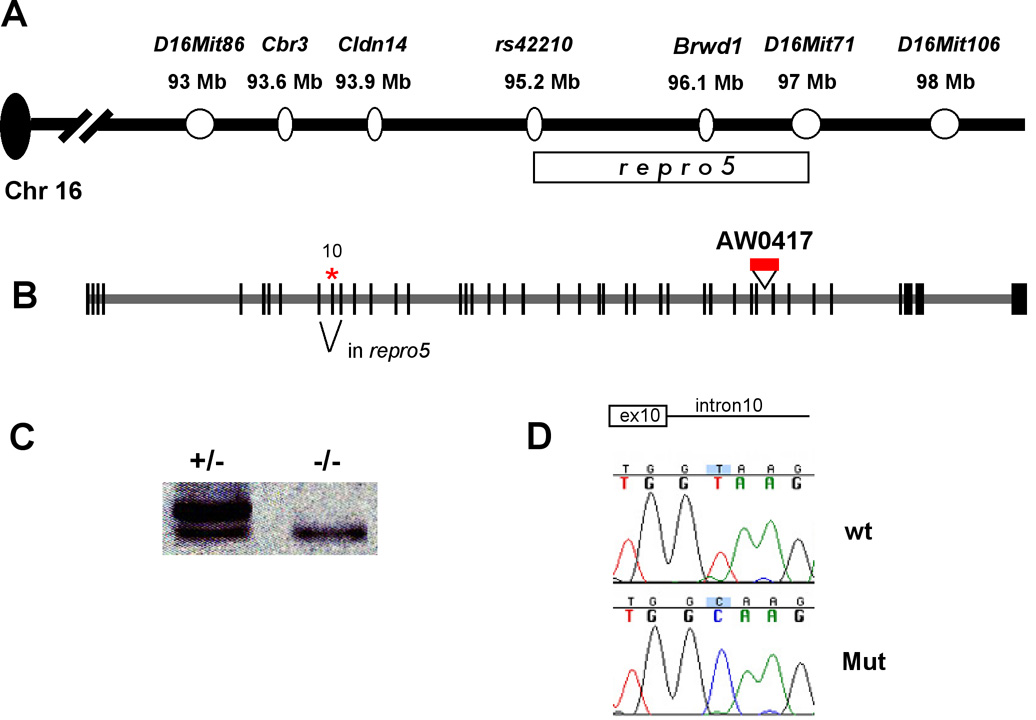
(A) Map of distal mouse Chr 16, including markers used to genetically define the repro5 critical region (delineated by rectangle under the map). (B) Genomic structure of Brwd1 with asterisk indicating the location of the Repro5 point mutation (see “D”) in the exon 10 splice junction that causes skipping of it during splicing. Also shown is a gene trap insertion, AW0417, used for complementation analyses. Information on this cell line can be obtained at www.genetrap.org. (C) RT-PCR analysis of repro5/+ and repro5/repro5 testis RNA. Primers were situated in exons 8 and 12. Mutant testes only contain the smaller product, which is missing exon 10. (D) DNA sequence chromatograms from homozygous wild-type and mutant animals across the exon10/intron10 junction, as indicated. There is a C>T change in the repro5 allele.
This 1.8 Mb critical region contains 16 annotated genes. These are listed in Supplemental Table 1, along with gene descriptions and predicted or known functions of their encoded proteins. Targeted alleles have been reported for four of these genes, but none cause phenotypes resembling that of repro5. To identify the causative mutation, RT-PCR was performed on mutant and wild-type (B6) testes and ovary RNA for all of the genes. Amplification products were obtained for seven: Erg, Brwd1, AK007051, Hmgn1, Wrb, AK029554, and B3galt5. The coding regions of alleles from mutant animals were amplified using a combination of RT-PCR and PCR of genomic DNA, followed by sequencing of the products. None of the exonic regions sequenced contained mutations, in aggregate representing the majority of coding regions (data not shown). However, a smaller RT-PCR product was produced by one set of primers designed to amplify exons 8 through 12 of the Bromodomain and WD repeat domain containing 1 (Brwd1) gene (Fig. 5C). Sequencing of the aberrant RT-PCR band revealed splicing from exon 9 directly to exon 11. Genomic DNA sequence around exon 10 revealed a T to C mutation at the 5’ splice donor site in the repro5 allele (from TG/GT to TG/GC; Fig. 5D).
