Abstract
Phosphorylation of the polarity protein Par-3 by the serine/threonine kinases aPKCζ/ι and Par-1 (EMK1/MARK2) regulates various aspects of epithelial cell polarity, but little is known about the mechanisms by which these posttranslational modifications are reversed. We find that the serine/threonine protein phosphatase PP1 (predominantly the α isoform) binds Par-3, which localizes to tight junctions in MDCKII cells. PP1α can associate with multiple sites on Par-3 while retaining its phosphatase activity. By using a quantitative mass spectrometry-based technique, multiple reaction monitoring, we show that PP1α specifically dephosphorylates Ser-144 and Ser-824 of mouse Par-3, as well as a peptide encompassing Ser-885. Consistent with these observations, PP1α regulates the binding of 14-3-3 proteins and the atypical protein kinase C (aPKC) ζ to Par-3. Furthermore, the induced expression of a catalytically inactive mutant of PP1α severely delays the formation of functional tight junctions in MDCKII cells. Collectively, these results show that Par-3 functions as a scaffold, coordinating both serine/threonine kinases and the PP1α phosphatase, thereby providing dynamic control of the phosphorylation events that regulate the Par-3/aPKC complex.
Keywords: epithelial polarity, multiple reaction monitoring, tight junctions, phosphorylation dynamics
Polarity is a common trait of eukaryotic cells and is critical for tissue development and cellular diversity. Genetic and biochemical studies of the “partitioning-defective” (par) phenotype in the Caenorhabditis elegans embryo, and of polarity defects in Drosophila melanogaster and mammalian epithelia and neuroblasts, have revealed that cell polarization relies on the concerted activities of asymmetrically localized protein assemblies (1–3), one of which is the evolutionary conserved Par/atypical protein kinase C (aPKC) complex. The polarity protein Par-3, a functional component of this complex, directly binds to the PDZ domain-containing protein Par-6 that, in turn, regulates the activity of associated aPKC by engaging the small GTPase Cdc42 (4, 5). However, Par-3 has also been implicated in various aspects of cell polarity independent of its association with Par-6, including asymmetric cell division (6), dendritic and axonal development in neurons (7, 8), and directed cell migration (9). In mammalian epithelial cells Par-3 localizes to the apical junctional complex (10) and is required for the establishment of apical-basal polarity and the formation of tight junctions (TJs) (11, 12). Many of the signaling events controlling cell polarity and TJ formation are regulated by the activities of serine/threonine protein kinases, notably aPKC and Par-1 (EMK1/MARK2) (4–6, 13–18). For example, aPKC maintains the apical membrane domain by phosphorylating the basolateral determinants Lgl and Par-1, thereby releasing them from the cell cortex. Conversely, phosphorylation of Par-3 by Par-1 creates binding sites for 14-3-3 proteins that, in turn, antagonize the association of Par-3 with aPKC. Additionally, formation of an aPKC-Par-3 complex is negatively regulated by aPKC phosphorylation of Par-3. In seeking regulators that may counterbalance these phosphorylation events, we identified the serine/threonine protein phosphatase 1 (PP1) as a functional component of the Par-3 scaffold. We provide evidence that PP1α is required for the dephosphorylation of Par-3 at several key serine residues, thereby regulating its association with 14-3-3 proteins. Our data also suggest a role for PP1α in the formation of functional TJs by influencing the association of Par-3 and aPKC.
Results
PP1α Is a Component of the Par-3 Complex and Localizes to Tight Junctions.
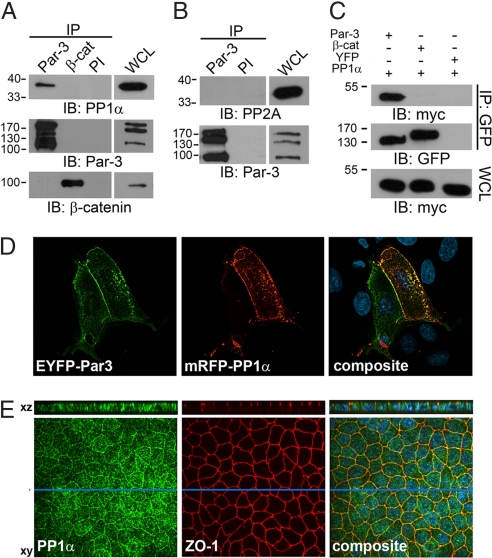
To pursue the identities of Par-3-associated proteins, we used an affinity purification/mass spectrometry approach to identify potential regulators of Par-3 function. Immunopurification of 3×FLAG-Par-3 from a stable 293T cell line followed by SDS/PAGE and staining with colloidal Coomassie blue detected several proteins that coimmunoprecipitated with the fusion protein and that were not present in the control sample [supporting information (SI) Fig. S1A]. By using tandem mass spectrometry we identified several known interaction partners of Par-3 (Fig. S1B), including Par-6, aPKCι (5), 14-3-3ε (13, 14), angiomotin (19), and DNA-PK (20). In addition, analysis of proteins migrating at a relative mass of 38,000 (Mr 38,000) revealed peptides