Summary
Structural, biochemical and genetic studies have led to proposals that a mobile element of multi-subunit RNA polymerases, the Trigger Loop (TL), plays a critical role in catalysis and can be targeted by antibiotic inhibitors. Here we present evidence that the Saccharomyces cerevisiae RNA Polymerase II (Pol II) TL participates in substrate selection. Amino acid substitutions within the Pol II TL preferentially alter substrate usage and enzyme fidelity, as does inhibition of transcription by α-amanitin. Finally, substitution of His1085 in the TL specifically renders Pol II highly resistant to α-amanitin, indicating a functional interaction between His1085 and α-amanitin that is supported by re-refinement of an α-amanitin-Pol II crystal structure. We propose that α-amanitin inhibited Pol II elongation, which is slow and exhibits reduced substrate selectivity, results from direct α-amanitin interference with the TL.
INTRODUCTION
Recent structural studies complement a large body of work that has strived to illuminate the mechanism of transcription by multi-subunit RNA polymerases. Three-dimensional X-ray structures of eukaryotic Pol II and prokaryotic RNA polymerase (RNAP) highlight the structural conservation of these enzymes, particularly within their active sites (Cramer et al., 2000; Cramer et al., 2001; Gnatt et al., 2001; Hirata et al., 2008; Murakami et al., 2002a; Murakami et al., 2002b; Vassylyev et al., 2002; Vassylyev et al., 2007a; Vassylyev et al., 2007b; Wang et al., 2006; Westover et al., 2004; Zhang et al., 1999). RNA polymerases must balance speed of transcription with fidelity, efficiently selecting correct NTP substrates specified by the DNA template. Additionally, RNA polymerases must select correct NTP substrates over 2’-dNTP substrates. Multi-subunit RNA polymerases are observed to make contacts with the 2’-OH and 3’-OH groups of incoming NTPs (Vassylyev et al., 2007b; Wang et al., 2006; Westover et al., 2004), but these contacts cannot fully explain the robust selection of NTPs over 2’-dNTPs.
Several conserved sequence blocks have been identified in eukaryotic Pol I, II and III and their prokaryotic counterpart, RNAP (Jokerst et al., 1989). One of these, homology block G, includes a highly conserved Trigger Loop (TL), named due to its mobile nature and possible relationship to the translocation mechanism (Vassylyev et al., 2002).
In the presence of template-specified “matched” NTP substrates, the TLs of both Pol II and RNAP are stabilized in a conformation that directly interacts with base-paired substrates (Vassylyev et al., 2007b; Wang et al., 2006). This orientation of the TL is hypothesized to promote catalysis of phosphodiester bond formation (Vassylyev et al., 2007b; Wang et al., 2006). The observed contacts between NTPs and TLs of Pol II and RNAP are similar but not identical. In both systems, an absolutely conserved histidine (His1085 in S. cerevisiae Rpo21, referred to as Rpb1 hereafter, His1242 in Thermus thermophilusβ', and His936 in E. coli β') within the TL appears to interact with NTP substrates and has been proposed to be critical for TL function based on the recent structures (Wang et al., 2006). In E. coli RNAP, this residue is essential for viability (Weilbaecher et al., 1994).
The exact role of the TL in transcription is unclear. The TL is not required for matched NTP base-pairing with the template DNA, but rather may function by stabilizing the reaction product or enabling a conformational change in the substrate that promotes catalysis. We have previously proposed that TL contacts with base-paired NTP substrates may be specifically tailored to such matched substrates, and thus the TL may function in transcriptional fidelity (Wang et al., 2006). The TL is not absolutely required for bond formation, as deletion or severe mutation of the TL in either E. coli or T. thermophilus RNAP does not inactivate the enzyme, but renders elongation very slow (Temiakov et al., 2005; Toulokhonov et al., 2007; Vassylyev et al., 2007b).
TL mutants alter elongation properties of RNAP and Pol II, both negatively and positively, and affect the ability of RNAP to respond to elongation factors (Bar-Nahum et al., 2005; Malagon et al., 2006; Svetlov et al., 2007; Temiakov et al., 2005; Toulokhonov et al., 2007; Vassylyev et al., 2007b; Weilbaecher et al., 1994). The TL of RNAP has been identified as essential for NTP-dependent aspects of the transcription reaction observed in kinetic experiments, which are proposed to relate to a step other than chemistry (Toulokhonov et al., 2007) and have been attributed to translocation or NTP binding to an allosteric site (Bar-Nahum et al., 2005; Foster et al., 2001; Nedialkov et al., 2003; Toulokhonov et al., 2007). The RNAP TL has also been implicated in active site rearrangements during transcriptional pausing, where the TL interacts with the 3’-end of the nascent RNA (Toulokhonov et al., 2007).
In bacterial RNAP, the TL is required for inhibition of transcription by Streptolydigin (Stl) (Temiakov et al., 2005; Tuske et al., 2005). Stl slows RNAP elongation and a crystal structure of the Tth RNAP EC with Stl shows the TL positioned away from a matched NTP substrate base-paired with DNA in the RNAP active site, suggesting Stl blocks TL interaction with substrate (Vassylyev et al., 2007b). In the eukaryotic enzyme, the mushroom toxin α-amanitin, a specific Pol II inhibitor, binds near the region traversed by TL movement, raising the possibility that α-amanitin inhibits Pol II by preventing or altering TL movement (Bushnell et al., 2002; Wang et al., 2006).
Genetic data concerning α-amanitin resistant Pol II mutants from several species and the crystal structure of α-amanitin bound to S. cerevisiae Pol II are in agreement that α-amanitin functionally binds the Pol II active site just below the “Bridge Helix” domain (Bushnell et al., 2002) (and references therein). α-amanitin appears not to alter substrate association with Pol II, but instead slows elongation considerably, thereby favoring pausing and backtracking events (Chafin et al., 1995; Rudd and Luse, 1996). α-amanitin inhibition of human Pol II in kinetic experiments has been interpreted as resulting from a block to Pol II translocation, possibly through restraint of Bridge Helix conformational changes (Gong et al., 2004) as had been previously proposed by (Bushnell et al., 2002). In the presence of bound α-amanitin, Pol II is able to add several nucleotides before encountering normal pause sites that are in turn exacerbated by α-amanitin, suggesting that any translocation block is not absolute (Chafin et al., 1995; Rudd and Luse, 1996).
In this work, we present evidence that the Pol II TL functions in substrate selectivity, promoting the fast addition of template-specified NTP substrates but not of mismatched NTPs or matched 2’-dNTPs. A loss-of-function substitution within the TL strongly compromises matched NTP usage but has more modest effects on 2’-dNTP incorporation or NTP misincorporation, indicating that slow addition of inappropriate substrates may represent a distinct, TL-independent mode of synthesis by Pol II. Other substitutions in the TL increase incorporation rate for either all substrates or for inappropriate substrates, illustrating the close relationship between the TL and correct substrate usage. We show that α-amanitin specifically restrains the Pol II TL through a functional interaction with TL residue His1085 and that the consequence of α-amanitin inhibition of transcription is conversion of Pol II from a fast, accurate, TL-dependent mode to a slow, inaccurate, TLindependent mode.
RESULTS
Rpb1 Trigger Loop (TL) Residue His1085 is Important for Pol II Function in vivo
Recent crystal structures of both Pol II and prokaryotic RNAP have revealed a new conformation of the TL, showing a direct interaction between the TL and a matched NTP positioned at the 3’-end of the nascent RNA (Figure 1A). In both enzymes, a conserved histidine residue within the TL is observed to directly interact with the NTP substrate. In order to probe the requirement for this histidine in TL function and transcription in general, we engineered different amino-acid substitutions in place of S. cerevisiae Rpb1 His1085 (Figure 1B). Substitution of alanine or phenylalanine at position 1085 results in inviability, while substitution of tyrosine for histidine (H1085Y) supports viability but confers a severe growth defect.
Figure 1. Substitution of Rpb1 His1085 confers severe growth defects or lethality in vivo.
