Abstract
Magnetic particles offer high technological potential since they can be conveniently collected with an external magnetic field. Magnetotactic bacteria synthesize bacterial magnetic particles (BacMPs) with well-controlled size and morphology. BacMPs are individually covered with thin organic membrane, which confers high and even dispersion in aqueous solutions compared with artificial magnetites, making them ideal biotechnological materials. Recent molecular studies including genome sequence, mutagenesis, gene expression and proteome analyses indicated a number of genes and proteins which play important roles for BacMP biomineralization. Some of the genes and proteins identified from these studies have allowed us to express functional proteins efficiently onto BacMPs, through genetic engineering, permitting the preservation of the protein activity, leading to a simple preparation of functional protein–magnetic particle complexes. They were applicable to high-sensitivity immunoassay, drug screening and cell separation. Furthermore, fully automated single nucleotide polymorphism discrimination and DNA recovery systems have been developed to use these functionalized BacMPs. The nano-sized fine magnetic particles offer vast potential in new nano-techniques.
Keywords: magnetotactic bacteria, bacterial magnetic particles, surface modification of magnetite, magnetic separation, protein display, fully automated system
1. Overview
Living organisms produce complex composite materials that exhibit an amazing hierarchical assembly of bioorganic molecules and inorganic ions ranging from the nano- to macroscopic scale (Mann & Ozin 1996; Addadi & Weiner 1997; Pouget et al. 2007). These highly organized structures often exhibit excellent physical and/or chemical properties that outperform artificial materials, and, in comparison with the usual synthetic methods, the intricate architectures of these biominerals can be formed under conditions that are incredibly mild. As a result, over the years, the biological and chemical principles of biomineralization have been studied in order to exploit these key principles for the development of advanced nanomaterials.
Magnetotactic bacteria synthesize intracellular magnetic particles comprising iron oxide, iron sulphides or both (Bazylinski et al. 1994, 1995). In order to distinguish these particles from artificially synthesized magnetic particles (AMPs), they are referred to as bacterial magnetic particles (BacMPs; Matsunaga et al. 2007c) or magnetosomes (Balkwill et al. 1980). BacMPs, which are aligned in chains within the bacterium, are postulated to function as biological compass needles that enable the bacterium to migrate along oxygen gradients in aquatic environments, under the influence of the Earth's geomagnetic field (Blakemore 1975). BacMPs can easily disperse in aqueous solutions because they are enveloped by an organic membrane that mainly consists of phospholipids and proteins (Gorby et al. 1988; Nakamura et al. 1991; Grünberg et al. 2004), and an individual BacMP contains a single magnetic domain or magnetite that yields superior magnetic property (Matsunaga & Kamiya 1987; Bazylinski et al. 1994). Based on the beneficial properties of BacMPs as presented above, functional BacMPs have been developed and investigated to use them for various biotechnological applications. In this review, we describe and introduce the current knowledge of the genes and proteins involved in BacMP biomineralization and the development of functional magnetic particles and their biotechnological applications based on their unique properties.
2. Diversity of magnetotactic bacteria
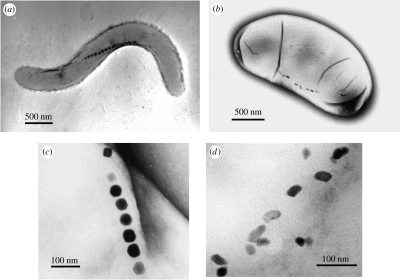
Since the first peer-reviewed report of magnetotactic bacteria in 1975 (Blakemore 1975), various morphological types including cocci, spirilla, vibrios, ovoid bacteria, rod-shaped bacteria and multicellular bacteria (Thornhill et al. 1994; Spring & Schleifer 1995) possessing unique characteristics have been identified and observed to inhabit various aquatic environments. Magnetotactic cocci, for example, have shown high diversity and distribution and have been frequently identified at the surface of aquatic sediments (Thornhill et al. 1995). The discovery of this bacterial type, including the only cultured magnetotactic coccus strain MC-1, suggested that they are microaerophilic (DeLong et al. 1993; Meldrum et al. 1993b). In the case of the vibrio bacterium, three facultative anaerobic marine vibrios—strains MV-1, MV-2 and MV-4—have been isolated from estuarine salt marshes. These bacteria have been classified as members of α-Proteobacteria, possibly belonging to the Rhodospirillaceae (Meldrum et al. 1993a) family, observed to synthesize BacMPs of a truncated hexa-octahedron shape and grow chemoorganoheterotrophically as well as chemolithoautotrophically (Bazylinski & Frankel 2004). The members of the family Magnetospirillaceae, on the other hand, can be found in fresh water sediments. With the use of growth mediums and magnetic isolation techniques established, a considerable number of the magnetotactic bacteria isolated to date have been found to be members of this family. The Magnetospirillum magnetotacticum strain MS-1 was the first member of the family to be isolated (Blakemore et al. 1979), while the Magnetospirillum gryphiswaldense strain MSR-1 is also well studied with regard to both its physiological and genetic characteristics (Schleifer et al. 1991; Schüler & Baeuerlein 1996, 1998). We have successfully isolated two facultative anaerobic magnetotactic spirilla, Magnetospirillum magneticum AMB-1 (figure 1a; ATCC 700264; Matsunaga et al. 1991) and MGT-1 (FERM P-16617; Matsunaga et al. 1990), and an obligate anaerobe, Desulfovibrio magneticus RS-1 (ATCC 700980; figure 1b; Sakaguchi et al. 1993, 2002). M. magneticum strains AMB-1 and MGT-1, both classified as α-Proteobacteria, are nitrate-reducing bacteria that synthesize cubo-octahedral-shaped BacMPs (figure 1c). They are capable of growing under both microaerobic and aerobic conditions in liquid or solid media (Matsunaga et al. 1991), which makes them ideal candidates for genetic manipulation (Matsunaga et al. 1992). The facultative anaerobic characteristic of this bacterium has enabled us to generate non-magnetic mutant strains of strain AMB-1 by transposon mutagenesis, providing valuable information on the genes involved in BacMP biomineralization (Nakamura et al. 1995b; Wahyudi et al. 2001, 2003). Techniques such as electroporation and conjugation have also been specifically designed for the AMB-1 and MGT-1 strains, in addition to protein display on BacMPs based on the gene fusion. Further details are provided in a subsequent section (see §4.1).
Figure 1.
TEM images of magnetotactic bacteria. (a) Magnetospirillum magneticum strain AMB-1 and (b) Desulfovibrio magneticus strain RS-1. BacMPs of (c) the AMB-1 strain and (d) the RS-1 strain.
Desulfovibrio magneticus RS-1 (figure 1b)—a dissimilatory sulphate-reducing bacterium belonging to the δ-Proteobacteria class—produces unique irregular bullet-shaped BacMPs (figure 1d; Sakaguchi et al. 1993, 2002; Kawaguchi et al. 1995). The production of BacMPs within the RS-1 strain is substrate dependent, whereby RS-1 cells produced an average of six BacMPs per individual cell when fumarate was used as an electron acceptor, while almost no BacMPs were produced when sulphate was used. This bacterium also produces extracellular magnetic iron sulphide (Sakaguchi et al. 1993) and hematite (Posfai et al. 2006) on its surface. Magnetotaxis in the RS-1 strain is distinctly different from that in the other strains of magnetotactic bacterial cells that swim or orient themselves along a single direction on applying a magnetic field. Although RS-1 cells respond to the direction of the magnetic field, their orientation or migration is not identical. The cells swim or migrate in random orientations when the direction of the magnetic field is changed. It is believed that the observed magnetic responses are related to the low number and disordered orientation of the BacMPs within the cell (figure 1d). Owing to their weak magnetic responses, the RS-1 strain was isolated based on a different strategy to that used for isolating other magnetotactic bacteria (Sakaguchi et al. 1996). The isolation strategy implemented includes incubation of natural sediments, bacterial enrichment in the medium and colony formation. This strategy allows isolation of non-motile and non-responsive or weakly responding magnetotactic bacteria, which cannot be isolated by magnetic force. Furthermore, systematic characterization of the RS-1 strain has also been examined (Sakaguchi et al. 2002), whereby phylogenetic analysis based on 16S rDNA gene sequences revealed that RS-1 is a member of the genus Desulfovibrio (Kawaguchi et al. 1995; figure 2). The RS-1 strain contains desulphoviridin, c-type cytochromes and, unlike other Desulfovibrio spp., possesses menaquinone MK-7(H2) instead of MK-6 or MK-6(H2). The cells use lactate, pyruvate, malate, oxaloacetate and glycerol as carbon sources, and sulphate, thiosulphate and fumarate as electron acceptors. As the cells grow, they also ferment pyruvate. The cellular fatty acid profile of the RS-1 strain indicated the presence of branched-chain saturated fatty acids as a major component. These physiological and biochemical characteristics are consistently found in the members of Desulfovibrio.
Figure 2.
16S rDNA-based phylogenetic relationships of magnetotactic bacteria. Magnetotactic bacteria and pure culture bacterial strains are indicated in bold characters and underlined, respectively.
In addition to the identification of currently known magnetotactic bacteria, 16S rDNA analyses of uncultured magnetotactic bacteria have been reported. Multicellular magnetotactic prokaryotes were shown to produce both iron sulphide (Farina et al. 1990; Mann et al. 1990) and iron oxide BacMPs (Lins & Farina 1999). Several strains of this organism have been identified as the members of δ-Proteobacteria (DeLong et al. 1993; Keim et al. 2004; Simmons et al. 2004). A barbell-shaped magnetotactic bacterium was recently reported and also classified into δ-Proteobacteria (Simmons et al. 2006). The presence of magnetotactic bacteria in another phylum was also reported. A large rod-shaped bacterium, tentatively termed Magnetobacterium bavaricum, was classified into the Nitrospira phylum (Spring et al. 1993). The cell contained up to a 1000 bullet-shaped BacMPs. Recently, a similar magnetotactic rod termed MHB-1 was identified and revealed to have 91% sequence similarity with M. bavaricum, based on 16S rDNA analysis (Flies et al. 2005).
3. Biomineralization of bacterial magnetic particles (magnetosomes)
The molecular mechanism of BacMP biomineralization is hypothesized to be a multistep process. Based on recent studies on genome, proteome and transcriptome analyses, proteins located on the BacMP membrane were elucidated to represent the key components of its biomineralization. An outline of the molecular mechanism has been illustrated. Justification and clarification of magnetite biomineralization pathways also contribute to the development of biotechnological applications for a broad range of research disciplines. Recent efforts directed towards the molecular analyses of M. magneticum AMB-1 have been summarized and discussed with respect to the role that genes and proteins may play in BacMP biomineralization.
3.1 Genome analyses of magnetotactic bacteria
The first complete genome sequence of magnetotactic bacteria was obtained for M. magneticum AMB-1 (Matsunaga et al. 2005); it comprised a single circular 4 967 148 bp chromosome and 4559 predicted open reading frames (ORFs). Within the genome sequence, a remarkable number of sensor and response domains, numerous vestiges of past exogenous gene transfers, such as insertion sequence (IS) elements, integrases and large regions containing phage-coding genes were identified. The IS-concentrated regions had lower GC contents than the average GC content of the entire genome, which suggests gene transfer from other bacteria or phages. During the genome analysis, we obtained a spontaneous non-magnetic mutant that lacks a 98 kb genomic region, which exhibited the characteristics of a genomic island (magnetosome island): it had a low GC content; was located between two 1.1 kb repetitive sequences; and contained an integrase in the flanking region of the first repetitive sequence. The spontaneous deletion of this 98 kb genomic region before and after excision from the chromosome was detected by PCR amplification (Fukuda et al. 2006). A spontaneous non-magnetic mutant that lacks the magnetosome island was also obtained with the M. gryphiswaldense MSR-1 strain (Schübbe et al. 2003). Various phenotypes showing altered numbers, sizes and alignment of BacMPs were isolated and analysed (Ullrich et al. 2005). The mutants revealed several deletions in different regions, suggesting the occurrence of frequent transposition events in the cell. Considering the typical structure of a magnetosome island and the dynamics after excision from the chromosome, these results suggested that the island was derived from lateral gene transfer. An ancestral magnetotactic bacterium came into contact with another bacterium harbouring the magnetosome island and the resulting generation eventually diverged to form the AMB-1, MS-1, MC-1 and MSR-1 strains.