from the catalytic subunit of the serine-threonine protein phosphatase PP1. We then immunoprecipitated endogenous Par-3 from a lysate prepared from mouse E13 embryonic tissue by using an antibody that detects the major Par-3 isoforms (180, 150, and 100 kDa) (8) and identified an association with endogenous PP1, but not with β-catenin used as a negative control (Fig. 1A). Similar results were obtained with 293T and MDCKII cells (data not shown). Furthermore, Par-3 did not detectably coprecipitate the catalytic subunit of the serine/threonine protein phosphatase PP2A (Fig. 1B). In addition, exogenously expressed 2xmyc-tagged PP1 (isoform α) specifically bound to yellow fluorescent protein (YFP) tagged Par-3 (100 kDa), but not to YFP-β-catenin or YFP alone (Fig. 1C). To assess the subcellular distribution of PP1α and Par-3 we transfected MDCKII cells with constructs encoding YFP-tagged Par-3 (180 kDa) and PP1α N-terminally tagged with a monomeric variant of red fluorescent protein (mRFP). As expected, fluorescence microscopy revealed YFP-Par-3 at areas of cell–cell contact (10) (Fig. 1D Left). Additionally, mRFP-PP1α strongly codistributed with YFP-Par-3 at the plasma membrane and within cytoplasmic aggregates (Fig. 1D Right). More importantly, spinning disk confocal microscopy revealed that endogenous PP1α codistributes with the tight junction protein ZO-1 at the apical aspect of the plasma membrane (Fig. 1E and Fig. S2A).
Fig. 1.
Par-3 interacts with protein phosphatase 1. (A and B) Endogenous Par-3 and β-catenin immunoprecipitated from mouse E13 lysates were probed with anti PP1α or anti-PP2A antibodies. Preimmune (PI) serum was used as a control. (C) YFP immunoprecipitates (IPs) from MDCKII cells expressing 2xmyc-PP1α and YFP-Par-3, YFP-β-catenin, or YFP were immunoblotted (IB) with anti-myc (Top) or anti-GFP (Middle) antibodies. Whole-cell lysates (WCLs) were probed for equal expression of 2xmyc-PP1α by using an anti-myc antibody (Bottom). (D) EYFP-Par-3 and mRFP-PP1α codistribute in MDCKII cells at the plasma membrane. (E) Endogenous PP1α codistributes with the tight junction marker protein ZO-1 at sites of cell–cell contact in MDCKII cells. Enface images and confocal XZ-sections (section indicated by blue bar) are shown.
The mammalian genome encodes four PP1 isoforms, PP1α, PP1β, PP1γ1, and PP1γ2, with the latter two arising through alternative splicing (21). Strikingly, probing endogenous Par-3 immunoprecipitates with isoform-specific antibodies revealed a strong association of Par-3 with PP1α, but not with PP1β and only a weak binding with PP1γ1 (Fig. S3A). Similarly, exogenously expressed YFP-Par-3 bound to 2xmyc-tagged PP1α, and to a much lesser extent to PP1γ1, but not to PP1β (Fig. S3B). Because the different PP1 isoforms reportedly localize to distinct subcellular compartments both during interphase and mitosis (22), we expressed PP1α, PP1β, and PP1γ1 fused to YFP in MDCKII cells and investigated their localization by spinning disk confocal microscopy. PP1α codistributed with endogenous Par-3, whereas PP1γ1 was much less evident at sites of cell–cell contact, and PP1β was not observable at the plasma membrane (Fig. S3C). Taken together, these data suggest that Par-3 predominantly interacts with PP1α at sites of cell–cell contact in mammalian epithelial cells.
PP1α Directly Binds to Multiple Regions of Par-3.
To dissect the regions of Par-3 that mediate its interaction with PP1α, we expressed various YFP-tagged fragments of Par-3 in 293T cells and tested for their ability to coimmunoprecipitate 2xmyc-tagged PP1α (Fig. S2B). Par-3 fragments containing the first PDZ domain (PDZ1; amino acid 205–477) and a C-terminal region (C-term2; amino acid 986-1333) strongly interacted with PP1α, whereas the N-terminal domain of Par-3 (N-term; amino acid 1–205), the second PDZ domain (PDZ2; amino acid 435–585), the aPKC-binding region (C-term1; amino acid 705-1020), or YFP alone did not. In addition, weak binding of the third PDZ domain of Par-3 (PDZ3; amino acid 570–720) to 2xmyc-tagged PP1α could be detected. To test whether these interactions were direct, we used pulldown assays by using purified regions of Par-3 fused to maltose-binding protein (MBP) and probed for their ability to bind purified PP1α. In these experiments, the fragments spanning Par-3 PDZ1 and C-term2-1 (amino acid 1020–1217) showed a robust interaction with PP1α, whereas PDZ3 bound more weakly and PDZ2, C-term2-2 (amino acid 1217–1333), or MBP alone did not bind (Fig. 2A).