Brwd1, formerly known as Wdr9, is a large gene consisting of 42 exons spanning over 89 kB (Fig. 5B). The mature mRNA corresponds to 8207 nucleotides (NM_145125) and encodes a 2304 amino acid protein (NP_660107). Exon 10 of Brwd1 is 70 nucleotides long; exclusion of this exon should result in a frameshift, generating a premature stop codon, thereby potentially subjecting the mutant transcripts to nonsense mediated mRNA decay (NMD; reviewed in (Chang et al., 2007)). Degradation of aberrant mRNA messages via the NMD pathway can initiate from both the 5’ and 3’ ends of the message (Conti and Izaurralde, 2005). Consistent with this, we noted that except for primers targeting the central parts of the Brwd1 transcript, RT-PCR of homozygous mutant RNA was weak or unsuccessful (data not shown). The presence of a deleted exon causing a premature stop, coupled with decreased message levels, indicates that Brwd1repro5 is a null allele or severe hypomorph.
To confirm that the Brwd1repro5 mutation is responsible for the observed phenotypes, we generated an additional allele from an embryonic stem (ES) cell line (AW0417) containing a gene trap insertion between exons 34 and 35 of Brwd1 (Fig. 5B; the allele will be abbreviated here as Brwd1Gt). A complementation test was performed by breeding Brwd1Gt/Brwd1repro5 animals. We examined testis histology and epididymal sperm of two such compound heterozygotes plus two Brwd1Gt/+ brothers. As with repro5 homozygotes, the seminiferous tubules of the Brwd1Gt/Brwd1repro5 males contained fewer spermatozoa than controls, and the spermatids exhibited abnormally shaped heads and acrosomes (Supplemental Fig. 1A, B). The epididymal sperm count was reduced >5 fold (Table 2). These sperm had poor motility, and most had morphological abnormalities similar to Brwd1repro5 homozygotes (Table 2; sperm images 15 not shown). Brwd1Gt/Brwd1Gt testis histology revealed similar though less pervasive anomalies (Supplemental Fig. 1C). The sperm count was ~3 fold lower than controls (Table 1). Most of these sperm were morphologically abnormal, but the proportion with dismorphologies was lower than in repro5 homozygotes (87.3% vs 97.5%; ANOVA p=0.042). It is possible that gene trap is not a complete null, or that the introduction of the 129-strain background from the AW0417 ES cells suppressed the phenotype slightly. Nevertheless, the failure of Brwd1Gt to complement repro5 provides proof that the mutation in Brwd1 is responsible for the observed phenotype. Furthermore, as described above and reported in Table 1, Brwd1Gt oocytes showed a severe failure in the oocyte-embryo transition as did repro5 oocytes. Therefore, we now designate the repro5 allele as Brwd1repro5.
DISCUSSION
Mutations affecting gametogenesis in both sexes (without causing additional phenotypes or altering sexual behavior) most often involve genes required for prophase of meiosis I, the establishment/expansion of primordial germ cells, or endocrine pathways (Matzuk and Lamb, 2002). The phenotype of Brwd1 mutants is unusual because gametogenesis in both sexes is affected after the chromosome pairing events of meiotic prophase. Normal oocyte and sperm development differ dramatically after reaching diplonema, both in terms of timing of meiotic progression and cellular events. Oocytes undergo dictyate arrest, during which stage they acquire competence for meiotic maturation and embryogenesis during follicular development and await ovulation and fertilization to complete meiosis. In contrast, spermatocytes complete meiosis without an arrest period and immediately enter the haploid phase of spermiogenic differentiation. While oocytes must acquire “competence” to support embryonic development, there is apparently no obviously analogous event in spermatogenesis. In fact, round spermatids electrofused with, or injected into, oocytes can produce normal offspring (Ogura et al., 1994; Tamashiro et al., 1999), although there is only very low success in the ability of spermatocytes to complete meiosis after injection into oocytes (Kimura et al., 1998).