Recent efforts directed towards genome sequencing of magnetotactic bacteria have provided genome information of several other strains. The draft sequence for the M. magnetotacticum strain MS-1 and the complete sequence for the magnetotactic coccus strain MC-1 were provided by the Joint Genome Institute (JGI; http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html), while the draft genome sequence of the M. gryphiswaldense strain MSR-1 was analysed in comparison with other magnetotactic bacterial genome sequences (Richter et al. 2007). The comparative study revealed that approximately 891 genes were shared by all four magnetotactic bacteria. In addition to a set of approximately 152 genus-specific genes shared by the three Magnetospirillum strains, 28 group-specific genes, which occur in all four magnetotactic bacteria but exhibit no similarity or only remote similarity to the genes from non-magnetotactic organisms, were identified. This study revealed the magnetotactic bacteria-specific set of genes that are most likely to be involved in BacMP biomineralization (Richter et al. 2007).
3.2 Gene expression analysis of iron-inducible genes
Magnetotactic bacteria take up approximately 100 times more iron than non-magnetic bacteria to synthesize intracellular BacMPs (Blakemore et al. 1979; Matsunaga et al. 1991). To identify global gene expression profiles, especially those of iron uptake genes under different iron concentrations, transcriptome analysis through microarray imprinted with DNA from the whole genome sequence of AMB-1 was investigated (Suzuki et al. 2006). Three ferrous and two ferric iron uptake systems encoded by specified genes were identified from the genome sequence, and two distinct patterns were observed in relation to the iron concentration in the growth medium and gene expression. High-affinity ferrous iron transport genes were upregulated in BacMP-forming cells grown under iron-rich conditions (20–300 μM), and ferric iron transport genes were downregulated under these conditions (Suzuki et al. 2006). In high-iron conditions wherein magnetite synthesis occurs, the ferrous iron uptake system is dominant, while ferric iron or siderophore transport genes were expressed after most of the iron ions were taken up by the magnetic cells. This is consistent with our previous report on siderophore production by M. magneticum AMB-1 grown in media containing different initial iron ion concentrations (Calugay et al. 2003). The initial high concentration of iron was rapidly assimilated from the medium within only 4 hours after inoculation, reaching levels comparable with that of iron-deficient cultures, thereby triggering siderophore production. Furthermore, enhancement of BacMP production was observed when ferrous iron was used as the iron source (Yang et al. 2001a). These results suggest that ferrous iron uptake systems mainly serve as iron supply lines for magnetite formation. Our results indicate that despite the unusual high iron requirement of M. magneticum AMB-1, it uses robust but simple iron uptake systems similar to those of other gram-negative bacteria (Andrews et al. 2003). By contrast, Schüler and Baeurlein observed high-velocity transport of ferric iron in M. gryphiswaldense MSR-1 (Schüler & Baeuerlein 1996). They also reported that the uptake of ferrous iron is a slow diffusion-like process. Although the results from the two studies are not consistent, iron ions are eventually accumulated within the cell in the form of ferrous iron and are used for magnetite biomineralization in membrane-invaginated vesicles.
On the other hand, Schüler and co-workers focused on studying the transcriptional regulation of mam and mms genes in the magnetosome island (Schübbe et al. 2006; Würdemann et al. 2006). The transcription levels of several genes, which varied in response to the iron and oxygen concentrations in the MSR-1 strain, were analysed by reverse transcriptase-polymerase chain reaction (RT-PCR) and microarray. Although results from the two methods were not consistent in certain areas, seven genes (mgI458, mgI460, mamG, mamD, mamM, mamA and mamB) were determined to be downregulated under the iron-limiting condition (less than 1 μM iron; Schübbe et al. 2006). Seven other genes (mgI458, mgI460, mamD, mamH, mamK, mamM and mamA) were downregulated under the aerobic (10 kPa O2) condition in both the methods. They stated that the various transcription levels of individual genes from the same transcriptional unit might be explained by differential internal stabilities within a polycistronic mRNA. Interestingly, mam genes including the mamC gene were found to be expressed throughout the growth period of the MSR-1 cells. The MamC protein was also detected in magnetic and non-magnetic cells cultured under iron-limiting and aerobic conditions, by Western blot analysis. In the protein analysis of the AMB-1 strain, Mms13 (identical to MamC) was found in both the BacMP and cytoplasmic membranes (Tanaka et al. 2006). Analysis of the deletion mutant of MamC in the MSR-1 strain suggested that the absence of this protein did not markedly affect BacMP size and the formation of an organic membrane (Scheffel et al. 2008). However, the expression of the protein in both BacMP and cytoplasmic membranes during most of the cell cycles and its function or contribution to the formation of BacMPs is still unknown.
3.3 Protein analyses of the BacMP membrane
Since the discovery of magnetotactic bacteria, BacMP surface proteins have been believed to play key roles in their biosynthesis (Gorby et al. 1988). To elucidate the molecular mechanism of BacMP biomineralization, proteome analyses of BacMP membrane proteins have currently been conducted in M. magneticum AMB-1 (Matsunaga et al. 2000b; Okamura et al. 2000, 2001; Arakaki et al. 2003; Tanaka et al. 2006), M. gryphiswaldense MSR-1 (Grünberg et al. 2001, 2004) and M. magnetotacticum MS-1 (Okuda et al. 1996), from which numerous novel proteins with potentially crucial roles in BacMP biomineralization have been discovered. In the analysis of BacMP membrane proteins from M. gryphiswaldense MSR-1, approximately 30 proteins were identified; a considerable number of these proteins were found to be assigned in gene clusters located within the magnetosome island (Grünberg et al. 2004). A similar examination was attempted with M. magneticum AMB-1, wherein a total of 78 proteins were identified from the surface of its BacMP (figure 3; Tanaka et al. 2006). In order to investigate the origin of the BacMP membrane, proteins acquired from AMB-1 cells were separated into the outer membrane, cytoplasmic membrane, BacMP membrane and cytoplasmic–periplasmic fractions. Although apparent contaminants were also observed in the BacMP membrane protein fraction of AMB-1, some of these, such as cytochromes, dehydrogenases and ATPase subunits, may possibly play a role in BacMP biomineralization. A high degree of similarity was observed between the protein profiles of the BacMP and cytoplasmic membranes of the AMB-1 strain. One surprising observation was the identification of the Mms13 protein—a protein highly associated with the BacMP surface—in the cytoplasmic as well as BacMP membrane fractions. Fatty acid comparative analysis also indicated that both these fractions showed similar profiles. These results suggest that the BacMP membrane could have been derived from the cytoplasmic membrane. Direct evidence of this phenomenon was also reported by Komeili et al. Invagination of the cytoplasmic membrane was clearly observed by electron cryotomography (Komeili et al. 2006).
Figure 3.
The two-dimensional electrophoresis proteomic analysis of the BacMP membrane in the following ranges of pIs: (a) 3–10, (b) 4–7 and (c) 6–11.
Functional analyses of individual proteins have been investigated. Five dominant proteins in the BacMP membrane of M. magneticum AMB-1 were identified by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE; Okamura et al. 2000). One of these is a 24 kDa protein designated as Mms24, which was observed in other Magnetospirillum spp. and corresponds to Mam22 in M. magnetotacticum MS-1 (Okuda et al. 1996) and MamA in M. gryphiswaldense MSR-1 (Grünberg et al. 2001). A mutant strain harbouring the defected gene encoding this protein showed lesser BacMP formation in comparison with its wild-type counterpart, suggesting that the protein may be required for the activation of BacMP vesicles (Komeili et al. 2004). Taoka et al. (2006) recently reported the presence of a novel matrix surrounding a chain of BacMPs. They investigated the precise localization of Mam22 and Mam12 (identical to Mms13 and MamC) and revealed that Mam22 and Mam12 exist in the matrix and the BacMP membrane, respectively. A 16 kDa protein (Mms16), a small GTPase, is speculated to prime the invagination of the cytoplasmic membrane for vesicle formation in M. magneticum AMB-1 (Okamura et al. 2001). MpsA shows homology with acetyl-CoA carboxylase (transferase) and the acyl-CoA-binding motif, which is considered to function as a mediator of cytoplasmic membrane invagination (Matsunaga et al. 2000b). Moreover, from the analysis of the magnetite crystal surface of BacMPs, four novel proteins tightly bound to the magnetite crystal, designated Mms5, Mms6, Mms7 (identical to MamD) and Mms13 (identical to MamC and Mam12), were identified (Arakaki et al. 2003). These proteins show the common amphiphilic features containing hydrophobic N-terminal and hydrophilic C-terminal regions. The N-terminal regions in Mms5, Mms6 and MamD possess a common leucine and glycine repetitive sequence—LGLGLGLGAWGPX(L/I)(L/V)GX(V/A)GXAGA. The genes encoding these proteins are located proximally in the AMB-1 genome (within a 3.2 kbp region). Another gene homologue (ORF1) was identified from the AMB-1 genome, which encoded a protein that shared a similar motif with this protein group. Similar sequences were reported in silk fibroin-like molluscan shell framework protein (Sudo et al. 1997) and collagen-like fish otolith structure protein (Murayama et al. 2002), which are considered to form a β-sheet and produce a self-assembled framework structure for carbonate crystal formation. Although the LG-rich region in BacMP proteins is rather too short to form the intermolecular β-sheet structure, this region may participate in the self-assembly of proteins for magnetite crystal formation. The C-terminal regions in Mms6 contain carboxyl and hydroxyl groups that are found to be iron-binding sites. The magnetite particles synthesized in the presence of Mms6 in vitro showed cuboidal morphology similar to the AMB-1 strain BacMPs with sizes ranging from 20 to 30 nm in diameter (see §4.4). These results suggest that Mms6 binds iron ions to initiate magnetite crystal formation and/or regulates the morphology by producing a self-assembled framework structure. Crucial proteins for BacMP alignment in the cell were also reported. MamK is a homologue of the bacterial actin-like protein, and is revealed to form filaments to establish the chain-like structure of BacMPs through appropriate subcellular targeting (Komeili et al. 2006). MamK nucleates at multiple sites to form long filaments that were confirmed via protein expression in Escherichia coli (Pradel et al. 2006). In vitro polymerization of recombinant MamK was examined, and formation of long filamentous bundles was observed (Taoka et al. 2007). MamJ is an acidic protein associated with the filamentous structure, and it directs the assembly of the BacMP chain (Scheffel et al. 2006). Direct interaction between MamJ and MamK has been revealed, and it is proposed that MamJ plays a role in chain assembly and maintenance (Scheffel & Schuler 2007).
Several genes involved in BacMP biomineralization were also identified through transposon mutagenesis (Nakamura et al. 1995a; Wahyudi et al. 2001, 2003; Calugay et al. 2004; Suzuki et al. 2007). The magA gene encodes a proton-driving H+/Fe (II) antiporter protein (Nakamura et al. 1995a). Intracellular localization of the MagA protein was examined using a MagA–luciferase fusion protein, and it was indicated that this protein is localized in both the cytoplasmic and BacMP membranes (Nakamura et al. 1995b). Interestingly, the MagA topology is inversely oriented between the cytoplasmic and BacMP membranes. The MagA appears to play a role in iron efflux in the former and iron influx in the latter. A tungsten-containing aldehyde ferredoxin oxidoreductase (AOR), which plays a role in aldehyde oxidation, was also isolated (Wahyudi et al. 2003). The AOR was found to be expressed under microaerobic conditions and localized in the cytoplasm of AMB-1. Transmission electron microscopy (TEM) of the mutant revealed that no BacMPs were completely synthesized, but polyhydroxybutyrate (PHB)-like granules were persistently produced. The AOR is considered to contribute to ferric iron reduction during BacMP synthesis under microaerobic respiration (Wahyudi et al. 2003; Calugay et al. 2004). A cytoplasmic ATPase essential for BacMP biomineralization was considered to be involved in iron trafficking within the cell (Suzuki et al. 2007).
The experimental data obtained from these studies with Magnetospirillum spp. allowed us to postulate that BacMP biomineralization comprises three major stages (figure 4). The first stage entails the invagination of the cytoplasmic membrane, and the vesicle formed serves as the precursor of the BacMP membrane. The mechanism of envelope formation, however, still remains unclear. It is most probable that magnetotactic bacteria have similar mechanisms for vesicle formation to most eukaryotes and that a specific GTPase mediates the priming of the invagination. The formed vesicles were assembled into a linear chain along with cytoskeletal filaments. The second stage of BacMP biomineralization entails the accumulation of ferrous ions into the vesicles by the transmembrane iron transporters. External iron is internalized by transport proteins and siderophores. The internal iron is controlled strictly by an oxidation–reduction system. In the final stage, tightly bound BacMP proteins trigger magnetite crystal nucleation and/or regulate morphology. Various proteins associated with the BacMP membrane could play functional roles involved in magnetite generation. These include the accumulation of supersaturating iron concentrations, maintenance of reductive conditions and the oxidation of iron to induce mineralization, or the partial reduction and dehydration of ferrihydrite to magnetite.
Figure 4.
Scheme of the hypothesized mechanism of BacMP biomineralization.