Fig. 2.
PP1 binds directly to multiple regions of Par-3. (A) Purified PP1α was incubated with amylose resin-immobilized MBP-fusion proteins as indicated. MBP alone was used as a control. Bound PP1α was detected by immunoblotting with anti-PP1α antibody. (B) 293T cells coexpressing wild-type Par-3-PDZ1 or RVXF-mutated YFP-Par-3-PDZ1 (Top) and 2xmyc-PP1α were lysed and YFP-fusion proteins were immunoprecipitated with an anti-GFP antibody. Immunoprecipitates (IP) were probed with anti-myc or anti-GFP antibodies and whole-cell lysates (WCLs) with anti-myc antibody.
A common feature of many PP1-interacting proteins is the RVXF motif, [RK]-X0,1-[V/I]-[F/W], which mediates the association with a hydrophobic groove on the surface of PP1 (21). Par-3 harbors a single degenerate RVXF motif located at the C-terminal border of PDZ1, spanning residues 351–355 (RVIWF). Mutation of key residues to alanine (RVIWF to AVIWA) significantly decreased but did not completely abrogate binding of the first PDZ domain to PP1α (Fig. 2B). However, immunoprecipitating YFP-tagged Par-3 containing the RVXF mutation from 293T cells coexpressing 2xmyc PP1α did not reveal a substantial difference in binding when compared with wild-type YFP-Par-3, most likely because of the secondary binding sites present in Par-3 (Fig. S2C). Collectively, these results indicate that PP1α directly binds multiple regions of Par-3, including the first PDZ domain, which contains a degenerate RVXF motif.
Par-3 Does Not Inhibit The Enzymatic Activity of PP1α.
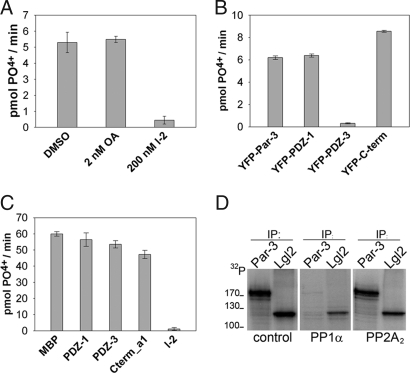
To address whether Par-3 influences the activity of PP1α, Par-3 immunocomplexes (Fig. S4A), isolated from MDCKII cells, were monitored for their ability to dephosphorylate a phosphopeptide known to be a substrate for both PP1 and PP2A (Fig. 3A, DMSO). Low doses of okadaic acid (OA), which predominantly inhibit PP2A (23), had little effect on the Par-3-associated phosphatase activity, whereas the addition of the PP1-specific inhibitor I-2 almost abolished the release of free phosphate (Fig. 3A). Subsequently, we tested whether any of the PP1-binding regions of Par-3 had an impact on PP1 activity. As shown in Fig. 3B, 2xmyc-PP1α coimmunoprecipitating with either YFP-Par-3, YFP-PDZ1, YFP-PDZ3, or YFP-C-term2 (Fig. S4B) was able to dephosphorylate the substrate, with the PDZ3-complexes displaying the lowest phosphatase activity. However, this is most likely because of the low amounts of PP1α that coimmunoprecipitate with the third PDZ domain of Par-3 (Fig. S2B) and is not a consequence of an active inhibition of PP1α by this region. These results are further substantiated by experiments demonstrating that the addition of purified binding regions of Par-3 fused to MBP (as described in Fig. 2A Top), or MBP alone, did not significantly lower the enzymatic activity of purified PP1α, even if applied in a 10-fold higher concentration (Fig. 3C). In contrast, the addition of a 1:1 ratio of I-2 abolished the activity of PP1α (Fig. 3C, I-2). Collectively, these data indicate that PP1α remains active when bound to Par-3.
Fig. 3.
Par-3-bound PP1α is active and dephosphorylates Par-3 in vitro. (A) Endogenous Par-3 immunocomplexes collected from MDCKII cells were tested for the release of free phosphate from a substrate in the presence of okadaic acid (2 nM), the PP1-specific inhibitor Inhibitor-2 (200 nM), or DMSO. (B) Lysates prepared from 293T cells coexpressing 2xmyc-PP1α and various regions of Par-3 fused to YFP were incubated with anti-GFP antibody and resulting complexes were monitored for phosphatase activity. (C) Purified PP1α (100 ng) was incubated with 1 μg of various regions of Par-3 purified as MBP-fusion proteins and phosphatase activity was monitored. Each value is the mean of triplicate measurements (n = 3). Error bars represent 1 SD. (D) Purified PP1α, but not PP2A2, dephosphorylates aPKC-phosphorylated Par-3 in vitro (see Methods for details). Additionally, PP1α is less reactive toward the polarity protein Lgl2.
PP1 Dephosphorylates Specific Serine Residues of Mouse Par-3.