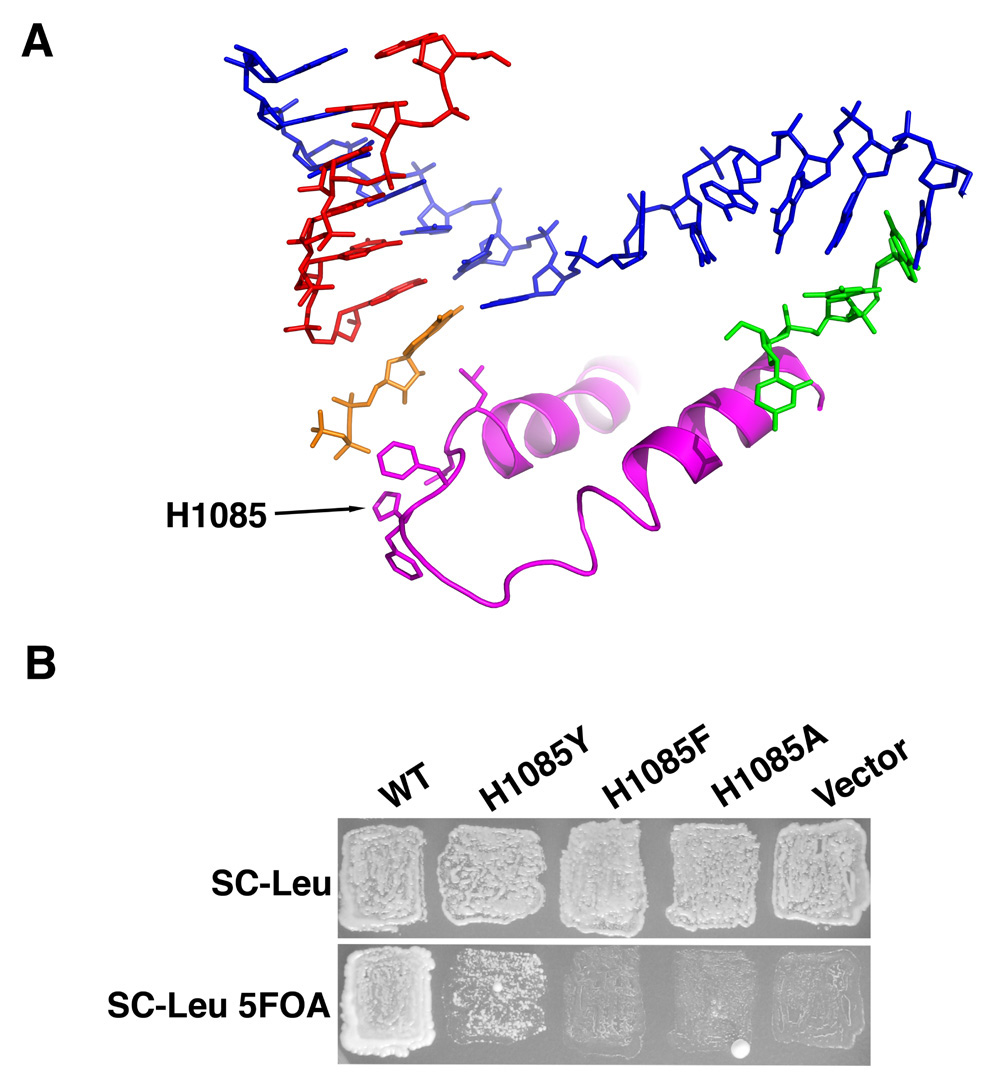
A. Structural model of TL interaction with matched GTP in Pol II active site (PDB 2E2H)(Wang et al., 2006). TL residues Rpb1 1070–1105 (magenta) are shown together with a GTP substrate (orange), template DNA (blue), downstream non-template DNA (green) and RNA (red). Sidechains for some residues in the TL (Rpb1 1081–1082,1084–1086) are also shown. His1085 is positioned to interact with the β-phosphate of the GTP. B. Phenotypes of Rpb1 His1085 substitutions. In the absence of functional Rpb1, cells cannot lose an RPB1 plasmid marked with URA3. rpb1-H1085A and rpb1-H1085F plasmids fail to confer viability to an rpb1Δ strain, rendering cells sensitive to the drug 5-FOA because they have been forced to maintain an RPB1-URA3 plasmid for Rpb1 function (bottom row). rpb1-H1085Y can provide the sole source of Rpb1 function, but exhibits a severe growth defect.
Substitution of Rpb1 His1085 Causes Defects in Pol II Elongation and Reduces Selection of NTP Substrates over 2’-dNTP and Mismatched NTP Substrates
In order to understand the molecular defects underlying the severe growth defect caused by the H1085Y substitution, we purified this enzyme from yeast and examined its activity with a variety of substrates (Figure 2)(Supplemental Figure 1). Wild type (WT) and H1085Y enzymes were assembled onto nucleic acid scaffolds similar to those used for structural studies of Pol II and their ability to utilize various substrates examined. H1085Y exhibited a strong elongation defect for run-off transcription at saturating NTP concentration (Figure 2A)(Supplemental Figure 1A). Compared to its defect with NTPs, H1085Y was less impaired for the addition of 2’-dNTPs (Figure 2B)(Supplemental Figure 1B). Similarly, H1085Y exhibited a milder defect for misincorporation of GTP at a position where ATP was specified than it did for NTP addition (Figure 2C)(Supplemental Figure 1D). WT and H1085Y were also characterized in a modified single nucleotide addition assay (Wang et al., 2006). In this assay substrate concentration is lowered significantly to allow a single incorporation to be observed. Strong H1085Y effects on ATP or GTP incorporation were observed using the same templates where H1085Y had only mild defects in 2’-dATP or 2’-dGTP incorporation or GTP misincorporation (Supplemental Figure 1E). The defect in H1085Y is primarily on catalysis, as KMs of H1085Y for the substrates tested do not significantly differ from WT (Supplemental Table 1). Comparing kcat/KM values for different substrates from Supplemental Table 1, WT Pol II shows a 300-fold selection of NTPs over 2’-dGTP and a 5000-fold selection of NTPs over 2’-dATP. H1085Y shows only a 60-fold selection for NTPs over 2’-dGTP and a 600-fold selection for NTPs over 2’-dATP. These results suggest that His1085 promotes selection of correct NTP substrates over incorrect NTPs and matched 2’-dNTPs.
Figure 2. Elongation Defects and Altered Substrate Selection by rpb1 H1085Y Pol II.
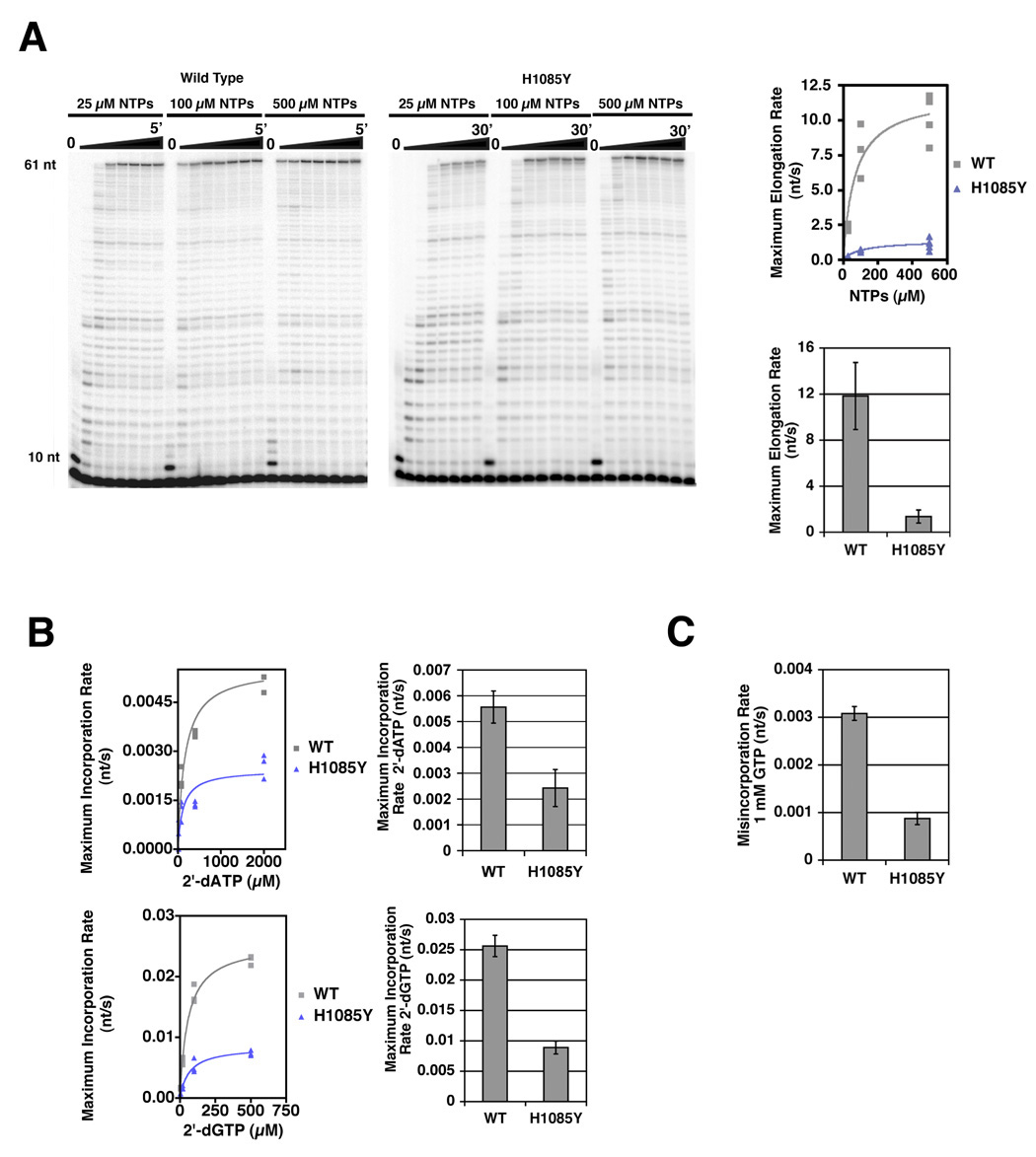
A. H1085Y exhibits reduced elongation rate using NTP substrates. Run-off transcription of an oligonucleotide scaffold template generates a 61 nt RNA product. Representative experiments for WT and H1085Y Pol II are shown in the left and right panels, respectively. Average elongation rates for each NTP concentration were measured as the length of the transcribed region (51 nt) divided by the time of half-maximal accumulation of run-off product (61 nt). Average elongation rates were then plotted versus NTP concentration to infer maximum average elongation rate (see Experimental Procedures for details)(top right graph). Inferred maximum average elongation rates are shown in the bottom right graph with error bars representing the 95% confidence interval (See Experimental Procedures for details). B. H1085Y Pol II exhibits only modest defects for 2’-dNTP incorporation. WT and H1085Y Pol II ECs were formed on oligonucleotide scaffolds, containing 10-mer RNAs with templates specifying addition of different NTPs at position 11. Average incorporation rates for different template-specified 2’-dNTPs were measured as 1/t1/2 for maximal accumulation of 11-mer RNA. Incorporation rates were then plotted versus 2’-dNTP concentration and maximum incorporation rate for either 2’-dATP or 2’-dGTP was inferred (left panels). Maximum incorporation rates for WT Pol II and H1085Y are shown in the right panels with error bars representing the 95% confidence interval (See Experimental Procedures for details). C. H1085Y Pol II exhibits modest defects in GTP misincorporation. WT and H1085Y ECs were formed and labeled as in (B) with templates specifying incorporation of ATP at the position being measured, but were challenged with 1 mM GTP and misincorporation rate measured as the 1/t1/2 for maximal incorporation. Mean misincorporation rate from at least three experiments is represented in the bar graph (error bars represent +/− SD).