4. Development of functional magnetic particles
Magnetic iron oxide particles, such as magnetite (Fe3O4) or maghemite (γ-Fe2O3), are widely used in the development of medical and diagnostic applications such as magnetic resonance imaging (MRI; Gleich & Weizenecker 2005), cell separation (Miltenyi et al. 1990), drug delivery (Plank et al. 2003) and hyperthermia (Pardoe et al. 2003). To use these particles for the biotechnological applications, it is important to consider surface modification of magnetic particles with functional molecules such as proteins, antibodies, peptides and DNA. We have developed several methods to modify and assemble these functional organic molecules over the BacMP surface using chemical and genetic techniques. Moreover, production strategies for the magnetic particles by in vivo and in vitro methods have been investigated.
4.1 Protein assembly on the surface of magnetic particles
The assembly of functional proteins and peptides on the surface of solid particles facilitates the separation and detection of diverse molecules by the specific recognition function of target molecules. The technique is often used for the identification of therapeutics, detection of environmental pollutants, affinity purification of proteins or cells, and imaging agents. It also requires the assembly of sufficient functional proteins to perform specific bioassays on matrices that provide stable environments for the attached proteins. Because the amount and stability of the assembled proteins are strongly dependent on the method used, protein assembly on the particle surface is a key process. In order to assemble functional proteins and antibodies on BacMPs with high efficiency, several approaches have been exploited.
BacMPs are covered by a lipid bilayer membrane that mainly consists of phospholipids (comprising 58–65% of the total lipids), 50% of which is phosphatidylethanolamine (Nakamura & Matsunaga 1993; Frankel et al. 1998). The outward-oriented amine groups from phospholipids facilitate the immobilization of functional molecules on the BacMP surface through a cross-linking reaction. In order to immobilize antibodies and oligonucleotides, various cross linkers were used. Homofunctional cross linkers, which contain two aldehydes or N-hydroxysuccinimide (NHS) esters reacted with amines on BacMPs and antibodies or oligonucleotides (Matsunaga et al. 2003; Tanaka et al. 2004b; Ceyhan et al. 2006; Wacker et al. 2007). A heterofunctional cross linker, N-succinimidyl 3-(2-pyridyldithio)propionate, which contains one NHS ester, was used for the thiolation of BacMPs (Nakamura & Matsunaga 1993; Nakamura et al. 1993; Matsunaga et al. 1996a). Dithiothreitol was reacted with antibodies for the reaction. Two heterofunctional cross linkers were used for the preservation of antibody specificity after chemical conjugation. Sulphosuccinimidyl 4-(N-maleimidomethyl)-cyclohexane-1-carboxylate, which contains the NHS ester and maleimide, was reacted with antibodies (Matsunaga et al. 1996a). Streptavidin immobilized on BacMPs was prepared using the same principle. Sulpho-NHS-LC-LC-biotin was used to modify the magnetic particles with biotin (Amemiya et al. 2005; Ceyhan et al. 2006; Wacker et al. 2007). The biotin was then used for immobilization of streptavidin. The streptavidin-modified BacMPs were designed to immobilize biotin-modified antibodies or oligonucleotides.
Protein display on BacMPs was developed by a fusion technique involving anchor proteins isolated from magnetotactic bacteria. Several proteins identified from the proteome analysis of the BacMP membrane were used to display functional molecules on BacMPs. The MagA (47 kDa; Nakamura et al. 1995b; Matsunaga et al. 1999, 2000a, 2002), Mms16 (16 kDa; Yoshino et al. 2004, 2005) and Mms13 (13 kDa; Yoshino & Matsunaga 2005, 2006) proteins were used as anchor molecules. The fusion proteins usually become highly dependent on the structural properties of the inserted foreign protein domain, since the protein will be more constrained when inserted into a permissive site of an anchor molecule. Selection of an anchor protein and fusion sites are very crucial for the protein display. It is also known that expression properties sometimes differed, despite employing the same vector construct and promoter. The MagA is a transmembrane protein identified from a M. magneticum AMB-1 mutant strain generated by transposon mutagenesis (Nakamura et al. 1995a). This was used as an anchor molecule for the expression of foreign proteins on the surface of BacMPs. Soluble proteins, such as luciferase (Nakamura et al. 1995b), protein A (Matsunaga et al. 1999) and acetate kinase (Matsunaga et al. 2000a) were expressed on BacMPs. However, MagA maintains a large hydrophobic domain, which renders it unsuitable for assembling membrane proteins. Mms16 was found to be a smaller protein that was abundantly expressed in the BacMP membrane (Okamura et al. 2001). It is tightly anchored in the membrane by myristoylation. This characteristic was considered to be useful for the expression of large transmembrane proteins, since it affects the expression of foreign proteins to a lesser extent by dimensional obstruction. For example, G protein-coupled receptors (GPCRs) belong to the group of seven-transmembrane proteins involved in signal transduction cascades contributing to numerous biological activities (figure 5). Mms16 was therefore selected as an anchor for the assembly of a GPCR, and its efficient expression on BacMPs was confirmed (Yoshino et al. 2004). A recently discovered integral BacMP membrane protein, Mms13 (Arakaki et al. 2003), was also investigated for its efficiency in its role as an anchor molecule for displaying the functional proteins on BacMPs. The anchoring properties of Mms13 were confirmed by luciferase fusion studies. The C-terminal of Mms13 was shown to be expressed on the surface of BacMPs (Yoshino & Matsunaga 2006). Moreover, Mms13 was found to be bound tightly on the magnetite surface, permitting stable localization of a large protein-like luciferase (61 kDa) on BacMPs. Consequently, the luminescence intensity obtained from BacMPs using Mms13 as an anchor molecule was 400–1000 times higher than that of Mms16 or MagA. Based on these investigations, an efficient technique for the protein display on BacMPs was developed.
Figure 5.
Schematic for the preparation of functional BacMPs for protein display. The figure is modified from Matsunaga et al. (2007a–c).
To establish a high level of expression of proteins displayed on BacMPs, strong promoters were identified based on the AMB-1 genome and proteome databases (Yoshino & Matsunaga 2005). Upstream DNA sequences of ORFs that code for highly expressed proteins may contain strong promoters. Therefore, highly expressed proteins in AMB-1 were isolated. Several proteins that were highly expressed on the cell membrane and also localized on the BacMP membrane were selected. The N-terminal amino acids were determined based on the protein database of AMB-1. The sequences present before the start codons (ATG) of the identified ORFs were analysed using a computer program, and the promoter regions were predicted. To evaluate the activity of each promoter, the plasmids to be used in a luciferase-reporter gene assay were constructed. Consequently, luminescence intensity obtained using the msp3 promoter showed approximately 400 times higher activity than that with the magA promoter previously used.
A 3.7 kb cryptic plasmid designated pMGT, which is found in M. magneticum MGT-1, was also characterized and used for the development of an improved expression system in the AMB-1 strain through the construction of a shuttle vector, namely pUMG (Okamura et al. 2003). An electroporation method for magnetotactic bacteria that use the cryptic plasmid was also developed. The recombinant plasmid, pUMG, was constructed by ligating pUC19 and pMGT. pUMG containing the replicon of pMGT in AMB-1 yielded approximately 39 copies per cell, whereas the vector pRK415 yielded only three copies per cell. In addition, a luciferase-reporter expression system revealed that pUMG as an expression vector exhibited five times higher activity than the pKML previously used. These results are useful in improving the mass production of BacMPs and protein A displayed on BacMPs in fed-batch cultures, as we have described previously.
Mass production of BacMPs was also investigated in order to obtain functional BacMPs for biotechnological use. In our previous work, a set-up for large-scale cultivation with a 1000 l fermentor was prepared, and it yielded approximately 2.6 mg of BacMPs per litre of culture (Matsunaga et al. 1990). To enhance the productivity, growth conditions for mass production of luciferase–BacMPs by a recombinant AMB-1 were investigated in a pH-regulated fed-batch culture (Matsunaga et al. 2000a). Recombinant cells harbouring plasmids with magA–luc gene fusion were cultured. The culture conditions including iron source, nutrients and reducing agents in the medium were investigated (Matsunaga et al. 1996b). Fed-batch culture of AMB-1 harbouring a stably maintained expression plasmid in the cells for recombinant protein was carried out in a 10 l fermentor under microaerobic conditions. The addition of fresh nutrients was feedback controlled as a function of the pH of the culture. The yield of BacMPs was optimized by adjusting the rate of ferric iron addition. Feeding ferric quinate at 15.4 μg min−1 yielded 7.5 μg l−1 BacMPs (Yang et al. 2001b). Stable expression of the magA–luc fusion gene was indicated. BacMP biomineralization in M. gryphiswaldense was also investigated using an oxygen-controlled fermentor. Although the value of the amount of BacMPs is obtained by calculation, a significant BacMP yield (6.3 mg l−1 d−1) was reported (Heyen & Schüler 2003). Enrichment of the growth medium with l-cysteine, yeast extract and polypeptone enhanced both bacterial growth and BacMP production (Yang et al. 2001a). The presence of l-cysteine in the medium was useful for the induction of cell growth. Strict anaerobic conditions led to a prolonged lag phase and limited the final cell density. With regard to the iron sources, ferrous sulphate and ferric gallate dramatically enhanced the BacMP yield when compared with ferric quinate, an iron chelate that is conventionally used. The optimized conditions increased the cell density to 0.59±0.03 g cells (dry weight) per litre, and the BacMP production to 14.8±0.5 g cells (dry weight) per litre in a fed-batch culture for 4 days (Yang et al. 2001a). The optimized culture condition is applicable for the production of other functional BacMPs.
As an alternative method for the protein display on BacMPs, in vitro reconstitution of fusion proteins with integral membrane proteins or peptides into lipid vesicles was investigated. Artificial integration of useful proteins is a powerful method for high-efficiency assembly of the soluble proteins on BacMPs. Electrostatic and hydrophobic interactions are considered to be the driving force for spontaneous insertion of proteins and peptides into lipid membranes (Knol et al. 1998). MagA–luciferase fusion proteins prepared from recombinant E. coli membranes were integrated by sonication (Matsunaga et al. 2002). Maximum luminescence was obtained, which was 18 times higher than that of the recombinant luciferase–MagA displayed on BacMPs generated using the gene fusion techniques. Furthermore, an antimicrobial peptide, temporin L, and its derivative were also employed as anchor peptides and immobilized streptavidin on BacMPs (Tanaka et al. 2004a). Temporin L is a cationic linear peptide with varied structure, length and orientation (Rinaldi et al. 2002). It forms an amphiphilic structure with the hydrophobic part that is organized in a helix when associated with a membrane. This spontaneous integration mechanism was applied for protein assembly onto the BacMPs. The C-terminal of temporin L was incorporated into a BacMP membrane, and the N-terminal was located on the BacMP membrane surface. The conjugation of biotin to the N-terminal of temporin L leads to the binding of streptavidin on a BacMP membrane. Improved efficiency of the integration of functional proteins is obtained by selecting the most suitable anchor molecule under optimized conditions.
4.2 In vitro chemical synthesis of magnetite particles
Organic molecules tightly or directly associated with biominerals often play key roles in the initiation or the growth regulation of crystal sizes and morphologies (Belcher et al. 1996; Shimizu et al. 1998; Lakshminarayanan et al. 2002). Detailed analyses of the biological processes have allowed us to use or mimic them for creating a new generation of synthetic materials (Sugawara et al. 2003; Nuraje et al. 2006; Sano et al. 2006). The Mms6 represents a class of proteins that are tightly associated with the BacMP surface in M. magneticum AMB-1 (Arakaki et al. 2003). The Mms6 amino acid sequence is amphiphilic and consists of an N-terminal LG-rich hydrophobic region and a C-terminal hydrophilic region containing multiplets of acidic amino acids. The Mms6 shows a high degree of aggregation in aqueous solution. The hydrophobic domain of Mms6 is considered to contribute to self-aggregation through hydrophobic interactions. Moreover, the Mms6 shows large negative net charges under neutral conditions. Following competitive iron-binding analysis with other inorganic cations, it has been suggested that the C-terminal region is an iron-binding site. In order to elucidate the role of Mms6 in magnetite crystal formation and use the function for biomimetic synthesis of magnetic particles, two synthetic methods were employed and investigated.
Co-precipitation of ferrous and ferric ions in the presence of Mms6 produced uniform magnetite crystals with sizes ranging from 20 to 30 nm, while the absence of this protein resulted in the formation of the magnetic particles of irregular shapes and sizes (Arakaki et al. 2003). Furthermore, the particles produced in the presence of Mms6 showed cuboidal morphology similar to the BacMPs formed in Magnetospirillum spp. Prozorov et al. (2007a) also investigated magnetite production with Mms6, ferritin and bovine serum albumin (BSA) in a solution containing non-ionic surfactants. They reported that only Mms6 had a size and shape that could have a regulatory effect on the magnetite crystals. The magnetite particles synthesized in the presence of Mms6 showed the largest magnetization values above the blocking temperature and magnetic susceptibility, compared with those of the particles synthesized with the other proteins. The synthetic method was also applied for the production of cobalt ferrite (CoFe2O3) particles (Prozorov et al. 2007b).