Because the binding of Par-3 does not inhibit the activity of PP1α, we investigated whether it serves as a substrate for PP1. To this end, we performed in vitro kinase assays on 3×FLAG-tagged Par-3 isolated from 293T lysates by using recombinantly purified aPKCζ (5). Subsequently, 32P-labeled Par-3 was specifically dephosphorylated when treated with purified PP1α, but not PP2A2 (Fig. 3D). In parallel, both catalytic subunits showed strong activities if tested in a malachite green assay by using a PP1/PP2A-specific phosphopeptide as substrate (data not shown). Furthermore, when compared with phosphorylated Par-3, purified PP1α had a significantly lower activity toward the polarity-related phosphoprotein Lgl2 (Fig. 3D).
Several residues of Par-3 have been shown to be phosphorylated by serine/threonine kinases, including the 14-3-3 binding sites S144/S885, which are substrates for Par-1 (EMK1/MARK2), and S824 of mouse Par-3, which is phosphorylated by aPKC (13, 14, 16). We therefore investigated whether the phosphorylation of these residues is reversed by PP1 by employing multiple reaction monitoring (MRM) (24), a mass spectrometric approach designed to quantitatively assay any selected peptides or modified peptides. The treatment of immunopurified Par-3 with PP1 resulted in a complete loss of phosphorylated S144 (Fig. 4A and Fig. S5) and S824 (Fig. 4B). MRM signals were also used to trigger an MS/MS scan, which confirmed the sequences of the analyzed pS144- and pS824-peptides (Fig. S6 A and B). Evidence of peptides containing phosphorylated S885 was below our limits of detection. Therefore, to gather the spectral information needed to build an MRM method we analyzed an in vitro MARK2-phosphorylated GST-fusion protein spanning residues 688–973 of Par-3 by LC-MS/MS and MRM. We obtained strong MRM signals and confirmatory MS/MS spectra for the phosphorylated and nonphosphorylated S885 peptides. However, MRM detection of the in vivo phosphorylated S885 (SSpSLESLQTAVAEVTLNGNIPFHRPR) peptide using immunopurified Par-3 remained unsuccessful, possibly because of additional phosphorylated residues that were not present in the in vitro MARK2-treated sample. Such multiple phosphorylated sites in the S885 peptide might be coordinately removed by PP1α because of their proximity to one another, and we therefore monitored for an increase in the nonphosphorylated S885 peptide. Indeed, after PP1α treatment, MRM analysis revealed a strong increase in the signal for multiple ion pair transitions (see Table S1) of the nonphosphorylated tryptic peptide containing S885, and the respective sequence was confirmed by MS/MS (Fig. S7). This indicates that PP1α dephosphorylates one or more of the serine/threonine residues in this peptide of mouse Par-3.
Fig. 4.
PP1α dephosphorylates S144 and S824, but not S25 of mouse Par-3. (A) Shown are MRM elution profiles of the peptides containing phosphorylated S144 with (Left) or without (Right) PP1α treatment. (B) The phosphorylated peptide containing S824 is not detectable after phosphatase-treatment (Left) as evidenced by multiple reaction monitoring. (C) Shown are the extracted ion current profiles (XIC) of the nonphosphorylated S25-peptides (Left) and the phosphorylated S25-peptides (Right) before and after treatment with PP1α. (D) The change in phosphorylation of S25 is calculated as the ratio of the integrated ion current profiles of nonphosphorylated/phosphorylated peptides. Shown are the ratios of phosphatase-treated (right-hand bar) and untreated samples (left-hand bar). Error bars represent 1 SD (n = 2).
To investigate whether PP1 is specific for these sites, as opposed to other phosphorylated Par-3 residues, we calculated the relative change in phosphorylation of a peptide containing S25, which we found to be phosphorylated during the initial analysis of Par-3. Strikingly, as shown in Fig. 4D, there was no relative change in the amount of phosphorylated peptide containing S25 before and after treatment with PP1α, when compared with the corresponding nonphosphorylated S25-peptide (see Fig. 4C for MRM elution profiles). Again, MRM-triggered MS/MS scans were performed, verifying the nonphosphorylated and phosphorylated S25-peptides (Fig. S6 C and D). In summary, our findings demonstrate that PP1α selectively dephosphorylates S144 and S824 and a peptide that encompasses S885 of mouse Par-3.
PP1 Regulates the Association of Par-3 with 14-3-3 and aPKCζ.
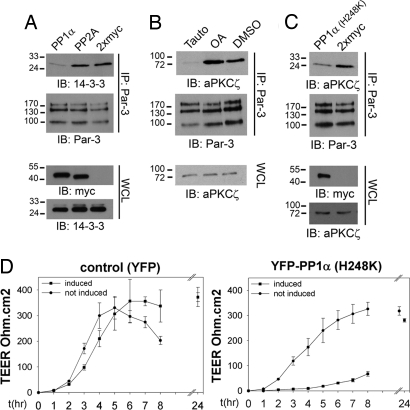
Because the binding of 14-3-3 to Par-3 depends on phosphorylation of S144 and S885 (14), we tested whether perturbing PP1 activity could influence interactions between the Par-3 scaffold and 14-3-3 proteins. Interestingly, we found that the overexpression of 2xmyc-tagged PP1α, but not PP2A, markedly reduced the binding of endogenous Par-3 to 14-3-3 (Fig. 5A), consistent with a model in which PP1α regulates the association of Par-3 and 14-3-3.