Resistance of TL Mutant H1085Y to α-amanitin and Substrate-Specific Pol II Inhibition by α-amanitin
To probe His1085 and TL function, we assessed the effect of α-amanitin treatment on usage of matched and mismatched NTP substrates and matched 2’-dNTP substrates by WT and H1085Y forms of Pol II. Our results above suggested that TL residue His1085 functions in efficient usage of matched NTP substrates but is less involved in utilization of inefficient substrates such as 2’-dNTPs or mismatched NTPs. We reasoned that if α-amanitin inhibition targets TL function, then it might exhibit substrate-selective effects on elongation, similar to those observed for H1085Y (Figure 3 and Supplemental Figure 2). Treatment of WT Pol II with α-amanitin resulted in a severe reduction in elongation using NTP substrates. The elongation activity of H1085Y, while reduced compared to WT, was comparatively much less affected by α-amanitin (35 fold inhibition for WT, less than two fold of H1085Y)(Figure 3A)(Supplemental Figure 2A).
Figure 3. α-amanitin inhibition of WT Pol II is substrate selective, while H1085Y Pol II is mostly resistant to α-amanitin.
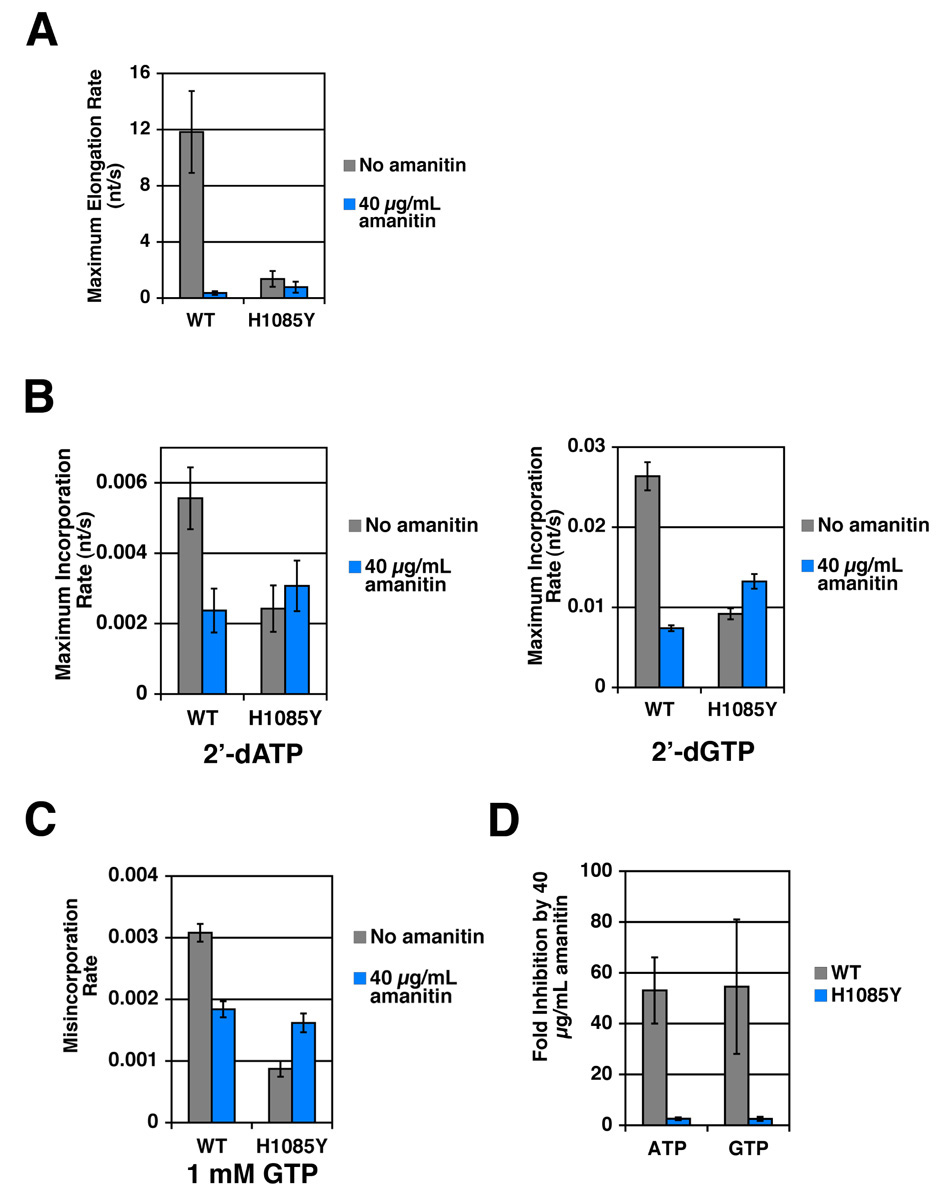
A. α-amanitin inhibition of elongation by WT and H1085Y Pol II elongation complexes. Average elongation rate with NTP substrates for α-amanitin treated WT and H1085Y Pol II ECs determined exactly as in Figure 2A. RNA 10-mer containing ECs were formed as in Figure 2A but chased with equimolar NTPs at various concentrations in the presence of 40 µg/mL α-amanitin. Average elongation rates were then plotted versus NTP concentration and maximum average elongation rate inferred (Supplemental Figure 1A). Maximum average elongation rates are shown with error bars representing the 95% confidence interval of the mean. Untreated values are from Figure 2A and shown for comparison. B. Incorporation rate for 2’-dNTPs in the presence of α-amanitin measured for WT and H1085Y Pol II. Elongation complexes formed exactly as in Figure 2B, but treated with various concentrations of 2’-dATP or 2’-dGTP as specified by appropriate DNA template in the presence of 40 µg/mL α-amanitin (Supplemental Figure 1B). Maximum average incorporation rates for 2’-dATP or 2’-dGTP for WT Pol II and H1085Y are shown with error bars representing the 95% confidence interval of the mean. Untreated values are from Figure 2B and shown for comparison. C. α-amanitin inhibition of GTP misincorporation by WT and H1085Y Pol II elongation complexes. Experiment is the same as Figure 2C, save for the addition of 40 µg/mL α-amanitin in treated samples (Supplemental Figure 1C). Untreated values are from Figure 2C and the graph represents mean misincorporation rate with error bars representing the standard deviation of the mean. D. α-amanitin inhibits addition of a single NTP by WT Pol II while H1085Y is mostly resistant. WT and H1085Y Pol II enzymes were prepared as in Figure 2B and aliquots were incubated with a titration of either ATP or GTP as specified by the DNA template, for 5 minutes. The values shown are the mean fold inhibition by 40 µg/mL α-amanitin from several experiments, as determined by concentration of substrate that gives half-maximal incorporation in the presence of 40 µg/mL α-amanitin divided by the concentration that gives half-maximal incorporation with no α-amanitin. Error bars represent +/− SD.