Alternatively, a method involving partial oxidation of ferrous hydroxide was employed to synthesize particulate magnetite with the addition of Mms6 (Amemiya et al. 2007). Because this method generates larger, uniform and well-defined rectangular magnetite crystals (octahedron), it facilitates the determination of the morphological and size changes of magnetite particles by the addition of Mms6. The ferrous solution containing Mms6 was incubated under ambient conditions to produce Mms6–iron complexes prior to magnetite crystal formation. During the reaction, the colour of the ferrous solution changed from transparent blue to bluish green very rapidly, suggesting the formation of ferrous hydroxide. The Mms6 interacts with iron ions and/or the iron hydroxide precursor. This process may define the size and the morphology of iron oxide crystals. Black magnetite precipitates were obtained by heating the solution. The heat treatment accelerates the crystallization of iron oxide. TEM images of the crystals prepared without Mms6 show the expected octahedral shape (figure 6d), while crystals formed in the presence of Mms6 showed cuboidal morphologies (figure 6a). The size distribution of the synthesized magnetic crystals was statistically analysed by measuring the crystal sizes in typical TEM images. The average size of the magnetite crystals synthesized in the presence of Mms6 was 20.2±4.0 nm. In the absence of Mms6, the synthesized magnetite crystals were 32.4±9.1 nm in size. The crystals synthesized with Mms6 are smaller than crystals produced without Mms6 and have similar dimensions to the co-precipitated particles formed in the presence of Mms6 (21.2±8.3 nm). Furthermore, the crystals formed in the presence of Mms6 are distributed over a narrower range than crystals synthesized in the absence of the protein. The presence of BSA had no significant effect on the size and the morphology of the magnetite crystals synthesized. A close comparison of the crystals formed with and without Mms6 was performed by high-resolution TEM (HRTEM) analysis. The images clearly show lattice fringe patterns, indicating a single crystal structure. The [110] zone axis of the crystal synthesized in the presence of Mms6 possessed a hexagonal face with interior angles of 110 and 125°. These angles correspond to the intersection of two {111} faces, and a {111} face with a {100} face of magnetite, as described for a cubo-octahedral shape (figure 6b,c). The presence of these intersections was also observed in the BacMP of M. magneticum AMB-1 as previously reported (figure 6h,i; Amemiya et al. 2007). The HRTEM image, along with the [110] crystal zone axis, revealed only {111} rhombic faces for crystals synthesized in the absence of protein (figure 6e,f). This is identical to the octahedron-shaped magnetite previously reported for this synthetic method. The formation of the magnetite {100} face could be attributed to a face-specific interaction of Mms6. The selectivity of Mms6 onto particular crystallographic faces could be considered to be due to the dissimilarity of the surface structures of magnetite {100} and magnetite {111}, as reported previously. When comparing the two magnetite formation methods of co-precipitation and partial oxidation of ferrous hydroxide, crystals of different quality, size and morphology in the absence of Mms6 (various small crystallites and larger octahedral crystals, respectively) were observed. However, both synthetic methods yield very similar crystals in the presence of Mms6. The crystals exhibit similar sizes (20 nm) and morphologies (cubo-octahedral), as opposed to the crystals formed in the absence of Mms6. This preparation process may provide an alternative method for the simple regulation of nano-sized magnetic particles.
Figure 6.
Magnetic particles synthesized by partial oxidation in (a–c) the presence and (d–f) absence of Mms6. (g–i) Extracted magnetite from M. magneticum AMB-1. (a,d,g) Low-magnification TEM images, (b,e,h) HRTEM images observed in the [110] zone axis and (c,f,i) ideal morphology of magnetic particles. Pictures are redrawn/modified from Amemiya et al. (2007).
4.3 Surface modification of BacMPs with amine-terminated organic compounds
Surface modification of magnetic nanoparticles is an important issue with regard to using them for various biotechnological applications (Osaka et al. 2006). With the aim of developing magnetic nanoparticles for DNA extraction, the BacMP surface was modified with aminosilane compounds. The use of magnetic particles as a solid-phase adsorbent is well suited for DNA extraction techniques because they can be easily manipulated through simple application of a magnet. Although artificially synthesized iron oxide has the capacity to adsorb DNA, aggregation is prominent due to magnetic attraction, reducing the usable surface area (Yoza et al. 2002). Magnetic particles can be formed synthetically, but they are often non-uniform, incompletely crystalline and not compositionally homogeneous. In the preliminary examination, the DNA-binding capacity of BacMP was very low. Moreover, synthetically formed magnetic particles have the capability to adsorb DNA, but they form aggregates due to the reduction of usable surface area for adsorption by magnetic attractive forces. Organosilane compounds can be covalently bound to iron oxides, which introduce a positive ionic charge that is beneficial for restoring dispersion and facilitating ionic interactions between DNA molecules. The BacMPs and synthetic magnetite were covalently modified with three aminosilane compounds (APTES, 3-aminopropyltriethoxysilane; NTIC, N(trimethoxysilylpropyl)isothiouronium chloride; AEEA, 3-[2-(2-aminoethyl)-ethylamino]-propyltrimethoxysilane) with 1, 2, and 3 amine groups, respectively (Yoza et al. 2002; Nakagawa et al. 2006). Introduction of a primary amine ratio of approximately 1 : 2 : 1 (APTES : NTIC : AEEA) was determined. DNA extraction using AEEA-modified BacMPs showed higher rates of DNA recovery than that using APTES- and NTIC-modified particles and other commercially available kits. The DNA-binding efficiency increased with an increase in the number of amino groups.
A polyamidoamine (PAMAM) dendrimer can introduce a dense outer amine shell through cascade-type generation. The number of outer amines doubles with every layer generated and is limited only by steric interference (Tomalia et al. 1990). A PAMAM coating enables the reduction of particle agglomeration, and the terminal groups on the periphery can be tailored to control composite solubility. The BacMPs were therefore modified with PAMAM dendrimers to increase the number of amino groups, allowing enhanced extraction of DNA from fluid suspension (figure 7; Yoza et al. 2003b). A PAMAM dendrimer forms a dense outer amine shell through cascade-type polymer generation. The BacMPs at successive dendrimer generations were investigated for their DNA-binding abilities. The amine numbers double with every layer generated on the BacMP surface. On the other hand, the number of amino groups generated for a particular size of the artificially synthesized magnetic particles (AMPs) is one-tenth of that on the BacMP at the sixth generation. Modified magnetic particles were mixed with 25 μg of DNA. The amount of DNA binding by dendrimer-modified BacMPs increased with every generation and 24.83±1.61 μg of DNA was bound to 100 μg BacMPs at the sixth generation. Furthermore, as dendrimer generations increased, a linear correlation was found between the amount of DNA removed from solution and the increase in amino charge. This occurrence further confirms the removal of DNA from solution owing to cationic charge contribution. DNA was recovered by incubating the DNA complex at 50°C for 30 min, which resulted in 87% release (21.70±1.59 μg). DNA recovery with dendrimer-modified BacMPs is approximately six times higher than that with dendrimer-modified AMPs. Furthermore, small quantities of dendrimer-modified BacMPs were used to extract DNA from blood. The efficiency of DNA recovery was consistent at approximately 30 μg of DNA with 2–10 μg of dendrimer-modified BacMPs. This process was incorporated into a fully automated system using a newly developed liquid-handling system (Yoza et al. 2003a).
Figure 7.
Synthesis of polyamidoamine dendrimers on BacMPs used for DNA extraction. The dendrimers were generated with amine groups derived from AEEA-modified BacMPs. Stepwise growth was repeated until the desired number of generations was obtained.
4.4 Construction of nanomagnetic beads and micropolystyrene bead composites, ‘Beads on Beads’
To develop a novel platform for conventional bioassays, the BacMPs were assembled onto non-magnetic microbeads (polystyrene beads, 5.0–5.9 μm in diameter). Novel nano- and microbead composites were constructed by assembling BacMPs onto micropolystyrene beads termed ‘Beads on Beads’ (Matsunaga et al. 2007a). Figure 8a shows a schematic of the method for constructing Beads on Beads. BacMPs displaying the immunoglobulin G (IgG)-binding domain of protein A (ZZ domain) were modified with biotin and Cy3 by a cross-linking reaction. The assembly of protein A–BacMPs onto polystyrene microbeads was then performed via the streptavidin–biotin reaction. The construction of Beads on Beads was confirmed by observing the fluorescence of Cy3 on polystyrene microbeads at each stage by observing Cy3 fluorescence on polystyrene microbeads. The Beads on Beads construct was efficiently assembled by gradual addition of Cy3- and biotin-labelled BacMPs (15 min reactions, 10 times) onto streptavidin-coated polystyrene microbeads (figure 8b). To confirm whether Cy3- and biotin-labelled BacMPs were assembled onto all the microbeads, 104 of the constructed Beads on Beads were analysed by flow cytometry. The peak of the Beads on Beads fluorescence intensity histograms showed gradually increasing intensities following the eighth addition, and it became sharper following the ninth and tenth additions. The results showed that 10 sequential additions of the Cy3- and biotin-labelled BacMPs generated each Beads on Beads construct with similar levels of conjugation. The surface observation of Beads on Beads by scanning electron microscopy (SEM) indicated that the BacMPs were bound to the microbead surfaces as single particles and did not show obvious aggregation. From the 26 SEM images of the Beads on Beads surface (4.59 μm2), the average number of BacMPs was determined to be 22.4 μm−2. The surface area of a single microbead is 78.5 μm2. Therefore, approximately 2000 BacMPs were uniformly assembled on a single microbead without aggregation. The constructed Beads on Beads were magnetized and separated from the suspension by magnetic separation with a higher efficiency than that for BacMPs alone, considering the composites are suitable for use in a fully automated bioassay system. A fully automated sandwich immunoassay for the detection of human PSA was further examined using Beads on Beads. The detection limit for the assay was found to be 1.48 ng ml−1. Beads on Beads could be a powerful tool for the development of high-throughput, fully automated, multiplexed bioassays. The sandwich immunoassay and the fully automated system are described in detail below.
Figure 8.
Construction of antibody-binding ‘Beads on Beads’. (a) The assembly of antibody-binding BacMPs onto streptavidin-coated polystyrene microbeads was established through biotin–streptavidin interaction. (b) Fluorescence microscopy images and (c) fluorescence intensity histograms of Beads on Beads during each conjugation step. Fluorescence intensity histograms were obtained by flow cytometric analysis of more than 104 Beads on Beads particles.
5. Biotechnological applications of the functional magnetic particles
The use of functional magnetic particles in bioassays facilitates the separation of bound and free analytes by the application of a magnetic field. Because BacMPs disperse evenly throughout the reaction mixture, their use facilitated rapid reaction kinetics without the need for continuous mixing or shaking, enabling the coupling of antibodies. Furthermore, nano-sized BacMPs provide a large surface area for reactions. These characteristics have enabled the use of BacMPs as stable platforms for antibodies that are used in sensitive immunoassays and cell separation. BacMPs were also used for the development of high-performance DNA/mRNA recovery, DNA discrimination analysis, receptor-binding assay and magnetic markers. Moreover, fully automated systems were developed for precise, rapid and high-throughput processing of these assays. The applications of BacMPs developed by the authors are introduced in this section.