Fig. 5.
PP1α influences the binding of aPKCζ and 14-3-3 to Par-3 and impacts TJ formation. (A) Endogenous Par-3 was immunoprecipitated (IP) in the presence of Calyculin A from 293T cells transiently expressing 2xmyc-PP1α, 2xmyc-PP2A, or 2xmyc and the binding of endogenous 14-3-3 was monitored by immunoblotting (IB). Whole-cell lysates (WCLs) were analyzed for the expression of 14-3-3 and the exogenously expressed proteins. (B) Endogenous Par-3 was immunoprecipitated from 293T cells treated for 30 min with 5 μM Tautomycetin or 100 nM okadaic acid and bound aPKCζ was detected by immunoblotting (IB). Whole-cell lysates were monitored for endogenous aPKCζ expression levels. (C) 293T cells were transiently transfected with 2xmyc-PP1α (H248K) or 2xmyc expression constructs and endogenous Par-3-immunocomplexes were probed with an anti-aPKCζ antibody. The expression of myc-tagged proteins and endogenous aPKCζ was monitored on whole-cell lysates. (D) YFP-PP1α (H248K) expression (Right), or YFP-expression as a control (Left), was induced in MDCK-T23 cells by the removal of doxycycline and the kinetics of TEER establishment was monitored for 24 h after a Ca2+ switch. Three independent experiments using two different cell lines were performed. The means of one representative experiment are shown (n = 3). Error bars represent 1 SD.
Par-3 and aPKC form a dynamic enzyme–substrate complex that is only stable when S824 of Par-3 is not phosphorylated (16). We therefore investigated whether the inhibition of PP1α could decrease the binding of aPKC to Par-3. Indeed, although incubation of 293T or MDCKII cells with low doses of OA, known to predominantly inhibit PP2A (23), or with DMSO as a control had no effect, treatment with Tautomycetin (23), a predominantly PP1-selective inhibitor, severely reduced aPKCζ binding to Par-3 (Fig. 5B and Fig. S8A). To further substantiate this result, we used a dominant negative approach with a catalytically inactive mutant of PP1α (H248K) (25). As shown in Fig. 5C, the expression of PP1α (H248K), but not 2xmyc, markedly reduced the binding of aPKCζ to Par-3. Taken together, these results demonstrate that PP1α regulates the phosphorylation-dependent association of Par-3 with 14-3-3 and aPKCζ in mammalian epithelial cells.
Perturbation of PP1 Activity Delays Formation of Functional Tight Junctions.
Because the association of Par-3 with aPKC has a significant role in the assembly of functional tight junctions (TJs) in epithelial cells (11, 12, 18), we investigated whether silencing PP1α by shRNA-mediated knockdown influenced the formation TJs. However, after generating shRNA-MDCKII lines that show Tet-inducible suppression of PP1α (Fig. S8B), we observed functional compensation for the loss of PP1α by PP1γ1. Because the PP1-binding sites are not occupied by PP1α on shRNA-mediated suppression, PP1γ1 is potentially able to bind Par-3. In addition, the expression level of PP1γ1 was elevated after PP1α suppression (Fig. S8 C and D). These results may explain the observation that TJ formation appeared normal in these cells. Because coordinate silencing of both PP1α and PP1γ1 was toxic to the cells (data not shown), we generated inducible, stable MDCKII cell lines (T23) that express catalytically inactive YFP-PP1α (H248K), or YFP alone. As evidenced by transepithelial electrical resistance (TEER) measurements, the induced expression of YFP alone did not have an effect on TJ assembly over a period of 8 h, whereas the expression of YFP-PP1α (H248K) severely impaired TEER development in MDCKII cells (Fig. 5D). After 24 h, normal TEER values were recorded, indicating that the formation of functional TJs is delayed, but not absolutely inhibited, by the association of a phosphatase-dead mutant of PP1α with Par-3. Collectively, these findings demonstrate that PP1α influences the formation of functional tight junctions in mammalian epithelial cells.