α-amanitin treatment was also much less effective for inhibition of 2’-dNTP addition (Figure 3B)(Supplemental Figure 2B) or for GTP misincorporation (Figure 3C)(Supplemental Figure 2C) by WT Pol II. In contrast to WT, H1085Y was completely resistant to or slightly stimulated by α-amanitin for these substrates. Our experiments with 2’-dNTP substrates or NTP misincorporation are limited to addition of a single nucleotide due to an almost complete inability of Pol II to incorporate more than one 2’-dNTP, even if matched, and the slow rate of multiple misincorporations. To more directly compare α-amanitin inhibition of NTPs with suboptimal 2’-dNTP and mismatched substrates, we examined the effect of α-amanitin in our modified single nucleotide addition assay (Figure 3D). In this assay, α-amanitin greatly increased the concentration of ATP or GTP required for WT Pol II to achieve a particular elongation rate, indicating strong inhibition of ATP or GTP addition by α-amanitin on the identical templates where α-amanitin only weakly inhibited 2’-dNTP addition or GTP misincorporation. Furthermore, H1085Y was only weakly inhibited. These results indicate that α-amanitin is a substrate-selective inhibitor of Pol II, suggesting that a simple block to Pol II translocation is insufficient to explain the inhibition mechanism.
Pol II TL Mutants Exhibit Distinct in vivo Phenotypes, and Allele-Specific Modulation of Substrate Selection and Interaction with α-amanitin in vitro
One model for resistance of H1085Y Pol II to α-amanitin is that H1085Y and α-amanitin both strongly compromise TL function, therefore a combination of the two defects would not further impair transcription. To ascertain the specificity of H1085Y α-amanitin resistance, we examined the effect of α-amanitin on a number of other TL mutants that we isolated in a genetic screen for rpb1 mutants with in vivo transcription defects or by site-directed mutagenesis (Figure 4). S. cerevisiae rpb1 alleles with substitutions within the TL have been identified previously (Archambault et al., 1998; Archambault et al., 1992; Hekmatpanah and Young, 1991; Malagon et al., 2006) and we identified many of these alleles and a large number of other TL mutants with distinct phenotypes (Figure 4A and 4B)(Supplemental Figure 3).
Figure 4. TL mutants exhibit complex phenotypes in vivo and have distinct effects on Pol II substrate selectivity and α-amanitin sensitivity in vitro.
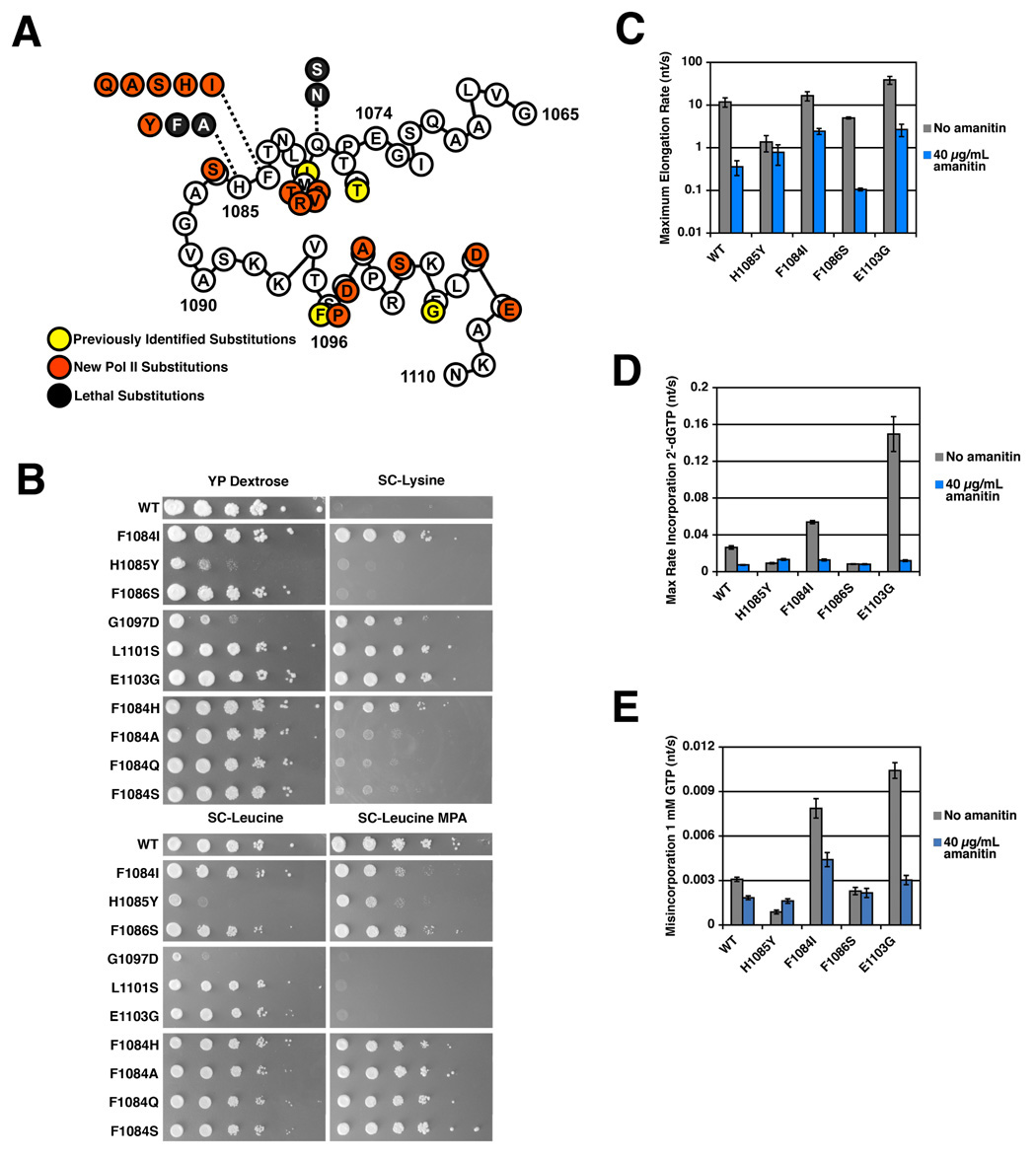
A. Schematic illustrating TL mutants isolated previously (Archambault et al., 1998; Hekmatpanah and Young, 1991; Malagon et al., 2006)(yellow filled circles) and those newly described in this work from genetic screens and site-directed mutagenesis (red or black filled circles). Numbers indicated amino acid position in S. cerevisiae Rpb1. Letters in unfilled circles indicate WT residues at those positions. Letters in filled circles indicated residue substitutions conferring mutant phenotypes in vivo. Black filled circles represent site-directed substitutions that are lethal. B. Phenotypes of selected TL mutants. TL mutants were transformed into a parent strain containing lys2-128∂, a marker allele for the Spt−phenotype (Simchen et al., 1984). 10-fold serial dilutions of TL mutant saturated liquid cultures were spotted onto several different media to illustrate growth differences of TL mutants compared to a WT strain. YP Dextrose plates illustrate growth differences on rich media with 2% Dextrose as carbon source. SC-Leu plates are minimal media lacking the amino acid leucine and illustrate differences in growth on minimal media and act as an untreated control for growth on SC-Leu in the presence of 20 µg/mL MPA. SC-Lys plates are minimal media lacking lysine, and assay the Spt- phenotype. A WT Pol II strain cannot grow on SC-Lys due to the lys2-128∂ allele, however mutants conferring the Spt− phenotype suppress this allele and allow growth. C. Altered elongation rate and sensitivity of NTP substrate usage to α-amanitin of selected TL mutants. Maximum elongation rates were measured as in Figure 2A and Figure 3A, with WT and H1085Y Pol II values shown for comparison. D. Altered 2’-dGTP substrate usage and α-amanitin sensitivity of selected TL mutants. Incorporation rates determined as in Figures 2B and 3B, with WT and H1085Y Pol II values shown for comparison. E. Altered GTP misincorporation and sensitivity to α-amanitin of selected TL mutants. Misincorporation rates determined as in Figure 2C and Figure 3C, with WT and H1085Y Pol II values shown for comparison.
Phenotypes proposed to be representative of transcription elongation defects include sensitivity to mycophenolic acid (MPA) a drug that lowers GTP pools in vivo via inhibition of inosine 5′-monophosphate dehydrogenase (IMPDH), the rate-limiting step for guanine synthesis (Sweeney, 1977). There are no known correlations between in vitro elongation defects and MPA sensitivity, likely indicating the etiologies behind this phenotype may be more complex than is generally recognized. Several newly isolated TL mutants confer MPA sensitivity, but notably not H1085Y, which has a clear in vitro elongation defect (Figure 4B and data not shown). TL mutants could also be distinguished from one another based on phenotypes in an in vivo transcriptional interference and 3’-end formation assay (Supplemental Figure 3).