5.1 High-throughput single nucleotide polymorphism detection system
Single nucleotide polymorphisms (SNPs) are the most common form of human genetic polymorphisms and serve as markers for identifying genes responsible for diseases such as cancer, hypertension and diabetes that are induced by environmental factors (Nyren et al. 1997; Tomita-Mitchell et al. 1998). In order to analyse the causes of disease, SNP databases containing several million SNPs have been constructed. In addition, clinical investigators have begun analysing SNP alleles in population-based studies in order to identify loci that are statistically associated with a particular disease or phenotype. In these clinical studies, the number of samples available for analysis is important for data reliability. High-throughput and accurate multiple assay systems are therefore required for SNP analysis. For DNA detection employing submicron-sized particles and fluorescence dyes, the wavelength from light scattering caused by small particles (1000–100 nm) overlaps fluorescence dye emissions, resulting in low sensitivity (Nakayama et al. 2003; Tanaka et al. 2003). This phenomenon is called Mie scattering of nanometre-sized particles in solution, and it disturbs accurate SNP detection in a BacMP system using fluorescence dye-labelled probes such as fluorescein isothiocyanate, Cy3 or Cy5, which show a Stokes' shift with narrow wavelengths. To avoid this interference of the fluorescence signal, we developed a protocol based on DNA thermal dissociation curve analysis for an automated system with BacMPs (Maruyama et al. 2004). The method comprises PCR amplification of the target gene region by biotinylated primers, recovery of the amplified PCR products by streptavidin-immobilized BacMPs (SA–BacMPs), denaturation of duplex DNA, magnetic separation of SA–BacMPs with the single-stranded target DNA, hybridization with fluorescence-labelled probes and, finally, detection. SNPs of aldehyde dehydrogenase-2 (ALDH2; Maruyama et al. 2004), epidermal growth factor receptor (EGFR; Maruyama et al. 2007) and transforming growth factor beta-1 (TGF-β1; Matsunaga et al. 2007b) genes have been successfully detected. Certain encumbrances are encountered in the SNP discrimination of each target gene. For example, TGF-β1 is known as a genetic marker associated with hereditary risks that lead to bone loss. However, the TGF-β1 gene region has a high GC content and contains short tandem repeat sequences, making it difficult to detect. By optimizing the target region and detection conditions, accurate SNP discrimination was achieved. The details of the experimental scheme are described below. Biotinylated PCR amplicons and SA–BacMPs were mixed and incubated at room temperature to capture the PCR amplicon on the BacMPs (figure 9). After incubation, the amplicons captured on the BacMPs were magnetically collected. Subsequently, sodium hydroxide was added in order to denature the amplicons. The mixture was then neutralized by adding a neutralization buffer. The prepared single-stranded target DNA on the BacMPs was mixed with two allele-specific probes (Cy3- and Cy5-labelled detection probes), and SA–BacMPs were simultaneously mixed. The mixture was denatured at 70°C and cooled slowly to 25°C. The DNA duplex and DNA–BacMP complex were then heated to 58°C, a temperature determined by dissociation curve analysis, and dissociated single-base mismatched detection probe was removed. The fluorescence signal of the detection probe was released into the supernatant from a completely matched duplex DNA–BacMP complex by heating to 80°C, and then measured. Based on this method, the genotypes of ALDH2, EGFR and TGF-β1 were successfully discriminated. Furthermore, this principle was applicable for the determination of microsatellite repeats in the human thyroid peroxidase gene (Nakagawa et al. 2007).
Figure 9.
Schematic of the SNP detection procedure. Amplified PCR products were targeted for SNP detection. SNP was determined by measuring the fluorescent intensity of the probes liberated from BacMP complexes by heating.
Furthermore, an automated SNP detection processor for BacMP-based SNP discrimination was designed based on the protocol for DNA thermal dissociation curve analysis (Maruyama et al. 2004). The designed processor is equipped with a 96-way automated pipette that collects and dispenses fluids as it moves in the vertical and horizontal directions. The platform contains a disposable tip rack, a reagent vessel and a reaction station with a magnetic separation unit for a 96-well microtitre plate. One pole of the magnet allows the application of a magnetic field for one well. Eight poles of the magnet were aligned on an iron rod, and 12 rods were set on the back side of the microtitre plate to apply magnetic fields to the 96 wells. The magnetic field is switched on and off by rotating the rods by 180°. The reaction station is combined with a heat block regulated in the temperature range of 4–99°C, configured to perform the hybridization step. This system permits simultaneous SNP discrimination of 96 samples per assay. In the case of the EGFR gene, the analysis required approximately 3.5 hours, which included DNA extraction for 40 min, DNA amplification for 60 min and detection for 105 min (Maruyama et al. 2007).
5.2 Automated DNA extraction
DNA extraction using magnetic particles is commonly accepted as an automation-friendly procedure. Many automated instruments have been developed for extraction of DNA from blood samples (Belgrader et al. 1995; Smit et al. 2000). However, the automation process for plant samples has not been achieved because conventional procedures are not readily amenable to automation. For example, conventional procedures for plant DNA extraction have employed caesium chloride density gradients to eliminate enzyme-inhibitory polysaccharides (Murray & Thompson 1980) as well as cetyltrimethylammonium bromide-based protocols (Rogers & Bendich 1985). These methods require centrifugation, which is the most difficult process for integration into the automated system.
The PNE-1080 is equipped with eight automated pestle units and a spectrophotometer that is interfaced with a photosensor amplifier (figure 10a,b; Ota et al. 2006). Each pestle unit contains a pestle with a magnetic pole that moves in both the vertical and horizontal directions; a disposable, pre-packaged reagent; and a disposable cuvette (top inner dimensions (i.d.), 7 mm; bottom i.d., 6 mm; and height, 12.5 mm). DNA concentrations and purities were measured in the cuvette using an absorbance spectrometer that was integrated with the PNE-1080. The light traversed the solution in the cuvette from the bottom to the top. The magnetic pole was composed of a neodymium–iron–boron (Nd–Fe–B) magnet (3 mm in diameter and 22 mm in height), which collected magnetic particles and transferred them to other wells. DNA extraction from dried maize is described in figure 10c. The maize powder was lysed in a buffer and incubated for 30 min. An aliquot of the maize lysate was transferred to a well and aminosilane-modified BacMPs were added to the same well. The mixture was further ground by vertical motion of the pestle, and the extracted DNA adhered to the surface of the aminosilane-modified BacMPs. The DNA–BacMP complexes were collected at the tip of the pestle and transferred to the washing well. After washing, the bound DNA was eluted from the aminosilane-modified BacMPs with phosphate buffer in a transparent plastic cuvette. Maize genomic DNA was extracted using aminosilane-modified BacMPs. The yield of genomic DNA was approximately 315±3 ng when 60 μg of BacMPs was used. The (A260−A320): (A280−A320) ratio of the extracted DNA was 1.9±0.1 (Ota et al. 2006). These results suggest that our proposed DNA extraction method allowed highly accurate DNA recovery for subsequent PCR processes. This method did not require laborious pretreatment using organic solvents and permitted rapid completion of DNA extraction within 30 min, from sample application to the measurement of extracted DNA concentration and purity. Although several automated instruments for DNA extraction from plant tissues are present, these methodologies require centrifugation before sample application in order to eliminate enzyme inhibitory polysaccharides. Our newly developed instrument facilitates DNA extraction from maize powder without the centrifugation steps.
Figure 10.
Overview of an automated plant DNA extraction system (PNE-1080). (a) Automated DNA extraction system, (b) reaction well and (c) schematic of the DNA extraction procedure.
5.3 Magnetic detection of biomolecular interaction using BacMPs as a label
Currently, fluorescence and chemiluminescence technologies have been used as markers for the detection of various biochemical assays due to their high sensitivity, dynamic range and multiplexing capabilities. However, a reduced labelling signal due to photodegradation is a common disadvantage. The magnetic particles conjugated with biological molecules, which are attractive materials for the assay system, have been proposed for use as a label. They are unaffected by the measurement process and the samples may be stored indefinitely (Kriz et al. 1998; Arakaki et al. 2004). Furthermore, the magnetic particles can be detected by measuring their magnetism. A relatively simple and compact instrument has been described for the detection of magnetic particles (Edelstein et al. 2000). We have developed a system for streptavidin detection using biotin conjugated to BacMPs as magnetic labels for magnetic force microscopy (MFM) imaging (figure 11a; Amemiya et al. 2005). Biotin–BacMPs were applied to biotin immobilized on the glass slide after treatment with various concentrations of streptavidin (0.001–100 pg ml−1). The magnetic signal was obtained from a single particle using MFM without application of an external magnetic field. In the MFM images, immobilized BacMPs are observed as small dark areas on the contrasting background. The relationship between streptavidin concentrations and BacMP numbers in a 20×20 μm area are shown in figure 11b,c. The number of biotin–BacMPs bound to streptavidin immobilized on the glass slides increased with streptavidin concentrations of up to 100 pg ml−1. Biotin–BacMP binding difference was not observed in the absence of streptavidin and with a streptavidin concentration of up to 0.1 pg ml−1. The minimum detection limit for streptavidin was 1 pg ml−1. For comparison, fluorescence detection of Cy3-labelled streptavidin binding onto a biotinylated glass slide was performed by a photomultiplier using a fluorescent scanner. The photomultiplier has the potential to detect 40 molecules of Cy3 or Cy5 in the same scan area. The minimum detection limit of Cy3–streptavidin was 100 pg ml−1 of streptavidin, which corresponds to approximately 2000 molecules of streptavidin in the same area, if all the streptavidin molecules were immobilized on their surface. The detection limit is consistent with that of a previous report (150 pg ml−1 of IgG; Wacker et al. 2004). Therefore, we concluded that, compared with fluorescent detection, the experimental results of the BacMP-based assay are 100 times more sensitive for the detection of streptavidin, suggesting that its use has potential advantages for extremely sensitive biomolecule detection. A high-sensitivity detection system for streptavidin using nano-sized single-domain magnetic particles and MFM was developed. Biotinylated DNA or antibodies can also be immobilized onto BacMPs by the same detection strategy. The functionalized BacMPs are also available as a label for highly sensitive detection of DNA hybridization or immunoassay on solid supports.
Figure 11.
Magnetic detection of streptavidin using biotin–BacMPs. (a) Schematic of the reaction of biotin on the substrate and streptavidin on BacMP. (b) AFM and (c) MFM images of BacMPs on the substrate.
5.4 Immunoassay and receptor-binding assay
Competitive chemiluminescence enzyme immunoassays using antibodies immobilized onto BacMPs were developed for the rapid and sensitive detection of small molecules, such as environmental pollutants, hormone and toxic detergents (Matsunaga et al. 2003; Tanaka et al. 2004b). Xenoestrogens, such as alkylphenol ethoxylates, bisphenol A (BPA) and linear alkylbenzene sulphonates (LAS) were detectable using monoclonal antibodies immobilized onto BacMPs, based on the competitive reaction of xenoestrogens. The entire procedure was completed in 15 min, while typical plate methods take more than 2.5 hours. This method provided a wider range and lower detection limit than ELISA, in which the same antibodies were used for detection. Furthermore, the detection limits of LAS and BPA were similar to those obtained through gas chromatography–mass spectrometry (GC–MS). These experiments suggest that BacMP-based immunoassay systems have superior advantages due to their high sensitivity and rapid measurement of samples.
The Z domain of protein A in Staphylococcus aureus has the ability to bind to the Fc region of IgG (Lowenadler et al. 1987). A technique for the assembly of the Z domain on the BacMP surface by gene fusion using a protein A–magA hybrid gene was constructed (Matsunaga et al. 1999). The antibody was accurately oriented on the BacMP due to its association with protein A, unlike that in immobilization by chemical conjugation. When a chemiluminescence sandwich immunoassay was performed using antibody–protein A–BacMP complexes and BacMPs chemically immobilized with antibodies, the antigen-binding activity per microgram of antibodies for antibody–protein A–BacMP complexes was two times higher than that for antibody–BacMP conjugates prepared by chemical immobilization (Tanaka & Matsunaga 2000). Human insulin concentrations in blood serum were measured by a fully automated sandwich immunoassay using antibody–protein A–BacMP complexes and alkaline phosphatase-conjugated antibodies as primary and secondary immunoreactants, respectively. Dose–response curves were obtained from the luminescence intensity for human insulin concentrations. A detection limit of 2 μU ml−1 and a linear correlation between the signal and the concentration was apparent over the range of 19–254 μU ml−1 (Tanaka & Matsunaga 2000). On the other hand, Wacker et al. (2007) developed the magneto-immuno-PCR method using antibodies immobilized onto BacMPs as the capture phase. Using this method, a detection limit of 320 pg ml−1 for the hepatitis B surface antigen was reported.
BacMPs displaying receptors were developed and used for receptor-binding assays. GPCR represents one of the most predominant families of transmembrane proteins and is a prime target for drug discovery (Mirzabekov et al. 2000). Various challenges with regard to ligand screening of GPCRs have been undertaken; however, these proteins are generally expressed at very low levels in the cell and are extremely hydrophobic, rendering the analysis of ligand interaction very difficult. Moreover, the purification of GPCRs from cells is frequently time consuming and typically results in the loss of native conformation. In BacMPs displaying D1 dopamine receptor (D1R), GPCR was used as a model for a ligand-binding assay (Yoshino et al. 2004). Efficient assembly of D1R into the lipid membrane of nano-sized BacMPs was accomplished, and this was used for competitive dopamine-binding assays. This system conveniently refines the native conformation of GPCRs without the need for detergent solubilization, purification and reconstitution after cell disruption. This novel system provides advantages for studying various membrane proteins, which are usually difficult to assay.