One important similarity, however, is that gametocytes of both sexes undergo periods of dramatic nuclear remodeling during their differentiation. Until oocytes are almost fully-grown, they are incompetent to resume meiosis because they lack the cell cycle molecules necessary to drive entry into metaphase I. During the growth phase, oocyte nuclei exhibit increased transcriptional activity until the antral stage, at which time all of the necessary molecules to support meiotic resumption have been synthesized. Meiotic arrest of these maturation-competent oocytes is maintained by interactions with follicular somatic cells. Shortly thereafter, the nuclei undergo “epigenetic maturation,” in which unique large scale chromatin modifications result in condensation of the chromatin and global transcriptional silencing (reviewed by (De La Fuente, 2006). Fully grown oocytes resume meiosis in response to the preovulatory LH surge, as indicated by breakdown of the germinal vesicle (GV; i.e, the nuclear membrane).
Similar, but even more extreme chromatin remodeling events occur during male gametogenesis (Govin et al., 2004; Meistrich et al., 2003). After the completion of meiosis, chromatin is packaged with histones and maintained in a somewhat diffuse array in the round spermatid nucleus, perhaps in specific chromosome territories. Subsequently, during spermatid elongation, the histones become hyperacetylated before being replaced by transition proteins and ultimately by protamines to achieve dramatic reduction in volume in packaging chromatin (Hazzouri et al., 2000). Whether the sex-specific nuclear remodeling events in oocytes and spermatids are related mechanistically is not known, but, as discussed below, BRWD1 may play a role. Experiments in which cultured cells were induced to a hyperacetylated state by treatment with the histone deacetylase inhibitor TSA and transfected with the dual bromodomain containing gene Brdt caused dramatic chromatin condensation (Pivot-Pajot et al., 2003).
Functional and comparative data suggest that BRWD1 functions as a transcriptional regulator or co-regulator. A key signature of BRWD1 is the presence of two tandem bromodomains. Bromodomains are known to interact with acetylated lysine residues, particularly in the C terminal tails of histones (Dey et al., 2003; Pivot-Pajot et al., 2003; Trousdale and Wolgemuth, 2004). Lysine acetylation plays an important role in chromatin remodeling and transcriptional regulation, and the only known protein motif that recognizes acetyl lysine is the bromodomain (Zeng and Zhou, 2002). BRWD1 also contains a polyglutamine stretch that was found to have transcriptional activation activity via luciferase activation in vitro, and BRWD1 appears to be excluded from nuclear heterochromatic regions (Huang et al., 2003). BRWD1 was found to interact with BRG1, the ATPase component of two mammalian SWI/SNF chromatin-remodeling complexes (Huang et al., 2003).
BRWD1 is a member of a novel protein family that has both WD40 repeat domains (seven) and dual bromodomains. WD40 repeats are present in diverse eukaryotic proteins including G proteins, phosphatases and transcription regulators (Smith et al., 1999). These ~40 amino acid repeats form structures that interact with other proteins or ligands. The double bromodomain arrangement present in BRWD1 is less common than single bromodomains, but this configuration also exists in TAFII250, the largest subunit of TFIID. In this protein, the bromodomains bind acetylated histone H4 residues, and are hypothesized to mediate targeting of TFIID to chromatin-bound promoters (Jacobson et al., 2000). Together, these lines of evidence are highly suggestive of a role for BRWD1 in transcriptional regulation via chromatin remodeling, which is a key feature of both oocyte differentiation and a hallmark feature of spermiogenesis (Kimmins and Sassone-Corsi, 2005; Sassone-Corsi, 2002). Studies are currently under way to explore this possibility.
In contrast to the gamete morphogenesis defects in males, Brwd1 mutant oocytes appear morphologically normal, but are defective in the oocyte-embryo transition. This phenotype is similar to that of Zar1 knockout eggs, which also develop to the 2-pronuclear stage where ≥ 80% become arrested. However, unlike Brwd1repro5 oocytes, Zar1 mutant oocytes mature to the MII stage (Wu et al., 2003). BRDW1 contains both bromo- and WD domains, while ZAR1 contains PHD (finger) domains. Bromo- and PHD domain-containing proteins both affect transcription, presumably by modulating hetero/euchromatin configuration. The transition of a fully-grown oocyte to a 2-cell mouse embryo occurs during a transcriptionally silent period. Fully-grown, GV-stage mouse oocytes contain all of the transcripts necessary to support the oocyte-to-embryo transition since culture of inseminated eggs with the potent transcription inhibitor α-amanitin does not prevent cleavage to the 2-cell stage (Evsikov et al., 2006; Flach et al., 1982). Because fully-grown mutant oocytes are morphologically indistinguishable from WT oocytes, BRWD1 and ZAR1 may promote or suppress transcriptional patterns in developing oocytes, or control transcript degradation patterns during the GV-to-MII transition in ways that are specifically related to the successful oocyte-to-embryo transition.