One previously isolated allele rpb1-E1103G was identified as increasing Pol II elongation rate in vitro (Malagon et al., 2006) and was reisolated in our screens. Here we identify the rpb1-E1103G allele and several closely positioned substitutions as conferring the Suppression of Ty (Spt−) phenotype (Figure 4B). The Spt− phenotype in our screens relates to suppression of a transcription defect caused by a specific insertion of a Ty1 retroelement long terminal repeat (LTR) into the LYS2 gene (Simchen et al., 1984). A large number of transcription related mutants, including those in elongation factors, coactivators, TBP and histones, cause various spectra of Spt− phenotypes (Winston and Sudarsanam, 1998). Previously identified Spt− alleles of Pol II subunits cluster in and around the TL (Hekmatpanah and Young, 1991). Isolation of new Spt− mutants from our screens of several Pol II subunits identified substitutions exclusively within the TL, suggesting a specific defect might underlie this Spt− phenotype (Figure 4B and data not shown).
Elongation assays show that TL mutants have different transcriptional defects, consistent with their distinct growth profiles in various plate assays (Figure 4C–4E)(Supplemental Figure 4). Substitutions in the TL can increase (E1103G)(Malagon et al., 2006) or decrease elongation rate (H1085Y, F1086S) (Figure 2A and Figure 4C)(Supplemental Figure 4A). F1086S shows a loss of function, albeit milder than H1085Y, for NTP substrates but remains sensitive to α-amanitin. The confinement of strong α-amanitin resistance to H1085Y (Figure 3A and Figure 4C) among tested alleles for usage of NTP substrates fails to support a model for H1085Y α-amanitin resistance wherein H1085Y phenotypes are simply redundant with the α-amanitin-inhibited state, allowing no further inhibition by α-amanitin. When incorporation of 2’-dGTP or misincorporation of GTP by TL mutants was examined (Figure 4D–4E, Supplemental Figure 4B–4C), the Spt−alleles (F1084I, E1103G) showed increased incorporation of inappropriate substrates, with the increased activity of E1103G for such substrates being mostly α-amanitin sensitive. F1086S was resistant to weak α-amanitin inhibition of 2’-dGTP incorporation or GTP misincorporation.
α-amanitin Inhibition of Wild Type and H1085Y Pol II Usage of NTP and 2’-dNTP Substrates is Distinct, While Substrate Specific α-amanitin Inhibition is Conserved
While sensitive to α-amanitin, S. cerevisiae Pol II requires much higher concentrations of α-amanitin for inhibition than do Pol II enzymes from other species (Wieland and Faulstich, 1991). We determined whether H1085Y’s resistance to α-amanitin could be overcome by increased α-amanitin concentration, consistent with a previously unobserved role for His1085 in α-amanitin binding (Figure 5). α-amanitin inhibition of WT Pol II usage of NTP substrates is not complete even at 1 mg/mL, a concentration several orders of magnitude higher than the Kd of mammalian Pol II for α-amanitin (Figure 5A). It is clear that α-amanitin resistance of H1085Y cannot be overcome by increased concentration of the drug, suggesting that α-amanitin-Pol II association is not the primary defect of H1085Y. Other TL substitutions alter the inhibition profile of α-amanitin, increasing sensitivity (F1086S) or decreasing it (F1084I, E1103G) (but not to the extent of H1085Y). H1085Y, which is weakly inhibited for NTP usage by α-amanitin, is entirely resistant for 2’-dATP usage, while 2’-dATP usage by WT Pol II has a different inhibition profile than for NTPs, underscoring α-amanitin’s substrate selectivity (Figure 5B). To determine if substrate-selective effects of α-amanitin apply to other Pol II enzymes that form much tighter enzyme-inhibitor complexes, we examined the α-amanitin inhibition of different substrate usage by calf thymus Pol II (Figure 5C–5E)(Supplemental Figure 5). The calf thymus enzyme responds to α-amanitin as does yeast Pol II suggesting that substrate-selective effects are inherent to the α-amanitin mechanism.
Figure 5. Resistance of H1085Y Pol II to α-amanitin and conservation of α-amanitin substrate-selective inhibition.
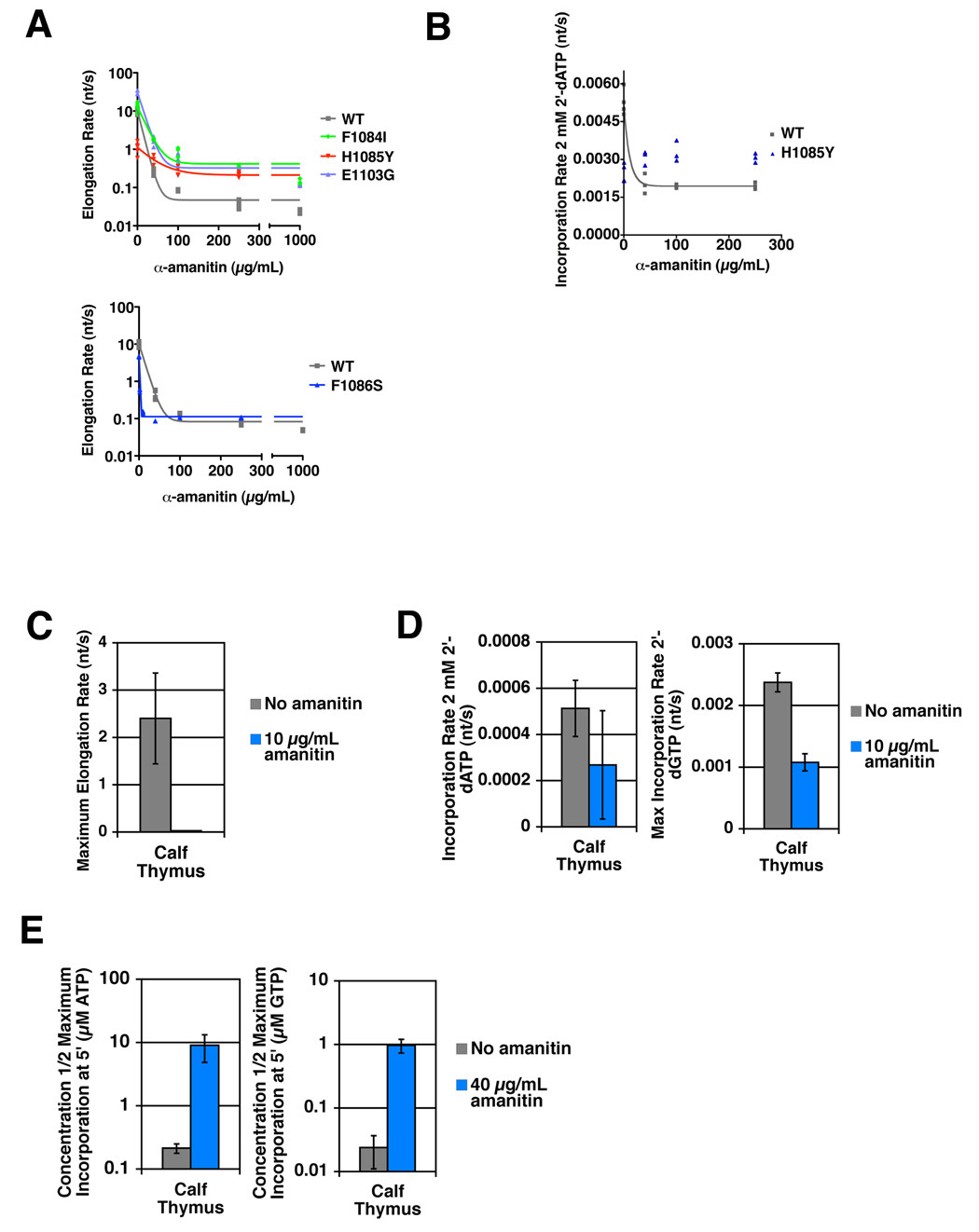
A. Elongation rates at 500 µM NTPs for WT Pol II and TL mutant enzymes in the presence of increasing amounts of α-amanitin determined as in Figure 2A. Top panel plots elongation rates determined from production of run off product (61 nt) as in Figure 2A. F1086S Pol II was too slow in the presence of higher concentrations of α-amanitin to accurately quantify accumulation of 61 nt run off product, therefore elongation rates in the presence of α-amanitin were determined from accumulation of 23 nt and higher products for F1086S and WT Pol II for comparison (bottom panel). Note log scale on y-axes. B. α-amanitin inhibition of 2’-dATP usage by wild type Pol II is different than inhibition of NTP usage, while α-amanitin has no effect on H1085Y. Incorporation rate at 2 mM 2’-dATP in the presence of increasing amounts of α-amanitin determined as in Figure 2B. Values from three experiments are plotted. C. α-amanitin strongly inhibits elongation by Calf Thymus Pol II. Elongation assay and rate calculation in the absence and presence of 10 µg/mL α-amanitin exactly as in Figure 2A. For untreated samples, elongation over 48 nt was used to determine average elongation rate. For treated samples, elongation over 3 nt was used to determine average elongation rate. Inferred maximum elongation rates are shown with error bars representing the 95% confidence interval. D. α-amanitin weakly inhibits 2’- dNTP usage by Calf Thymus Pol II. Assay performed and rates determined exactly as in Figure 2B with α-amanitin treatment as in Figure 3B, except α-amanitin was at 10 µg/mL. For 2’-dGTP experiments, inferred maximal incorporation rate is shown with error bars representing the 95% confidence interval. For 2’-dATP experiments, incorporation rate for 2 mM 2’-dATP is shown as the mean of several experiments +/− standard deviation of the mean. E. α-amanitin inhibits single nucleotide addition by Calf Thymus Pol II. Experiment performed exactly as in Figure 3D. Values shown are the concentration of substrate that gives half-maximal incorporation over a 5-minute incubation. Lower values represent lower concentration of substrate required for a specified elongation rate, and thus a faster elongating enzyme. Values are the mean of at least three experiments +/− standard deviation of the mean. Note that y-axes are logarithmic scale.