5.5 Cell separation
Immunomagnetic particles have been used preferentially in target cell separation from leucocytes for in vitro diagnosis because the methodology is more rapid and simple than cell sorting using flow cytometry (Parra et al. 1997). The magnetic bead technology facilitates simple, rapid and efficient enrichment of target cell populations. In general, nano-sized magnetic particles rather than microsized beads are preferred for cell separation because the cells that are separated using the latter are subsequently useful for flow cytometric analysis. Microsized magnetic particles on the other hand have inhibitory effects on cell growth and differentiation after magnetic separation. However, commercially available nanoparticles are superparamagnetic and require the use of specially designed magnetic columns for cell separation to produce a high magnetic field gradient. Because BacMPs consist of ferromagnetic iron oxide, they are easily separated from cell suspensions using a permanent magnet with no special column. We have therefore applied BacMPs to develop highly efficient magnetic cell separation (Kuhara et al. 2004). BacMPs expressing protein A (protein A–BacMPs) bound to the Fc fragment of anti-mouse IgG antibodies were used to separate mononuclear cells from peripheral blood. The procedure for positive selection involves incubation of mononuclear cells and mouse monoclonal antibodies against different cell surface antigens (CD8, CD14, CD19 and CD20) prior to treatment with protein A–BacMP complexes with rabbit anti-mouse IgG secondary antibodies. The average purities of the separated mononuclear cells of CD19+ and CD20+ were 97.5 and 97.6%, respectively (Kuhara et al. 2004). Furthermore, more than 95% of the recovery ratio was obtained for these cells. Stem cell marker (CD34)-positive cells were also separated using the binding of protein A–BacMP complexes with the antibody. CD34 is the best identified surface antigen expressed on haematopoietic stem cells, which are known to be rare. The stem cells separated using BacMPs maintained their capability of colony formation as haematopoietic stem cells without inhibition of their proliferation and differentiation abilities. Specific separation of target cells using BacMPs was achieved by simple magnetic separation from cell suspensions using a permanent magnet with no special magnetic column.
To enhance the separation efficiencies, protein A was coexpressed with the Mms13 protein on BacMPs, and these results were compared with those obtained when MagA was used as an anchor molecule (Matsunaga et al. 2006). The evaluations of the number of bound antibody molecules and the binding capabilities of the protein A–BacMPs using Mms13 indicated that the antibodies efficiently bound to the protein A–BacMPs using Mms13 in comparison with MagA. In addition, the recovery ratio of the target cells of the magnetic cell separation system was enhanced using protein A–BacMPs with Mms13. Positive selection against peripheral blood mononuclear cells was used to achieve separation of the CD14+ cells at a purity of more than 99% by protein A–BacMPs using Mms13 (figure 12a). Furthermore, in the evaluation of the influence of protein A–BacMPs on the separated cells, the CD14+ cells separated using protein A–BacMPs were successfully differentiated into dendritic cells (figure 12b).
Figure 12.
Separation of CD14+ cells from PBMCs using protein A–BacMPs bound with antibodies. (a) Fluorescence histograms of the cells separated by flow cytometry from PBMCs (i) before and (ii) after magnetic separation. Upon magnetic separation, the unstained (negative fraction) and stained (positive fraction) fractions were analysed. (b) Microscopic photographs of the (i) separated CD14+ cells (ii) before (immature DC) and (iii) after (mature DC) differentiation.
6. Future prospects
Comprehensive molecular studies on the genes and proteins involved in BacMP biomineralization have led to the postulation of the step-by-step events of magnetite biomineralization. Furthermore, the results obtained from these basic studies have allowed us to develop novel functional constructs on the BacMP surface. The functional BacMPs generated by both chemical processes and genetic manipulations are useful for various biotechnological applications in immunoassays, receptor-binding assays, DNA extraction and cell separation. The in vitro magnetite synthesis and Beads on Beads technique could provide alternative materials that could be useful for such applications. Moreover, the application of functional BacMPs in a fully automated system provides precise, rapid and high-throughput analyses. Further studies will facilitate the development of new biomaterials.
Acknowledgements
This work was funded in part by a Grant-in-Aid for Scientific Research (A; no. 18206084) and Scientific Research on Priority Areas ‘Applied Genomics’ (no. 18018013) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also funded in part by the Industrial Technology Research Grant Program (no. 06A05005a) in 2006 from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
References
- Addadi L, Weiner S. Biomineralization—a pavement of pearl. Nature. 1997;389:912. doi: 10.1038/40010. [DOI] [Google Scholar]
- Amemiya Y, Tanaka T, Yoza B, Matsunaga T. Novel detection system for biomolecules using nano-sized bacterial magnetic particles and magnetic force microscopy. J. Biotechnol. 2005;120:308–314. doi: 10.1016/j.jbiotec.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Amemiya Y, Arakaki A, Staniland S.S, Tanaka T, Matsunaga T. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials. 2007;28:5381–5389. doi: 10.1016/j.biomaterials.2007.07.051. [DOI] [PubMed] [Google Scholar]
- Andrews S.C, Robinson A.K, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Arakaki A, Webb J, Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 2003;278:8745–8750. doi: 10.1074/jbc.M211729200. [DOI] [PubMed] [Google Scholar]
- Arakaki A, Hideshima S, Nakagawa T, Niwa D, Tanaka T, Matsunaga T, Osaka T. Detection of biomolecular interaction between biotin and streptavidin on a self-assembled monolayer using magnetic nanoparticles. Biotechnol. Bioeng. 2004;88:543–546. doi: 10.1002/bit.20262. [DOI] [PubMed] [Google Scholar]
- Balkwill D.L, Maratea D, Blakemore R.P. Ultrastructure of a magnetotactic spirillum. J. Bacteriol. 1980;141:1399–1408. doi: 10.1128/jb.141.3.1399-1408.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazylinski D.A, Frankel R.B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- Bazylinski D.A, Garratt-Reed A.J, Frankel R.B. Electron microscopic studies of magnetosomes in magnetotactic bacteria. Microsc. Res. Technol. 1994;27:389–401. doi: 10.1002/jemt.1070270505. [DOI] [PubMed] [Google Scholar]
- Bazylinski D.A, Frankel R.B, Heywood B.R, Mann S, King J.W, Donaghay P.L, Hanson A.K. Controlled biomineralization of magnetite (Fe3O4) and greigite (Fe3S4) in a magnetotactic bacterium. Appl. Environ. Microbiol. 1995;61:3232–3239. doi: 10.1128/aem.61.9.3232-3239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher A.M, Wu X.H, Christensen R.J, Hansma P.K, Stucky G.D, Morse D.E. Control of crystal phase switching and orientation by soluble mollusc-shell proteins. Nature. 1996;381:56–58. doi: 10.1038/381056a0. [DOI] [Google Scholar]
- Belgrader P, Del Ri S.A, Turner K.A, Marino M.A, Weaver K.R, Williams P.E. Automated DNA purification and amplification from blood-stained cards using a robotic workstation. BioTechniques. 1995;19:426–432. [PubMed] [Google Scholar]
- Blakemore R.P. Magnetotactic bacteria. Science. 1975;190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- Blakemore R.P, Maratea D, Wolfe R.S. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 1979;140:720–729. doi: 10.1128/jb.140.2.720-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calugay R.J, Miyashita H, Okamura Y, Matsunaga T. Siderophore production by the magnetic bacterium Magnetospirillum magneticum AMB-1. FEMS Microbiol. Lett. 2003;218:371–375. doi: 10.1016/S0378-1097(02)01188-6. [DOI] [PubMed] [Google Scholar]
- Calugay R.J, Okamura Y, Wahyudi A.T, Takeyama H, Matsunaga T. Siderophore production of a periplasmic transport binding protein kinase gene defective mutant of Magnetospirillum magneticum AMB-1. Biochem. Biophys. Res. Commun. 2004;323:852–857. doi: 10.1016/j.bbrc.2004.08.179. [DOI] [PubMed] [Google Scholar]
- Ceyhan B, Alhorn P, Lang C, Schuler D, Niemeyer C.M. Semisynthetic biogenic magnetosome nanoparticles for the detection of proteins and nucleic acids. Small. 2006;2:1251–1255. doi: 10.1002/smll.200600282. [DOI] [PubMed] [Google Scholar]
- DeLong E.F, Frankel R.B, Bazylinski D.A. Multiple evolutionary origins of magnetotaxis in bacteria. Science. 1993;259:803–806. doi: 10.1126/science.259.5096.803. [DOI] [PubMed] [Google Scholar]
- Edelstein R.L, Tamanaha C.R, Sheehan P.E, Miller M.M, Baselt D.R, Whitman L.J, Colton R.J. The BARC biosensor applied to the detection of biological warfare agents. Biosens. Bioelectron. 2000;14:805–813. doi: 10.1016/S0956-5663(99)00054-8. [DOI] [PubMed] [Google Scholar]
- Farina M, Esquivel D.M.S, Debarros H.G.P.L. Magnetic iron–sulfur crystals from a magnetotactic microorganism. Nature. 1990;343:256–258. doi: 10.1038/343256a0. [DOI] [Google Scholar]
- Flies C.B, Peplies J, Schuler D. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 2005;71:2723–2731. doi: 10.1128/AEM.71.5.2723-2731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel R.B, Bazylinski D.A, Schüeler D. Biomineralization of magnetic iron minerals in bacteria. Supramol. Sci. 1998;5:383–390. doi: 10.1016/S0968-5677(98)00036-4. [DOI] [Google Scholar]
- Fukuda Y, Okamura Y, Takeyama H, Matsunaga T. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett. 2006;580:801–812. doi: 10.1016/j.febslet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Gleich B, Weizenecker R. Tomographic imaging using the nonlinear response of magnetic particles. Nature. 2005;435:1214–1217. doi: 10.1038/nature03808. [DOI] [PubMed] [Google Scholar]
- Gorby Y.A, Beveridge T.J, Blakemore R.P. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 1988;170:834–841. doi: 10.1128/jb.170.2.834-841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K, Wawer C, Tebo B.M, Schüler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K, Muller E.C, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 2004;70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyen U, Schüler D. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 2003;61:536–544. doi: 10.1007/s00253-002-1219-x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Burgess J.G, Sakaguchi T, Takeyama H, Thornhill R.H, Matsunaga T. Phylogenetic analysis of a novel sulfate-reducing magnetic bacterium, RS-1, demonstrates its membership of the delta-Proteobacteria. FEMS Microbiol. Lett. 1995;126:277–282. doi: 10.1111/j.1574-6968.1995.tb07430.x. [DOI] [PubMed] [Google Scholar]
- Keim C.N, Martins J.L, Abreu F, Rosado A.S, de Barros H.L, Borojevic R, Lins U, Farina M. Multicellular life cycle of magnetotactic prokaryotes. FEMS Microbiol. Lett. 2004;240:203–208. doi: 10.1016/j.femsle.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Knol J, Sjollema K, Poolman B. Detergent-mediated reconstitution of membrane proteins. Biochemistry. 1998;37:16 410–16 415. doi: 10.1021/bi981596u. [DOI] [PubMed] [Google Scholar]
- Komeili A, Vali H, Beveridge T.J, Newman D.K. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl Acad. Sci. USA. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman D.K, Jensen G.J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Kriz K, Gehrke J, Kriz D. Advancements toward magneto immunoassays. Biosens. Bioelectron. 1998;13:817–823. doi: 10.1016/S0956-5663(98)00047-5. [DOI] [PubMed] [Google Scholar]
- Kuhara M, Takeyama H, Tanaka T, Matsunaga T. Magnetic cell separation using antibody binding with protein A expressed on bacterial magnetic particles. Anal. Chem. 2004;76:6207–6213. doi: 10.1021/ac0493727. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R, Kini R, Valiyaveettil S. Investigation of the role of ansocalcin in the biomineralization in goose eggshell matrix. Proc. Natl Acad. Sci. USA. 2002;99:5155–5159. doi: 10.1073/pnas.072658899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins U, Farina M. Organization of cells in magnetotactic multicellular aggregates. Microbiol. Res. 1999;154:9–13. [Google Scholar]