Among the known transitional processes that could be affected are those controlling the progression of meiotic maturation, egg activation, or initiating zygotic DNA synthesis. Bromodomain proteins can function as either activators or suppressors of transcription. As oocytes near the end of their growth phase in antral follicles there is a global silencing of transcription. If BRWD1 normally functions as a suppressor of transcription in mouse oocytes, it is possible that this silencing does not occur in Brwd1repro5 mutant oocytes and that overproduction of some transcripts occurs. This could be deleterious to the oocyte-embryo transition. For example, using Dicer conditional knockout oocytes, it was shown that prevention of transcript degradation during meiotic maturation dramatically disrupts the oocyte-embryo transition (Murchison et al., 2007; Tang et al., 2007). Alternatively, BRWD1 might promote the transcription of genes involved in the oocyte-embryo transition before global silencing of transcription. Such genes might be required to progress beyond the 2-pronuclear stage, or to enable DNA replication; inhibition of DNA synthesis in inseminated eggs causes arrest at the 2-pronuclear stage (Howlett, 1986). Finally, BRWD1 might affect oocyte chromatin organization in a way that is permissive for further development.
BRWD1 is not the only bromo domain-containing protein required for gametogenesis. BRDT, a member of the BET family of dual bromodomain-containing proteins, is involved in large scale chromatin remodeling of acetylated histones during spermatogenesis (Pivot-Pajot et al., 2003). Its expression is restricted to testis and oocytes (Paillisson et al., 2007; Shang et al., 2007). Homozygosity for a Brdt allele deleted for one of the Bromo domains caused male (but not female) infertility, with affected animals exhibiting decreased numbers of sperm with morphological abnormalites remarkably similar to those of Brwd1 mutant mice (Shang et al., 2007). Because transcription of H1t was altered (elevated) in the mutant, it was hypothesized that the main role of BRDT may be in regulating the expression of key genes in spermatogenesis. The expression of one of its 3 paralogs, Brd2, appears to correlate with various stages of oocyte maturation (Trousdale and Wolgemuth, 2004) and is also expressed in diplotene spermatocytes and round spermatids. However, embryonic lethality associated with a knockout of Brd2 precludes analysis of functional consequences on fertility (Ph.D. dissertation of Enyuan Shang, Columbia, Univ.; Debra Wolgemuth, personal communication). Brd3 is strongly expressed in round spermatids (Shang et al., 2004). A gene trap-induced mutation of the Brd4 gene also causes embryonic lethality in the homozygous state (Houzelstein et al., 2002). It will be of interest to elucidate the potential roles of all these bromodomain proteins in spermatogenesis.
Interestingly, human BRWD1 lies on Chr 21, in a region implicated in Down syndrome phenotypes (Ramos et al., 2002). According to gene expression data available from GEO Profiles (http://www.ncbi.nlm.nih.gov/geo), UniGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene), GNF SymAtlas (http://symatlas.gnf.org/SymAtlas) and Ramos et al (Ramos et al., 2002), it is transcribed at most developmental timepoints and in a variety of adult tissues including testis and ovary. In mice, Huang et al (Huang et al., 2003) observed expression of both Brwd1 mRNA and protein in many somatic tissues and developmental stages (analysis of gonads was not conducted). The widespread expression in development suggests that BRWD1 may have some developmental function, but it is not absolutely essential for viability or morphogenesis. The lack of a phenotype may be due to the existence of two highly similar paralogs in the mouse genome, pleckstrin homology interacting protein (Phip) and Brwd3. Each protein contains WD40 repeats and dual Bromo domains in essentially identical positions relative to their N termini, and they share over 60% amino acid identity and 73% similarity over most of the N terminal ~1400 amino acids (Fig. 5 and Supplemental Fig. 2). While there is a gene annotated as Brwd2 in the mouse and human genomes, this unfortunately is an erroneous designation; the BRWD2 predicted protein has WD40 but no Bromo domains, and there is no significant homology to BRWD1. Phip and Brwd3 are expressed broadly according to data represented in Unigene and SymAtlas, overlapping the Brwd1 expression pattern, which is consistent with the idea that one or both provide redundant function.