Rpb1 TL Mutant H1085Y is Resistant to α-amanitin Inhibition of TFIIS-Dependent RNA Cleavage
α-amanitin also blocks non-synthesis related functions of Pol II such as TFIIS-assisted RNA cleavage (Izban and Luse, 1992). To determine if H1085Y α-amanitin resistance extends to relieving α-amanitin inhibition of TFIIS cleavage, we assayed the ability of α-amanitin to block TFIIS function in two ways. First, we extended an RNA by transcription, stalling elongation complexes (ECs) at a downstream position by omission of UTP. These stalled ECs are prone to backtracking and were immobilized on beads via an affinity tag on Pol II, incubated with an excess of TFIIS and the shortening of RNA was analyzed. As has been previously shown, α-amanitin blocks TFIIS cleavage of stalled, wild type Pol II ECs. α-amanitin, however, fails to strongly inhibit TFIIS cleavage of H1085Y stalled ECs (Figure 6A). To determine whether the α-amanitin effect on TFIIS relates to a block in RNA cleavage as opposed to a block in Pol II backtracking (stalled ECs must backtrack to be cleaved), we also assembled ECs containing a 13-mer RNA complementary to the non-template DNA in its 5’ sequence, but mismatched at the last two positions of the 3’ end (Figure 6B). Such complexes should mimic the backtracked state, and be efficient substrates for TFIIS-mediated RNA cleavage. The mismatched scaffold supports efficient cleavage by TFIIS of WT elongation complexes, which is inhibited by α-amanitin. In contrast, α-amanitin did not inhibit cleavage of H1085Y, indicating H1085Y is resistant to α-amanitin for more than just transcription inhibition.
Figure 6. Resistance of H1085Y Pol II to α-amanitin inhibition of TFIIS-mediated RNA cleavage.
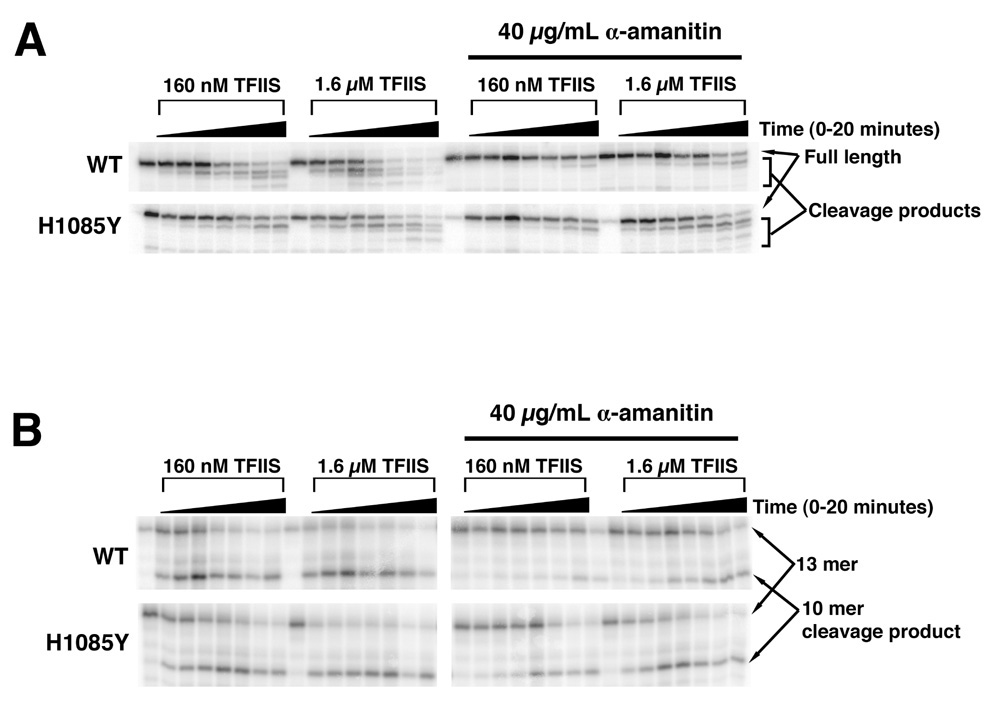
A. Purified WT and H1085Y stalled elongation complexes were treated with different amounts of TFIIS in the absence or presence of 40 µg/mL α-amanitin over a 20 minute time course followed by separation of RNA products by denaturing polyacrylamide gel electrophoresis. TFIIS-mediated RNA cleavage results in a shortening of transcript from the 3’-end, resulting in faster migrating RNA products. B. WT and H1085Y Pol II “backtracked” complexes were formed by incubation of polymerases with a nucleic acid scaffold containing a 13-mer RNA with two mismatches to the template at the 3’-end. These complexes were treated with TFIIS over a 20-minute time course in the absence or presence of 40 µg/mL α-amanitin. TFIIS-mediated RNA cleavage results in a shortening of transcript from the 3’-end.
α-amanitin Inhibition of Elongation by Direct Capture of Rpb1 TL
The results above are consistent with a model where α-amanitin inhibits Pol II transcription by compromising TL function. The specific resistance of H1085Y enzymes to α-amanitin suggests a possible direct functional interaction between α-amanitin and His1085. To examine this, we re-refined the Pol II-α-amanitin crystal structure (Bushnell et al., 2002) with specific attention to previously noted but not modeled electron density in the vicinity of α-amanitin and the partially built TL. The improved electron density maps allowed building of an ordered TL, directly interacting with the α-amanitin molecule (Figure 7). The position of the inhibitor is adjacent to the Bridge Helix and the TL is predominantly in a stable conformation away from the addition site (Figure 7A). The α-amanitin cyclic peptide backbone forms a pocket that allows specific complementary hydrogen bonds to the Rpb1 His1085 side chain imidazole ring, creating a point of restraint for the TL (Figure 7B). The specificity for imidazole by the α-amanitin pocket is achieved primarily through the interaction of amide NH of Gly7 to Rpb1 His1085 N(D1) and the carbonyl of Asn1 to Rpb1 His1085 NH (E2) (Figure 7C).
Figure 7. Direct Interaction Between Rpb1 His1085 and α-amanitin and TL Capture May Underlie α-amanitin Inhibition of Transcription.