- Lowenadler B, Jansson B, Paleus S, Holmgren E, Nilsson B, Moks T, Palm G, Josephson S, Philipson L, Uhlen M. A gene fusion system for generating antibodies against short peptides. Gene. 1987;58:87–97. doi: 10.1016/0378-1119(87)90032-1. [DOI] [PubMed] [Google Scholar]
- Mann S, Ozin G.A. Synthesis of inorganic materials with complex form. Nature. 1996;382:313–318. doi: 10.1038/382313a0. [DOI] [Google Scholar]
- Mann S, Sparks N.H.C, Frankel R.B, Bazylinski D.A, Jannasch H.W. Biomineralization of ferrimagnetic greigite (Fe3S4) and iron pyrite (FeS2) in a magnetotactic bacterium. Nature. 1990;343:258–261. doi: 10.1038/343258a0. [DOI] [Google Scholar]
- Maruyama K, Takeyama H, Nemoto E, Tanaka T, Yoda K, Matsunaga T. Single nucleotide polymorphism detection in aldehyde dehydrogenase 2 (ALDH2) gene using bacterial magnetic particles based on dissociation curve analysis. Biotechnol. Bioeng. 2004;87:687–694. doi: 10.1002/bit.20073. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Takeyama H, Mori T, Ohshima K, Ogura S, Mochizuki T, Matsunaga T. Detection of epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) using a fully automated system with a nano-scale engineered biomagnetite. Biosens. Bioelectron. 2007;22:2282–2288. doi: 10.1016/j.bios.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Kamiya S. Use of magnetic particles isolated from magnetotactic bacteria for enzyme immobilization. Appl. Microbiol. Biotechnol. 1987;26:328–332. doi: 10.1007/BF00256663. [DOI] [Google Scholar]
- Matsunaga T, Tadokoro F, Nakamura N. Mass culture of magnetic bacteria and their application to flow type immunoassays. IEEE Trans. Magnet. 1990;26:1557–1559. doi: 10.1109/20.104444. [DOI] [Google Scholar]
- Matsunaga T, Sakaguchi T, Tadokoro F. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biotechnol. 1991;35:651–655. doi: 10.1007/BF00169632. [DOI] [Google Scholar]
- Matsunaga T, Nakamura C, Burgess J.G, Sode K. Gene-transfer in magnetic bacteria—transposon mutagenesis and cloning of genomic DNA fragments required for magnetosome synthesis. J. Bacteriol. 1992;174:2748–2753. doi: 10.1128/jb.174.9.2748-2753.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Kawasaki M, Yu X, Tsujimura N, Nakamura N. Chemiluminescence enzyme immunoassay using bacterial magnetic particles. Anal. Chem. 1996a;68:3551–3554. doi: 10.1021/ac9603690. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Tsujimura N, Kamiya S. Enhancement of magnetic particle production by nitrate and succinate fed batch culture of Magnetospirillum sp. AMB-1. Biotechnol. Tech. 1996b;10:495–500. doi: 10.1007/BF00159513. [DOI] [Google Scholar]
- Matsunaga T, Sato R, Kamiya S, Tanaka T, Takeyama H. Chemiluminescence enzyme immunoassay using protein A-bacterial magnetite complex. J. Mag. Mater. 1999;194:126–134. doi: 10.1016/S0304-8853(98)00575-7. [DOI] [Google Scholar]
- Matsunaga T, Togo H, Kikuchi T, Tanaka T. Production of luciferase-magnetic particle complex by recombinant Magnetospirillum sp. AMB-1. Biotechnol. Bioeng. 2000a;70:704–709. doi: 10.1002/1097-0290(20001220)70:6%3C704::AID-BIT14%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Tsujimura N, Okamura Y, Takeyama H. Cloning and characterization of a gene, mpsA, encoding a protein associated with intracellular magnetic particles from Magnetospirillum sp. strain AMB-1. Biochem. Biophys. Res. Commun. 2000b;268:932–937. doi: 10.1006/bbrc.2000.2236. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Arakaki A, Takahoko M. Preparation of luciferase–bacterial magnetic particle complex by artificial integration of MagA–luciferase fusion protein into the bacterial magnetic particle membrane. Biotechnol. Bioeng. 2002;77:614–618. doi: 10.1002/bit.10114. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Ueki F, Obata K, Tajima H, Tanaka T, Takeyama H, Goda Y, Fujimoto S. Fully automated immunoassay system of endocrine disrupting chemicals using monoclonal antibodies chemically conjugated to bacterial magnetic particles. Anal. Chim. Acta. 2003;475:75–83. doi: 10.1016/S0003-2670(02)01036-X. [DOI] [Google Scholar]
- Matsunaga T, Okamura Y, Fukuda Y, Wahyudi A.T, Murase Y, Takeyama H. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 2005;12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Takahashi M, Yoshino T, Kuhara M, Takeyama H. Magnetic separation of CD14(+) cells using antibody binding with protein A expressed on bacterial magnetic particles for generating dendritic cells. Biochem. Biophys. Res. Commun. 2006;350:1019–1025. doi: 10.1016/j.bbrc.2006.09.145. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Maeda Y, Yoshino T, Takeyama H, Takahashi M, Ginya H, Asahina J, Tajima H. Fully automated immunoassay for detection of prostate-specific antigen using nano-magnetic beads and micro-polystylene bead composites, ‘Beads on Beads’. Anal. Chim. Acta. 2007a;597:331–339. doi: 10.1016/j.aca.2007.05.065. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Maruyama K, Takeyama H, Katoh T. High-throughput SNP detection using nano-scale engineered biomagnetite. Biosens. Bioelectron. 2007b;22:2315–2321. doi: 10.1016/j.bios.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Suzuki T, Tanaka M, Arakaki A. Molecular analysis of magnetotactic bacteria and development of functional bacterial magnetic particles for nano-biotechnology. Trends Biotechnol. 2007c;25:182–188. doi: 10.1016/j.tibtech.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Meldrum F.C, Mann S, Heywood B.R, Frankel R.B, Bazylinski D.A. Electron-microscopy study of magnetosomes in 2 cultured vibrioid magnetotactic bacteria. Proc. R. Soc. B. 1993a;251:237–242. doi: 10.1098/rspb.1993.0035. [DOI] [Google Scholar]
- Meldrum F.C, Mann S, Heywood B.R, Frankel R.B, Bazylinski D.A. Electron-microscopy study of magnetosomes in a cultured coccoid magnetotactic bacterium. Proc. R. Soc. B. 1993b;251:231–236. doi: 10.1098/rspb.1993.0034. [DOI] [Google Scholar]
- Miltenyi S, Muller W, Weichel W, Radbruch A. High-gradient magnetic cell-separation with macs. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Mirzabekov T, Kontos H, Farzan M, Marasco W, Sodroski J. Paramagnetic proteoliposomes containing a pure, native, and oriented seven-transmembrane segment protein, CCR5. Nat. Biotechnol. 2000;18:649–654. doi: 10.1038/76501. [DOI] [PubMed] [Google Scholar]
- Murayama E, Takagi Y, Ohira T, Davis J.G, Greene M.I, Nagasawa H. Fish otolith contains a unique structural protein, otolin-1. Eur. J. Biochem. 2002;269:688–696. doi: 10.1046/j.0014-2956.2001.02701.x. [DOI] [PubMed] [Google Scholar]
- Murray M.G, Thompson W.F. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Hashimoto R, Maruyama K, Tanaka T, Takeyama H, Matsunaga T. Capture and release of DNA using aminosilane-modified bacterial magnetic particles for automated detection system of single nucleotide polymorphisms. Biotechnol. Bioeng. 2006;94:862–868. doi: 10.1002/bit.20904. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Maruyama K, Takeyama H, Matsunaga T. Determination of microsatellite repeats in the human thyroid peroxidase (TPOX) gene using a automated gene analysis system with nanoscale engineered biomagnetite. Biosens. Bioelectron. 2007;22:2276–2281. doi: 10.1016/j.bios.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Matsunaga T. Highly sensitive detection of allergen using bacterial magnetic particles. Anal. Chim. Acta. 1993;281:585–589. doi: 10.1016/0003-2670(93)85018-F. [DOI] [Google Scholar]
- Nakamura N, Hashimoto K, Matsunaga T. Immunoassay method for the determination of immunoglobulin G using bacterial magnetic particles. Anal. Chem. 1991;63:268–272. doi: 10.1021/ac00003a015. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Burgess J.G, Yagiuda K, Kudo S, Sakaguchi T, Matsunaga T. Detection and removal of Escherichia coli using fluorescein isothiocyanate conjugated monoclonal antibody immobilized on bacterial magnetic particles. Anal. Chem. 1993;65:2036–2039. doi: 10.1021/ac00063a018. [DOI] [PubMed] [Google Scholar]
- Nakamura C, Burgess J.G, Sode K, Matsunaga T. An iron-regulated gene, magA, encoding an iron transport protein of Magnetospirillum sp. strain AMB-1. J. Biol. Chem. 1995a;270:28 392–28 396. doi: 10.1074/jbc.270.20.12319. [DOI] [PubMed] [Google Scholar]
- Nakamura C, Kikuchi T, Burgess J.G, Matsunaga T. Iron-regulated expression and membrane localization of the magA protein in Magnetospirillum sp. strain AMB-1. J. Biochem. (Tokyo) 1995b;118:23–27. doi: 10.1093/oxfordjournals.jbchem.a124884. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Arakaki A, Maruyama K, Takeyama H, Matsunaga T. Single-nucleotide polymorphism analysis using fluorescence resonance energy transfer between DNA-labeling fluorophore, fluorescein isothiocyanate, and DNA intercalator, POPO-3, on bacterial magnetic particles. Biotechnol. Bioeng. 2003;84:96–102. doi: 10.1002/bit.10755. [DOI] [PubMed] [Google Scholar]
- Nuraje N, Su K, Haboosheh A, Samson J, Manning E.P, Yang N.L, Matsui H. Room temperature synthesis of ferroelectric barium titanate nanoparticles using peptide nanorings as templates. Adv. Mater. 2006;18:807. doi: 10.1002/adma.200501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyren P, Karamohamed S, Ronaghi M. Detection of single-base changes using a bioluminometric primer extension assay. Anal. Biochem. 1997;244:367–373. doi: 10.1006/abio.1996.9913. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Takeyama H, Matsunaga T. Two-dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillum sp. AMB-1. Appl. Biochem. Biotechnol. 2000;84–86:441–446. doi: 10.1385/ABAB:84-86:1-9:441. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Takeyama H, Matsunaga T. A magnetosome-specific GTPase from the magnetic bacterium Magnetospirillum magneticum AMB-1. J. Biol. Chem. 2001;276:48 183–48 188. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Takeyama H, Sekine T, Sakaguchi T, Wahyudi A.T, Sato R, Kamiya S, Matsunaga T. Design and application of a new cryptic-plasmid-based shuttle vector for Magnetospirillum magneticum. Appl. Environ. Microbiol. 2003;69:4274–4277. doi: 10.1128/AEM.69.7.4274-4277.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y, Denda K, Fukumori Y. Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene. 1996;171:99–102. doi: 10.1016/0378-1119(95)00008-9. [DOI] [PubMed] [Google Scholar]
- Osaka T, Matsunaga T, Nakanishi T, Arakaki A, Niwa D, Iida H. Synthesis of magnetic nanoparticles and their application to bioassays. Anal. Bioanal. Chem. 2006;384:593–600. doi: 10.1007/s00216-005-0255-7. [DOI] [PubMed] [Google Scholar]
- Ota H, Lim T.K, Tanaka T, Yoshino T, Harada M, Matsunaga T. Automated DNA extraction from genetically modified maize using aminosilane-modified bacterial magnetic particles. J. Biotechnol. 2006;125:361–368. doi: 10.1016/j.jbiotec.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pardoe H, Clark P.R, St Pierre T.G, Moroz P, Jones S.K. A magnetic resonance imaging based method for measurement of tissue iron concentration in liver arterially embolized with ferrimagnetic particles designed for magnetic hyperthermia treatment of tumors. Mag. Resonan. Imag. 2003;21:483–488. doi: 10.1016/S0730-725X(03)00072-9. [DOI] [PubMed] [Google Scholar]
- Parra E, Wingren A.G, Hedlund G, Kalland T, Dohlsten M. The role of B7-1 and LFA-3 in costimulation of CD8+T cells. J. Immunol. 1997;158:637–642. [PubMed] [Google Scholar]
- Plank C, Schillinger U, Scherer F, Bergemann C, Remy J.S, Krotz F, Anton M, Lausier J, Rosenecker J. The magnetofection method: using magnetic force to enhance gene delivery. Biol. Chem. 2003;384:737–747. doi: 10.1515/BC.2003.082. [DOI] [PubMed] [Google Scholar]
- Posfai M, Moskowitz B.M, Arato B, Schuler D, Flies C, Bazylinski D.A, Frankel R.B. Properties of intracellular magnetite crystals produced by Desulfovibrio magneticus strain RS-1. Earth Planet. Sci. Lett. 2006;249:444–455. doi: 10.1016/j.epsl.2006.06.036. [DOI] [Google Scholar]
- Pouget E, et al. Hierarchical architectures by synergy between dynamical template self-assembly and biomineralization. Nat. Mater. 2007;6:434–439. doi: 10.1038/nmat1912. [DOI] [PubMed] [Google Scholar]