There are clear orthologs of all three genes in sequenced vertebrates (such as zebrafish, not shown), but invertebrates have a single Brwd gene. In Drosophila melanogaster, mutation of the Brwd1 ortholog, called ramshackle (ram), causes larval death (D'Costa et al., 2006). Milder alleles affect morphology of photoreceptor cells in the eye associated with nuclear mislocalization and other cellular dismorphologies (D'Costa et al., 2006).
One of the most intriguing aspects of the molecular role of BRWD1 is the extent to which its function in oogenesis and spermatogenesis potentially overlaps, even though the defects manifest themselves at different stages in spermatids vs oocytes. Thus, ultimate resolution of the molecular function of BRWD1 may reveal the expression of identical genes acting at different times in male and female gametogenesis, or similar features of chromatin remodeling required for different pathways in successful gametogenesis.
Supplementary Material
FIG. 6. Schematic alignment of mouse BRWD paralogs.
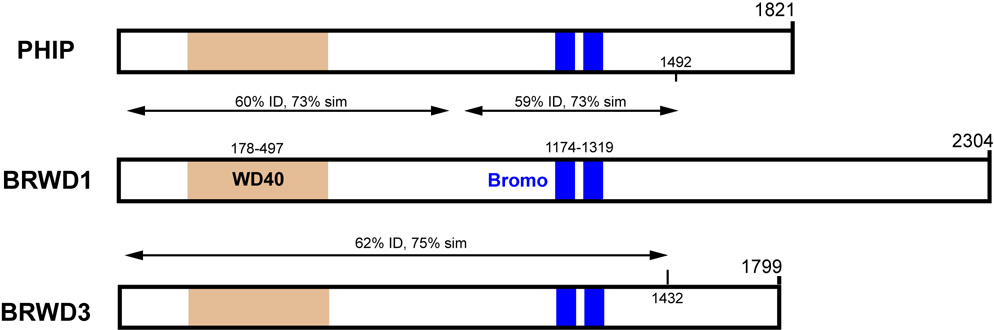
The diagrams depict alignments of predicted amino acid sequences encoded by the Brwd1, Phip, and Brwd3 genes. Locations of the WD and Bromo domains are shown. All numeric coordinates refer to amino acids. The indicated degrees of similarity refer to comparisons to BRWD1. There is a region in the center of PHIP that does not align well to BRWD1. The C-terminal ends of the proteins are divergent. Actual sequence alignments are presented in Supplemental Figure 2.
ACKNOWLEDGEMENTS
We thank Lucy Rowe, Rob Munroe, and Sheila Bornstein for technical contributions, and Drs. Carl Lessard and Janice Pendola for identification of repro5 mutants in the original ENU screen. This work was supported by Public Health Service grant P01HD42137 from the National Institute of Child Health and Human Development to JJE, MAH and JCS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bjorndahl L, et al. Evaluation of the one-step eosin-nigrosin staining technique for human sperm vitality assessment. Hum Reprod. 2003;18:813–816. doi: 10.1093/humrep/deg199. [DOI] [PubMed] [Google Scholar]
- Chang YF, et al. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- D'Costa A, et al. The Drosophila ramshackle gene encodes a chromatin-associated protein required for cell morphology in the developing eye. Mech Dev. 2006;123:591–604. doi: 10.1016/j.mod.2006.06.007. [DOI] [PubMed] [Google Scholar]
- De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292:1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Dey A, et al. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EP, et al. An Air-Drying Method for Meiotic Preparations from Mammalian Testes. Cytogenetics. 1964;15:289–294. doi: 10.1159/000129818. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, et al. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20:2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach G, et al. The transition from maternal to embryonic control in the 2-cell mouse embryo. Embo J. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianotten J, et al. Idiopathic impaired spermatogenesis: genetic epidemiology is unlikely to provide a short-cut to better understanding. Hum Reprod Update. 2004;10:533–539. doi: 10.1093/humupd/dmh045. [DOI] [PubMed] [Google Scholar]
- Govin J, et al. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Handel MA, et al. Mutagenesis as an unbiased approach to identify novel contraceptive targets. Mol Cell Endocrinol. 2006;250:201–205. doi: 10.1016/j.mce.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Hargreave TB. Genetic basis of male fertility. Br Med Bull. 2000;56:650–671. doi: 10.1258/0007142001903454. [DOI] [PubMed] [Google Scholar]
- Haynes SR, et al. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri M, et al. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol. 2000;79:950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett S. The effect of inhibiting DNA replication in the one-cell mouse embryo. Roux Arch Devel Biol. 1986;195:499–505. doi: 10.1007/BF00375890. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Expression of the Wdr9 gene and protein products during mouse development. Dev Dyn. 2003;227:608–614. doi: 10.1002/dvdy.10344. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, et al. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- Kimura Y, et al. Factors affecting meiotic and developmental competence of primary spermatocyte nuclei injected into mouse oocytes. Biol Reprod. 1998;59:871–877. doi: 10.1095/biolreprod59.4.871. [DOI] [PubMed] [Google Scholar]