Discussion
The establishment of apical-basal polarity and the formation of TJs rely in part on the assembly and asymmetric distribution of the Par/aPKC complex (1, 3). The spatial restriction of this complex to the apical domain depends on the actions of the serine/threonine kinase Par-1 (EMK1/MARK2). By creating docking sites for 14-3-3 proteins on Par-3, basolateral Par-1 activity disrupts the Par/aPKC complex, ultimately releasing it from the lateral cell cortex (13). Yet how these events are reversed to render this process dynamic has remained a puzzle. Our findings identify the serine/threonine protein phosphatase type 1 (PP1) as a functional component of the Par-3 scaffold. PP1 is an abundant serine/threonine phosphatase that has been identified in all eukaryotes analyzed to date and regulates multiple cellular functions (26). In mammals, the various PP1 isoforms associate with a diverse array of structurally distinct regulatory proteins, generating holoenzymes with different localizations and specificities (21). We demonstrate that multiple regions of Par-3 directly bind PP1α. However, in contrast to other scaffolds that associate with and inhibit PP1 through multiple binding surfaces (27), our results indicate that Par-3 does not markedly regulate PP1 activity but, instead, serves as a substrate. As a result, PP1 can release 14-3-3 by dephosphorylating S144 and S885 of mouse Par-3. Therefore, our results raise the possibility that PP1 balances the activities of Par-1 and the Par/aPKC complex by controlling the phosphorylation state of Par-3. We propose a model in which PP1α catalyzes the dephosphorylation of cytoplasmic Par-3 that has been released from the lateral cortex on phosphorylation by Par-1. As soon as phosphorylated Par-3 exits the active range of Par-1, PP1α allows the reestablishment of a functional Par/aPKC complex by mediating the release of 14-3-3 (Fig. 6). However, it is likely that additional members of the PPP (phosphoprotein phosphatases) family are active within the Par-3 complex and dephosphorylate Par-3-associated proteins such as aPKC (see below) (28). The analysis of additional protein phosphatases that may target Par-3 and its interacting proteins will provide further insight into the molecular mechanisms that allow the spatiotemporal regulation of the Par-3 complex.
Fig. 6.
Model for PP1 as a functional regulator of the Par-3 scaffold. Par-3 diffusing to the lateral cortex is phosphorylated by Par-1 (EMK1/MARK2), resulting in the recruitment of 14-3-3. aPKC subsequently dissociates from Par-3 and the complex is released from the cortex. As soon as Par-3 escapes the active range of Par-1 (indicated by gray area), PP1 dephosphorylates Par-3, allowing the re-formation of a functional Par/aPKC complex at the apical domain of epithelial cells. In addition, PP1 stabilizes the interaction between aPKC and Par-3 by dephosphorylating Par-3 at S824.
In mammals, the assembly of the Par/aPKC complex is required for the formation of TJs. TJs generally encircle the apical aspect of mammalian epithelial cells and are composed of transmembrane proteins that are linked to the cytoskeleton via a set of scaffolding and signaling molecules (29). By mediating a selective passage of solutes and by maintaining apical-basal plasma membrane asymmetry, TJs allow the physiological functioning of epithelia. Par-3 is important for the formation of TJs, because its overexpression results in a more rapid formation of monolayers with increased transepithelial electrical resistance (TEER), when associated with aPKC (12). In contrast, its suppression by RNA interference severely delays TJ biogenesis (11). Does Par-3-bound PP1 influence TJ formation? To date, the role of serine/threonine phosphatases in the modulation of TJ function has been mainly studied by using small-molecule inhibitors, including OA and calyculin A (30). However, because these toxins inhibit both PP1 and PP2A, the precise mechanisms underlying the induced TJ leakiness are not fully understood. By employing a dominant negative approach, we provide evidence that PP1 negatively regulates the formation of functional TJs in MDCKII cells, potentially by modulating the association of aPKC and Par-3 through dephosphorylation of S824 on Par-3. The situation is complex, however, because TJ function is likely regulated by several protein phosphatases. Previous studies have demonstrated that PP2A influences TJ biogenesis by negatively regulating the activity of aPKCζ and by controlling the phosphorylation state of TJ proteins (28, 31). Therefore, distinct serine/threonine phosphatases apparently target different TJ components, allowing the dynamic modulation of epithelial barriers in response to physiological or pathological stimuli (32).
Scaffold proteins often coordinately bind protein kinases and phosphatases at specific subcellular regions (33). In a similar fashion, our findings demonstrate that Par-3 acts as a spatially localized signaling platform for multiple serine/threonine kinases and the PP1 phosphatase, thereby creating a focal point of enzyme activities that dynamically regulate TJ formation. Finally, we show that the MRM MS-based approach is a powerful tool to dissect fluctuating posttranslational modifications associated with a complex biological behavior, such as TJ assembly and cell polarity.
Methods
Immunoprecipitation and in Vitro Binding Assays.
Immunoprecipitation experiments were carried out as described in SI Text and ref. 5. For mixing experiments, MBP-fusion proteins were purified from BL21 Escherichia coli as recommended by the manufacturer (NEB). Purified PP1α (100 ng; NEB) was incubated with 100 ng of the respective Par-3 MBP-fusion proteins bound to amylose resin in 500 μl of lysis buffer [20 mM Tris·Cl (pH 7.5), 150 mM NaCl, 5% glycerol, 1% Nonidet P-40, 10 μg·ml−1 Leupeptin, 10 μg·ml−1 Aprotinin, 10 μg·ml−1 Pepstatin A, 1 mM PMSF] for 1 h at 4°C, and after extensive washing the samples were analyzed by Western blot analysis and Coomassie brilliant blue staining.