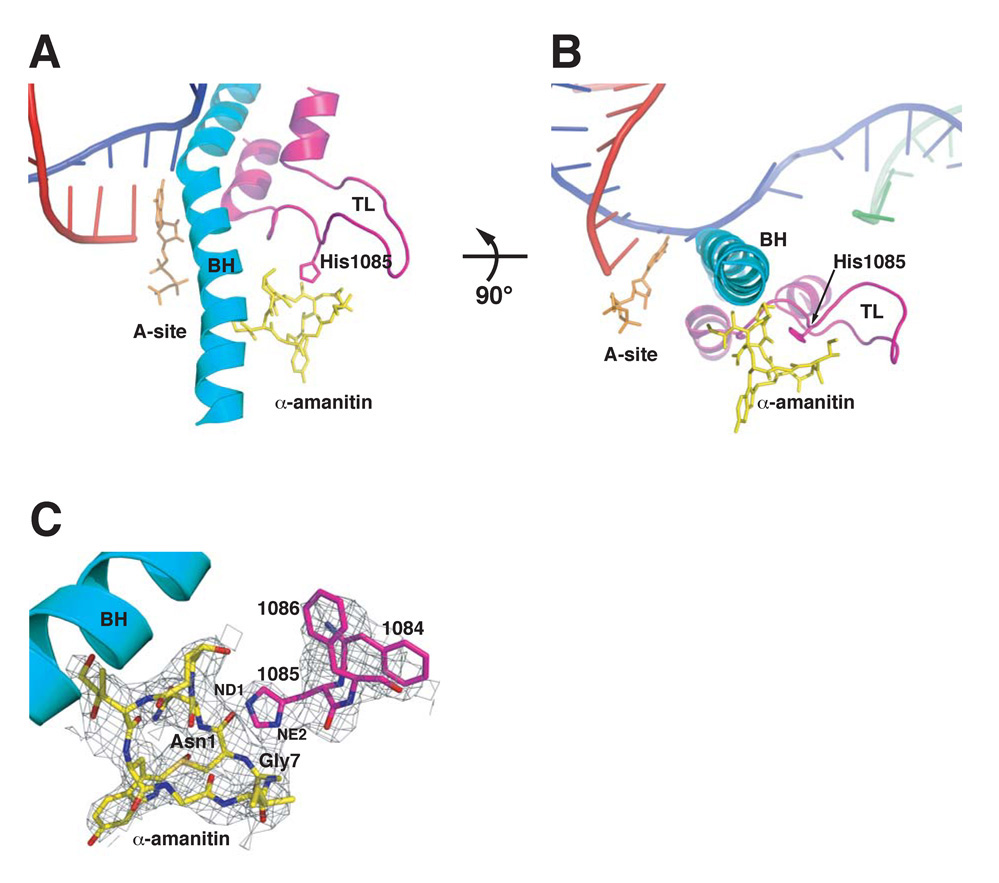
A. Overall view of .-amanitin and the new TL conformation and position in relation to the Bridge helix (BH). A superpositioned EC structure (PDB 2E2H) showing DNA (magenta), RNA (red), non-template DNA (green) and nucleotide GTP (orange) highlights the position of the inhibitor and TL in relation to EC components. B. A 90° rotation shows the α-amanitin position in relation to the Bridge helix (BH) and its capture of the TL Rpb1 His1085. C. TL residues Rpb1 1084–1086 and the entire α-amanitin modeled into electron density (dark grey mesh) from an initial unbiased 2Fo-Fc electron density map contoured at 0.6 σ
DISCUSSION
The TL of multisubunit RNA polymerases is emerging as a central element for the transcription mechanism and its regulation. The data presented here indicate that a function of the Pol II TL is promoting fast and accurate transcription by augmenting the selection of correct, templated NTP substrates over 2’-dNTP or mismatched NTP substrates. We present evidence that the natural product α-amanitin inhibits Pol II by a direct interaction with the TL, interfering with TL function and converting Pol II from a fast and accurate enzyme to a slow and inaccurate one.
Two Modes of Pol II Synthesis: Trigger Loop Dependent and Independent
Multi-subunit RNA polymerase-NTP interactions are sufficient to direct slow synthesis of RNA in the absence of a TL, as deletion of the RNAP TL does not fully abrogate transcription and likely not selection (Temiakov et al., 2005; Toulokhonov et al., 2007). This baseline TL-independent activity represents one mode of Pol II synthesis while activity promoted by TL function represents another, critical mechanism for NTP selection. Cooperating with NTP interactions via base-pairing and ribose contacts outside of the TL, the TL is proposed to fold or swing into position to interact with active site bound NTPs (Vassylyev et al., 2007b; Wang et al., 2006). In characterizing a number of TL mutants, we suggest that a major function of the TL is to promote catalysis of correctly positioned, matched NTP substrates, therefore acting as a critical component in the kinetic selection of matched NTPs over matched 2’-dNTPs that may also base-pair with the DNA template. A matched NTP is proposed to bind the A-site first, followed by subsequent stabilization or conformation change due to TL interaction, which would then promote phosphodiester bond formation. This two-step model is attractive for understanding how RNA polymerases select NTP substrates over 2’-dNTP substrates. TL movement and function may be similar in principle to a DNA polymerase mechanism, where a large conformational change of the DNA polymerase active site (movement of the “fingers” domain) closes the active site on appropriately matched substrates allowing catalysis. Mismatched NTPs or matched 2’-dNTPs may not be positioned appropriately in the active site, being sterically unfavorable for TL interaction, or have a long enough dwell time for productive TL interaction. In either case, it would be expected that alteration of TL function would have substrate-selective effects on polymerase activity as observed. Our work suggests that Pol II may specifically recognize some inherent distinction between matched NTPs and matched 2’-dNTPs base-paired with the template and does so via the TL.
Consistent with this model, we have identified TL mutants with decreased selectivity for NTPs versus 2’-dNTPs or mismatched NTPs relating to reduced activity for matched NTPs (H1085Y), TL mutants with reduced activity for all substrates (F1086S), increased activity for all substrates (E1103G), or decreased selectivity for NTPs relating to increased activity for inappropriate substrates (F1084I). Our model posits that the TL selects matched NTPs over matched 2’-dNTPs or mismatched NTPs by promoting catalysis of correctly positioned NTPs. We propose that the absolutely conserved TL histidine (Rpb1 His1085 in S. cerevisiae) contributes to this selection in the Pol II mechanism. Substitutions distal to the NTP interacting part of the TL such as E1103G may affect TL movement and alter substrate recognition. If the TL has an increased probability of being near the A site, this could result in increased activity with all substrates. The two mutants (E1103G, F1084I) that show loss of selectivity via higher relative increase in activity for inappropriate substrates both exhibit an Spt− phenotype in vivo. Spt− phenotypes result from altered promoter specificity and start-site selection. It is possible that altered affinity for initiating NTP alters start-site selection in these mutants, allowing normally disfavored start sites to be utilized, resulting in the observed suppression.
Mechanism of α-amanitin Inhibition
It has been proposed previously that α-amanitin inhibits Pol II transcription by blockage of a Pol II conformational change. Our results indicate a direct interaction between α-amanitin and TL residue His1085 that is required for α-amanitin inhibition of transcription. H1085Y Pol II is highly resistant to α-amanitin, and residual inhibition by α-amanitin of H1085Y has different properties from inhibition of WT Pol II. This residual inhibition may represent a basal level of transcription inhibition due to α-amanitin binding to Pol II independent of TL interaction, whereas full inhibition requires prevention of TL-substrate interactions due to α-amanitin capture of the TL.
The α-amanitin-Pol II crystal structure illustrates such a specific capture mechanism based on α-amanitin interaction with His1085, restraining the TL from substrate interaction. The α-amanitin resistance of H1085Y would be the result of incompatible H-bonding properties of tyrosine. The specificity of high α-amanitin resistance in H1085Y rationalized as this residue alone makes contacts with α-amanitin. Additionally, partial or complete capture of His1085 by α-amanitin is presumably still allowed in mutants with substitutions adjacent to His1085.
The proposed α-amanitin-TL complex represents a new conformation of the TL, likely distinct from either paused or TL-deleted states. α-amanitin capture of the TL through His1085 should abrogate TL function in nucleotide addition by blocking TL interaction with NTP substrates in the active site. The TL in the paused conformation is also blocked from nucleotide addition, but because it is constrained through an interaction with the 3’-end of the nascent RNA now presumed to occupy the A site (Toulokhonov et al., 2007). In the proposed α-amanitin-TL state, pausing is exacerbated, not attenuated as in RNAP deleted for the TL (Toulokhonov et al., 2007). Additionally, a backtracked EC can still accommodate α-amanitin binding, suggesting an α-amanitin-TL complex could also allow backtracking, another distinction from the paused state where backtracking is likely disfavored relative to that in ECs stalled at non-pause positions. Furthermore, forward translocation of α-amanitin-TL complexes appears to be allowed.
Interpretation of kinetic experiments have led to the proposal that α-amanitin blocks translocation and that a fraction of elongation complexes are resistant to α-amanitin because they reside in the post-translocated state (Gong et al., 2004). Our experiments cannot distinguish between fractions of pre- or post-translocated ECs, however α-amanitin has a differential ability to inhibit addition of various substrates to identical ECs. A model wherein α-amanitin solely blocks translocation, unless the translocation event is substrate-specific, cannot explain such substrate-specific effects. NTP-stimulated translocation has been proposed from kinetic and single molecule analyses (Nedialkov et al., 2003). Additionally, experiments with RNAP suggest that this reaction behavior could require the TL (Toulokhonov et al., 2007). It is conceivable, therefore, that some function of α-amanitin is to block NTP substrate-specific translocation. We do not observe an obvious fraction of α-amanitin resistant complexes, and thus we favor a model where α-amanitin constrains TL behavior that may include roles in both substrate-assisted translocation and catalysis.
Regulation of Pol II Via the Trigger Loop
The TL of multisubunit RNA polymerases such as RNAP and Pol II is emerging as a key element for both transcription and its regulation. The TL is targeted by numerous small molecule inhibitors of transcription as well as being important for regulation of RNAP by elongation factors. We expect that the Pol II trigger loop will also serve as the target for transcriptional regulators as alteration of its function can lead to both increased and decreased elongation.