- Lessard C, et al. New mouse genetic models for human contraceptive development. Cytogenet Genome Res. 2004;105:222–227. doi: 10.1159/000078192. [DOI] [PubMed] [Google Scholar]
- Mandon-Pepin B, et al. Human infertility: meiotic genes as potential candidates. Gynecol Obstet Fertil. 2002;30:817–821. doi: 10.1016/s1297-9589(02)00444-7. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4 suppl:s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, et al. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, et al. Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet. 2003;362:1714–1719. doi: 10.1016/S0140-6736(03)14845-3. [DOI] [PubMed] [Google Scholar]
- Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien MJ, et al. A revised protocol for in vitro development of mouse oocytes from primordial follicles dramatically improves their developmental competence. Biol Reprod. 2003;68:1682–1686. doi: 10.1095/biolreprod.102.013029. [DOI] [PubMed] [Google Scholar]
- Ogura A, et al. Birth of normal young after electrofusion of mouse oocytes with round spermatids. Proc Natl Acad Sci U S A. 1994;91:7460–7462. doi: 10.1073/pnas.91.16.7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillisson A, et al. Bromodomain testis-specific protein is expressed in mouse oocyte and evolves faster than its ubiquitously expressed paralogs BRD2, -3, and -4. Genomics. 2007;89:215–223. doi: 10.1016/j.ygeno.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C, et al. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol. 2003;23:5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A, Handel MA. Meiosis in male PL/J mice: a genetic model for gametic aneuploidy. Mol Reprod Dev. 2003;64:471–481. doi: 10.1002/mrd.10231. [DOI] [PubMed] [Google Scholar]
- Ramos VC, et al. Characterisation and expression analysis of the WDR9 gene, located in the Down critical region-2 of the human chromosome 21. Biochim Biophys Acta. 2002;1577:377–383. doi: 10.1016/s0167-4781(02)00421-9. [DOI] [PubMed] [Google Scholar]
- Reinholdt L, et al. Forward genetic screens for meiotic and mitotic recombination-defective mutants in mice. Methods Mol Biol. 2004;262:87–107. doi: 10.1385/1-59259-761-0:087. [DOI] [PubMed] [Google Scholar]
- Russell L, et al. Histological and histopathological evaluation of the testis. Clearwater: Cache River Press; 1990. [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Shang E, et al. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–3515. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- Shang E, et al. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr Patterns. 2004;4:513–519. doi: 10.1016/j.modgep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Smith TF, et al. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Tamashiro KL, et al. Bypassing spermiogenesis for several generations does not have detrimental consequences on the fertility and neurobehavior of offspring: a study using the mouse. J Assist Reprod Genet. 1999;16:315–324. doi: 10.1023/A:1020406016312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trousdale RK, Wolgemuth DJ. Bromodomain containing 2 (Brd2) is expressed in distinct patterns during ovarian folliculogenesis independent of FSH or GDF9 action. Mol Reprod Dev. 2004;68:261–268. doi: 10.1002/mrd.20059. [DOI] [PubMed] [Google Scholar]
- Ward JO, et al. Toward the genetics of mammalian reproduction: induction and mapping of gametogenesis mutants in mice. Biol Reprod. 2003;69:1615–1625. doi: 10.1095/biolreprod.103.019877. [DOI] [PubMed] [Google Scholar]
- Wilson L, et al. Random mutagenesis of proximal mouse Chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res. 2005;15:1095–1105. doi: 10.1101/gr.3826505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, et al. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