In Vitro Kinase and Phosphatase Assays.
Collected Par-3 or YFP-fusion protein complexes were used as a source for protein phosphatase 1 and were analyzed for the release of free phosphate using the Ser/Thr Phosphatase Assay Kit 1 (Upstate) according to the manufacturer's protocol. In brief, the collected immunocomplexes were incubated with the substrate (K-R-pT-I-R-R) for 2 h at 30°C and, after the addition of 100 μl of Malachite Green solution, the absorbance at 650 nm was determined. Results were normalized to values obtained from preimmune serum immunocomplexes or YFP immunoprecipitates collected from cells expressing 2xmyc-PP1α and YFP alone. To test the various Par-3 regions for their ability to influence the activity of PP1α, 100 ng of purified PP1α (NEB) were incubated with 1 μg of purified MBP, MBP-fusion proteins, or 100 ng of Inhibitor-2 (NEB) for 30 min in 20 μl of assay buffer [50 mM Tris·Cl (pH 7.5), 100 μM CaCl2]. Subsequently, the supplied phosphopeptide was added as a substrate and after 15 min the samples were analyzed as described earlier. All experiments were done in triplicate and error bars represent 1 SD.
For in vitro kinase assays, exogenously expressed proteins were immunoprecipitated (IP) from 293T cells stably expressing 3×FLAG-Par-3 or 3×FLAG-Lgl2 and were subjected to phosphorylation using purified aPKCζ (5, 17). The resulting 32P-labeled proteins were then incubated with the catalytic subunit of PP1α or PP2A2 in 30 μl of phosphatase buffer for 30 min at 30°C and the rate of dephosphorylation was monitored by SDS/PAGE and autoradiography.
LC-MS/MS and MRM Analysis.
Sample preparation and protein identification by LC-MS/MS was performed as described (19). For details on MRM sample preparation, see SI Text.
To establish MRM methods full-scan survey and MS/MS identification spectra were combined from a LTQ-Orbitrap (Thermo-Fisher) and a QSTAR-Elite (Applied Biosystems/Sciex) LC-MS/MS system to obtain optimal sequence coverage on in vivo phosphorylated Par-3. Several phosphorylation sites were detected including pS144, but no signals were observed for S824/S885 or pS824/pS885. To target and gather the spectral information needed to construct the MRM assays for S824/S885 and pS824/pS885, a fragment of Par-3 (amino acid 688–973) was purified from bacteria as a GST-fusion protein and an in vitro kinase assay using purified aPKCζ or the kinase domain of MARK2 was performed (17) to generate the phosphopeptides pS824 and pS885. After trypsin digestion the samples were analyzed by iterative MS/MS analyses by using the RepeatIDA function on the QSTAR Elite (Analyst 2.0, Applied Biosystems/Sciex). In brief, several injections of the Par-3 digest were introduced onto the QSTAR-Elite with each successive LC-MS/MS analysis automatically programmed to ignore any peptide signals that had been previously analyzed. Full-scan MS/MS sequence data and corresponding peptide sequences were automatically read into MRMPilot (Applied Biosystems/Sciex) and the MRM assays were built by extracting the charge state(s) and most intense MS/MS sequence ions of any given peptide to generate ion pairs for each individual peptide. On average at least 2 ion pairs were used per peptide to minimize the potential for false positive identification. MRMPilot automatically generated an acquisition method for a 4000QTRAP mass spectrometer (Applied Biosystems/Sciex) which was used to perform the analysis (Table S1). To ensure that low-level quantitative data were specific to our targeted peptides and were not contaminated with isobaric interferences, multiple ion pair transitions were used and a full-scan MS/MS was triggered from each MRM transition (MIDAS) (34). These full-scan data were then correlated to the appropriate mouse Par-3 phosphorylated peptides to ensure the MS/MS scan was consistent with the peptide sequence. A minimum of two repeats using immunoprecipitated mouse 3×FLAG-Par-3 from different cell passages were performed. For relative quantification of S25 the areas of the extracted ion curves (XIC) were integrated by using Analyst 1.4.2 software (Applied Biosystems/Sciex) and the ratio of nonphosphorylated/phosphorylated peptide was calculated for samples before and after PP1α treatment (error bars represent 1 SD; n = 2).
Transepithelial Electrical Resistance (TEER) Measurements.
Tet-inducible MDCK-T23 cells expressing either YFP or YFP-PP1α (H248K) were plated on polycarbonate treated Transwell inserts (Costar) in the presence of 20 ng·ml−1 doxycycline. Twenty-four hours later doxycycline was removed from the culture medium and after an additional 48 h the confluent monolayers were transferred to low Ca2+ medium for 12–16 h. After switching the cells back to normal growth medium, TEER was recorded by using an Epithelial Voltohmmeter (EVOM, World Precision Instruments) over a time period of 24 h. TEER values, expressed as Ω·cm2, were obtained by subtracting cell-free filter readings. Three independent filters for every time point and condition were measured by using two different stable cell lines. Error bars in plots represent 1 SD.