EXPERIMENTAL PROCEDURES
Enzymes, Reagents, Strains and Media
Enzymes for DNA manipulation were from New England Biolabs or Invitrogen. Radioactive nucleotides were from Perkin Elmer. DNA oligonucleotides were synthesized by IDT, while RNA oligos were from Dharmacon. Yeast media and manipulations were by standard procedures (Rose et al., 1990). All yeast strains are isogenic to a GAL2+ derivative of S288C (Winston et al., 1995).
Pol II and TFIIS purification
Pol II was purified via a modified tandem-affinity tag (TAP) procedure derived from (Puig et al., 2001; Wang et al., 2006). A cleavage competent N-terminal truncation of TFIIS (TFIIS.N2-146) was purified via a N-terminal 6-His-tag via standard procedures (Qiagen). Full details of these purifications are found in Supplemental Methods.
EC formation
EC formation for in vitro transcription assays were performed essentially as in (Wang et al., 2006; Westover et al., 2004). All transcription assays for all enzymes were performed at room temperature. Full details of transcription assays are available in Supplemental Methods.
Data analysis and elongation rate determination
All in vitro transcription reactions we analyzed by separation of RNA products by denaturing polyacrylamide gel electrophoresis. For run-off transcription assays with longer oligonucleotide templates, products were separated on 13.5% acrylamide:bisacrylamide (19:1) gels also containing 7 M Urea and 1X TBE (90 mM Tris base, 90 mM boric acid, 2 mM EDTA). Single-nucleotide addition RNA products and mismatched, backtrack simulating RNAs were separated on 18% acrylamide:bisacrylamide (19:1) gels also containing 7 M Urea and 1X TBE. Undried gels were frozen and exposed to PhosphorImager screens (GE Healthcare) for visualization of 32P-radiolabeled RNAs. Transcription assays were quantified using ImageQuant software and Microsoft Excel. Values for accumulation of particular RNA products were then plotted versus time for rate determination in GraphPad Prism for curve-fitting by non-linear regression. Non-linear regression was also used for curve-fitting of elongation rate data versus substrate concentration, extrapolation of maximal rates and calculation of 95% confidence intervals.
α-amanitin Treatment
α-amanitin (Roche) was stored as a 4 mg/mL stock in water at −20°C in the dark. For experiments with NTP substrates, α-amanitin was added to reactions with NTPs at 2X final concentration in an equal volume to that of Pol II complexes giving a final concentration of 1X α-amanitin and 1X NTPs. For experiments with 2’-dNTPs and NTP misincorporation, where α-amanitin has much weaker effects, we initially tried preincubation with 2X α-amanitin for 3 minutes prior to substrate addition as well as simultaneous addition of α-amanitin and substrate. We did not observe a qualitative difference in inhibition. All values reported for 2’-dNTP and NTP misincorporation were derived from experiments were Pol II complexes were first preincubated with 2X α-amanitin prior to addition of an equal volume of 2X substrate giving 1X substrate and 1X α-amanitin over a time course of incubation at room temperature.
TFIIS Cleavage Assays
For experiments involving artificially “backtracked” elongation complexes, a DNA template similar to those used in single nucleotide addition experiments (Wang et al., 2006), specifying addition of A at position 10 and U at position 11, was annealed with γ-32P-ATP -endlabeled 13-mer RNA complementary to the DNA template except for the two 3’ nucleotides. A non-template oligo complementary to the downstream end of the DNA template was also included in the annealing reaction, exactly as in (Wang et al., 2006). Complexes were captured by incubation with 10 µL of 1:1 slurry of Calmodulin Affinity Resin (Stratagene) pre-equilibrated in Transcription Buffer (TB, 20 mM Tris- HCl pH 8.0, 40 mM KCl, 5 mM MgCl2, 2 mM DTT) plus 0.05% NP-40 and 200 µM CaCl2 with mild agitation for 5 minutes at room temperature. Immobilized Pol II “backtracked” complexes were then washed 2–3 times in 1X TB plus 0.05% NP-40 and 200 µM CaCl2 to remove unbound hybrid and released RNA. “Backtracked” complexes were then aliquoted for treatment with TFIISΔ2-146 in the presence or absence of α-amanitin. For experiments involving natural backtracking, stalled elongation complexes were formed on CKO222/CKO223/RNA9 scaffolds as for elongation assays with NTP substrates (Supplemental Methods). 5 µL of CKO223/γ-32P-ATP-endlabeled RNA9 hybrid were incubated with 5 µg Pol II and 10 µL 1:1 slurry of Calmodulin Affinity Resin, pre-equilibrated in TB buffer plus 0.05% NP-40 and 200 µM CaCl2, with mild agitation for 5 minutes at room temperature. 300–600 pmol non-template oligo (CKO222) were added with mild agitation for 5 minutes at room temperature. RNA was extended 43 nt by incubation of complexes with 25 µM ATP, CTP and GTP in TB for 20 minutes at room temperature, creating Pol II complexes stalled by omission of UTP. These complexes were then washed 2–3 times in TB plus 0.05% NP-40 and 200 µM CaCl2 to remove unbound hybrid and released RNA transcripts. Stalled complexes were then aliquoted for treatment with TFIISΔ2-146 in the presence or absence of α-amanitin.
Refinement of Pol II-α-amanitin Structure
The 2.8 Å structure of Pol II in complex with α-amanitin (Bushnell et al., 2002), PDB 1K83, was re-refined using TLS refinement, omitting the α-amanitin and parts of Rpb1 comprising the proximal and distal TL. The new refinement resulted in connectivity improvement of the electron density map, allowing tracing of a main conformation of the entire TL. All tracing was performed in an initial unbiased 2Fo-Fc map contoured at 0.5– 0.6 sigma, which was followed by iterative model building using the program Coot (Emsley and Cowtan, 2004). The new coordinates have been deposited as PDB 3CQZ. All refinements employed TLS groups (4 TLS groups, comprising four parts of the Pol II, see PDB 3CQZ header for exact residue ranges) using Refmac5 (Winn et al., 2001). A Phenix (Adams et al., 2002) simulated annealing (SA) Fo-Fc omit map confirms the placement of the Rpb1 His1085. Besides the main conformation of the captured TL, electron density for one or several other, less ordered, states of the TL could be observed. The final model has an R/Rfree 22/29 with rmsd bond 0.018A and rmsd angles 2.0.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Avi Gnatt (U. Maryland at Baltimore) for the gift of Calf Thymus Pol II. This work was supported by NIH grants GM49985 and GM36659 to R.D.K. C.D.K. was supported by a Helen Hay Whitney Postdoctoral Fellowship and by a Ruth L. Kirschstein NRSA Postdoctoral Fellowship. K-M.L. was supported by a postdoctoral fellowship from The Wenner-Gren Foundations. We thank Jerry Kaplan, Dave Bushnell and Grant Hartzog for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Archambault J, Jansma DB, Kawasoe JH, Arndt KT, Greenblatt J, Friesen JD. Stimulation of transcription by mutations affecting conserved regions of RNA polymerase II. J Bacteriol. 1998;180:2590–2598. doi: 10.1128/jb.180.10.2590-2598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J, Lacroute F, Ruet A, Friesen JD. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci U S A. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafin DR, Guo H, Price DH. Action of alpha-amanitin during pyrophosphorolysis and elongation by RNA polymerase II. J Biol Chem. 1995;270:19114–19119. doi: 10.1074/jbc.270.32.19114. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Gong XQ, Nedialkov YA, Burton ZF. Alpha-amanitin blocks translocation by human RNA polymerase II. J Biol Chem. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- Hekmatpanah DS, Young RA. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′----5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Jokerst RS, Weeks JR, Zehring WA, Greenleaf AL. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002a;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002b;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, Yan H, Burton ZF. NTP-driven translocation by human RNA polymerase II. J Biol Chem. 2003;278:18303–18312. doi: 10.1074/jbc.M301103200. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rudd MD, Luse DS. Amanitin greatly reduces the rate of transcription by RNA polymerase II ternary complexes but fails to inhibit some transcript cleavage modes. J Biol Chem. 1996;271:21549–21558. doi: 10.1074/jbc.271.35.21549. [DOI] [PubMed] [Google Scholar]
- Simchen G, Winston F, Styles CA, Fink GR. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc Natl Acad Sci U S A. 1984;81:2431–2434. doi: 10.1073/pnas.81.8.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MJ. Mycophenolic acid and its mechanism of action in cancer and psoriasis. Jpn J Antibiot. 1977;30 Suppl:85–92. [PubMed] [Google Scholar]
- Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, Igarashi N, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD, et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007a;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007b;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Wang D, Bushnell D, Westover K, Kaplan C, Kornberg R. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta' subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Wieland T, Faulstich H. Fifty years of amanitin. Experientia. 1991;47:1186–1193. doi: 10.1007/BF01918382. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Winston F, Sudarsanam P. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harb Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