Additional details for materials and methods are discussed in the SI Text.
Supplementary Material
Acknowledgments.
We thank R. Colbran (Vanderbilt University Medical Center, Nashville, TN) for providing isoform-specific PP1 antibodies; J. Casanova (University of Virginia Health System, Charlottesville, VA) for providing the T23-MDCKII cells; K. Ebnet (Institute of Medical Biochemistry, ZMBE, Münster, Germany) for sharing the pEmU6pro-T vector; B. Larson (SLRI, Mount Sinai Hospital, Toronto, ON, Canada) for technical assistance with LC-MS/MS; J. Fawcett and members of the T.P. lab for reagents and helpful discussions. This work was supported by grants from the National Cancer Institute of Canada (NCIC) with funds made available by the Canadian Cancer Society (CCS), the Canadian Institutes for Health Research (Grants 6849 and 57793), Genome Canada through the Ontario Genomics Institute, the Ontario Research Foundation, and a Terry Fox Program Project grant from the NCIC/CCS. T.P. is a Distinguished Scientist of the CIHR. A.T. was supported by an Erwin Schrödinger Stipendium from the Austrian Science Fund (FWF), and G.W. was supported by fellowships from the European Molecular Biology Organization and NCIC.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804102105/DCSupplemental.
References
- 1.Goldstein B, Macara IG. The par proteins: Fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Intercellular junctions and cellular polarity: The PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- 4.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 5.Lin D, et al. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- 6.Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 7.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Izumi Y, et al. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 12.Hirose T, et al. Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J Cell Sci. 2002;115:2485–2495. doi: 10.1242/jcs.115.12.2485. [DOI] [PubMed] [Google Scholar]
- 13.Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- 14.Hurd TW, et al. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 17.Plant PJ, et al. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki A, et al. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells CD, et al. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 20.Fang L, et al. Cell polarity protein Par3 complexes with DNA-PK via Ku70 and regulates DNA double-strand break repair. Cell Res. 2007;17:100–116. doi: 10.1038/sj.cr.7310145. [DOI] [PubMed] [Google Scholar]
- 21.Cohen PT. Protein phosphatase 1-targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 22.Andreassen PR, Lacroix FB, Villa-Moruzzi E, Margolis RL. Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J Cell Biol. 1998;141:1207–1215. doi: 10.1083/jcb.141.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuhashi S, et al. Usage of tautomycetin, a novel inhibitor of protein phosphatase 1 (PP1), reveals that PP1 is a positive regulator of Raf-1 in vivo. J Biol Chem. 2003;278:82–88. doi: 10.1074/jbc.M208888200. [DOI] [PubMed] [Google Scholar]
- 24.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Zhang Z, Brew K, Lee EY. Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry. 1996;35:6276–6282. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- 26.Lin Q, Buckler ESt, Muse SV, Walker JC. Molecular evolution of type 1 serine/threonine protein phosphatases. Mol Phylogenet Evol. 1999;12:57–66. doi: 10.1006/mpev.1998.0560. [DOI] [PubMed] [Google Scholar]
- 27.Schillace RV, Voltz JW, Sim AT, Shenolikar S, Scott JD. Multiple interactions within the AKAP220 signaling complex contribute to protein phosphatase 1 regulation. J Biol Chem. 2001;276:12128–12134. doi: 10.1074/jbc.M010398200. [DOI] [PubMed] [Google Scholar]
- 28.Nunbhakdi-Craig V, et al. Protein phosphatase 2A associates with and regulates atypical PKC and the epithelial tight junction complex. J Cell Biol. 2002;158:967–978. doi: 10.1083/jcb.200206114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 30.Okada T, Narai A, Matsunaga S, Fusetani N, Shimizu M. Assessment of the marine toxins by monitoring the integrity of human intestinal Caco-2 cell monolayers. Toxicol in Vitro. 2000;14:219–226. doi: 10.1016/s0887-2333(00)00014-x. [DOI] [PubMed] [Google Scholar]
- 31.Seth A, Sheth P, Elias BC, Rao R. Protein phosphatases 2A and 1 interact with occludin and negatively regulate the assembly of tight junctions in the CACO-2 cell monolayer. J Biol Chem. 2007;282:11487–11498. doi: 10.1074/jbc.M610597200. [DOI] [PubMed] [Google Scholar]
- 32.Sontag JM, Sontag E. Regulation of cell adhesion by PP2A and SV40 small tumor antigen: An important link to cell transformation. Cell Mol Life Sci. 2006;63:2979–2991. doi: 10.1007/s00018-006-6300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong W, Scott JD. AKAP signalling complexes: Focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 34.Unwin RD, et al. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol Cell Proteomics. 2005;4:1134–1144. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.