- Pradel N, Santini C.L, Bernadac A, Fukumori Y, Wu L.F. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc. Natl Acad. Sci. USA. 2006;103:17 485–17 489. doi: 10.1073/pnas.0603760103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozorov T, et al. Protein-mediated synthesis of uniform superparamagnetic magnetite nanocrystals. Adv. Funct. Mater. 2007a;17:951–957. doi: 10.1002/adfm.200600448. [DOI] [Google Scholar]
- Prozorov T, et al. Cobalt ferrite nanocrystals: out-performing magnetotactic bacteria. ACS Nano. 2007b;1:228–233. doi: 10.1021/nn700194h. [DOI] [PubMed] [Google Scholar]
- Richter M, Kube M, Bazylinski D.A, Lombardot T, Glockner F.O, Reinhardt R, Schuler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi A.C, et al. Temporin L: antimicrobial, haemolytic and cytotoxic activities, and effects on membrane permeabilization in lipid vesicles. Biochem. J. 2002;368:91–100. doi: 10.1042/BJ20020806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.O, Bendich A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant-tissues. Plant Mol. Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Burgess J.G, Matsunaga T. Magnetite formation by a sulphate-reducing bacterium. Nature (Lond.) 1993;365:47–49. doi: 10.1038/365047a0. [DOI] [Google Scholar]
- Sakaguchi T, Tsujimura N, Matsunaga T. A novel method for isolation of magnetic bacteria without magnetic collection using magnetotaxis. J. Microbiol. Methods. 1996;26:139–145. doi: 10.1016/0167-7012(96)00905-0. [DOI] [Google Scholar]
- Sakaguchi T, Arakaki A, Matsunaga T. Desulfovibrio magneticus sp. nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. Int. J. Syst. Evol. Microbiol. 2002;52:215–221. doi: 10.1099/00207713-52-1-215. [DOI] [PubMed] [Google Scholar]
- Sano K, Sasaki H, Shiba K. Utilization of the pleiotropy of a peptidic aptamer to fabricate heterogeneous nanodot-containing multilayer nanostructures. J. Am. Chem. Soc. 2006;128:1717–1722. doi: 10.1021/ja057262r. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Schuler D. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J. Bacteriol. 2007;189:6437–6446. doi: 10.1128/JB.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko J.M, Schüler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gardes A, Grunberg K, Wanner G, Schuler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K.H, Schuler D, Spring S, Weizenegger M, Amann R, Ludwig W, Kohler M. The genus Magnetospirillum gen-nov - description of Magnetospirillum gryphiswaldense sp-nov and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb-nov. Syst. Appl. Microbiol. 1991;14:379–385. [Google Scholar]
- Schübbe S, et al. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 2003;185:5779–5790. doi: 10.1128/JB.185.19.5779-5790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübbe S, Würdemann C, Peplies J, Heyen U, Wawer C, Glockner F.O, Schüler D. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 2006;72:5757–5765. doi: 10.1128/AEM.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler D, Baeuerlein E. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 1996;166:301–307. doi: 10.1007/s002030050387. [DOI] [PubMed] [Google Scholar]
- Schüler D, Baeuerlein E. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 1998;180:159–162. doi: 10.1128/jb.180.1.159-162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Cha J, Stucky G.D, Morse D.E. Silicatein alpha: cathepsin L-like protein in sponge biosilica. Proc. Natl Acad. Sci. USA. 1998;95:6234–6238. doi: 10.1073/pnas.95.11.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons S.L, Sievert S.M, Frankel R.B, Bazylinski D.A, Edwards K.J. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl. Environ. Microbiol. 2004;70:6230–6239. doi: 10.1128/AEM.70.10.6230-6239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons S.L, Bazylinski D.A, Edwards K.J. South-seeking magnetotactic bacteria in the Northern Hemisphere. Science. 2006;311:371–374. doi: 10.1126/science.1122843. [DOI] [PubMed] [Google Scholar]
- Smit M.L, Giesendorf B.A.J, Heil S.G, Vet J.A.M, Trijbels F.J.M, Blom H.J. Automated extraction and amplification of DNA from whole blood using a robotic workstation and an integrated thermocycler. Biotechnol. Appl. Biochem. 2000;32:121–125. doi: 10.1042/BA20000043. [DOI] [PubMed] [Google Scholar]
- Spring S, Schleifer K.H. Diversity of magnetotactic bacteria. Syst. Appl. Microbiol. 1995;18:147–153. [Google Scholar]
- Spring S, Amann R, Ludwig W, Schleifer K.H, Vangemerden H, Petersen N. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a fresh-water sediment. Appl. Environ. Microbiol. 1993;59:2397–2403. doi: 10.1128/aem.59.8.2397-2403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakaguchi K, Tanaka M, Nakashima K, Takahashi T. Structures of mollusc shell framework proteins. Nature. 1997;387:563–564. doi: 10.1038/42391. [DOI] [PubMed] [Google Scholar]
- Sugawara A, Ishii T, Kato T. Self-organized calcium carbonate with regular surface-relief structures. Angew. Chem. Int. Edn. 2003;42:5299–5303. doi: 10.1002/anie.200351541. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Okamura Y, Calugay R.J, Takeyama H, Matsunaga T. Global gene expression analysis of iron-inducible genes in Magnetospirillum magneticum AMB-1. J. Bacteriol. 2006;188:2275–2279. doi: 10.1128/JB.188.6.2275-2279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Okamura Y, Arakaki A, Takeyama H, Matsunaga T. Cytoplasmic ATPase involved in ferrous ion uptake from magnetotactic bacterium Magnetospirillum magneticum AMB-1. FEBS Lett. 2007;581:3443–3448. doi: 10.1016/j.febslet.2007.06.047. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Matsunaga T. Fully automated chemiluminescence immunoassay of insulin using antibody-protein A-bacterial magnetic particle complexes. Anal. Chem. 2000;72:3518–3522. doi: 10.1021/ac9912505. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Maruyama K, Yoda K, Nemoto E, Udagawa Y, Nakayama H, Takeyama H, Matsunaga T. Development and evaluation of an automated workstation for single nucleotide polymorphism discrimination using bacterial magnetic particles. Biosens. Bioelectron. 2003;19:325–330. doi: 10.1016/S0956-5663(03)00189-1. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Takeda H, Kokuryu Y, Matsunaga T. Spontaneous integration of transmembrane peptides into a bacterial magnetic particle membrane and its application to display of useful proteins. Anal. Chem. 2004a;76:3764–3769. doi: 10.1021/ac035361m. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Takeda H, Ueki F, Obata K, Tajima H, Takeyama H, Goda Y, Fujimoto S, Matsunaga T. Rapid and sensitive detection of 17beta-estradiol in environmental water using automated immunoassay system with bacterial magnetic particles. J. Biotechnol. 2004b;108:153–159. doi: 10.1016/j.jbiotec.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Okamura Y, Arakaki A, Tanaka T, Takeyama H, Matsunaga T. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics. 2006;6:5234–5247. doi: 10.1002/pmic.200500887. [DOI] [PubMed] [Google Scholar]
- Taoka A, Asada R, Sasaki H, Anzawa K, Wu L.F, Fukumori Y. Spatial localizations of Mam22 and Mam12 in the magnetosomes of Magnetospirillum magnetotacticum. J. Bacteriol. 2006;188:3805–3812. doi: 10.1128/JB.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka A, Asada R, Wu L.F, Fukumori Y. Polymerization of the actin-like protein MamK, which is associated with magnetosomes. J. Bacteriol. 2007;189:8737–8740. doi: 10.1128/JB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill R.H, Burgess J.G, Sakaguchi T, Matsunaga T. A morphological classification of bacteria containing bullet-shaped magnetic particles. FEMS Microbiol. Lett. 1994;115:169–176. doi: 10.1111/j.1574-6968.1994.tb06633.x. [DOI] [Google Scholar]
- Thornhill R.H, Burgess J.G, Matsunaga T. PCR for direct detection of indigenous uncultured magnetic cocci in sediment and phylogenetic analysis of amplified 16S ribosomal DNA. Appl. Environ. Microbiol. 1995;61:495–500. doi: 10.1128/aem.61.2.495-500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomalia D.A, Naylor A.M, Goddard W.A. Starburst dendrimers—molecular-level control of size, shape, surface-chemistry, topology, and flexibility from atoms to macroscopic matter. Angew. Chem. Int. Edn Engl. 1990;29:138–175. doi: 10.1002/anie.199001381. [DOI] [Google Scholar]
- Tomita-Mitchell A, Muniappan B.P, Herrero-Jimenez P, Zarbl H, Thilly W.G. Single nucleotide polymorphism spectra in newborns and centenarians: identification of genes coding for rise of mortal disease. Gene. 1998;223:381–391. doi: 10.1016/S0378-1119(98)00408-9. [DOI] [PubMed] [Google Scholar]
- Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker R, Schroder H, Niemeyer C.M. Performance of antibody microarrays fabricated by either DNA-directed immobilization, direct spotting, or streptavidin-biotin attachment: a comparative study. Anal. Biochem. 2004;330:281–287. doi: 10.1016/j.ab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Wacker R, Ceyhan B, Alhorn P, Schüeler D, Lang C, Niemeyer C.M. Magneto Immuno-MR: a novel immunoassay based on biogenic magnetosome nanoparticles. Biochem. Biophys. Res. Commun. 2007;357:391–396. doi: 10.1016/j.bbrc.2007.03.156. [DOI] [PubMed] [Google Scholar]
- Wahyudi A.T, Takeyama H, Matsunaga T. Isolation of Magnetospirillum magneticum AMB-1 mutants defective in bacterial magnetic particle synthesis by transposon mutagenesis. Appl. Biochem. Biotechnol. 2001;91–93:147–154. doi: 10.1385/ABAB:91-93:1-9:147. [DOI] [PubMed] [Google Scholar]
- Wahyudi A.T, Takeyama H, Okamura Y, Fukuda Y, Matsunaga T. Characterization of aldehyde ferredoxin oxidoreductase gene defective mutant in Magnetospirillum magneticum AMB-1. Biochem. Biophys. Res. Commun. 2003;303:223–229. doi: 10.1016/S0006-291X(03)00303-6. [DOI] [PubMed] [Google Scholar]
- Würdemann C, Peplies J, Schübbe S, Ellrott A, Schüler D, Glockner F.O. Evaluation of gene expression analysis using RNA-targeted partial genome arrays. Syst. Appl. Microbiol. 2006;29:349–357. doi: 10.1016/j.syapm.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Yang C, Takeyama H, Tanaka T, Matsunaga T. Effects of growth medium composition, iron sources and atmospheric oxygen concentrations on production of luciferase-bacterial magnetic particle complex by a recombinant Magnetospirillum magneticum AMB-1. Enzyme Microb. Technol. 2001a;29:13–19. doi: 10.1016/S0141-0229(01)00343-X. [DOI] [PubMed] [Google Scholar]
- Yang C.D, Takeyama H, Matsunaga T. Iron feeding optimization and plasmid stability in production of recombinant bacterial magnetic particles by Magnetospirillum magneticum AMB-1 in fed-batch culture. J. Biosci. Bioeng. 2001b;91:213–216. doi: 10.1263/jbb.91.213. [DOI] [PubMed] [Google Scholar]
- Yoshino T, Matsunaga T. Development of efficient expression system for protein display on bacterial magnetic particles. Biochem. Biophys. Res. Commun. 2005;338:1678–1681. doi: 10.1016/j.bbrc.2005.10.148. [DOI] [PubMed] [Google Scholar]
- Yoshino T, Matsunaga T. Efficient and stable display of functional proteins on bacterial magnetic particles using mms13 as a novel anchor molecule. Appl. Environ. Microbiol. 2006;72:465–471. doi: 10.1128/AEM.72.1.465-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino T, Takahashi M, Takeyama H, Okamura Y, Kato F, Matsunaga T. Assembly of G protein-coupled receptors onto nanosized bacterial magnetic particles using Mms16 as an anchor molecule. Appl. Environ. Microbiol. 2004;70:2880–2885. doi: 10.1128/AEM.70.5.2880-2885.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino T, Kato F, Takeyama H, Nakai M, Yakabe Y, Matsunaga T. Development of a novel method for screening of estrogenic compounds using nano-sized bacterial magnetic particles displaying estrogen receptor. Anal. Chim. Acta. 2005;532:105–111. doi: 10.1016/j.aca.2004.10.074. [DOI] [Google Scholar]
- Yoza B, Matsumoto M, Matsunaga T. DNA extraction using modified bacterial magnetic particles in the presence of amino silane compound. J. Biotechnol. 2002;94:217–224. doi: 10.1016/S0168-1656(01)00427-8. [DOI] [PubMed] [Google Scholar]
- Yoza B, Arakaki A, Maruyama K, Takeyama H, Matsunaga T. Fully automated DNA extraction from blood using magnetic particles modified with hyperbranched polyamidoamine dendrimer. J. Biosci. Bioeng. 2003a;95:21–26. doi: 10.1016/S1389-1723(03)80143-3. [DOI] [PubMed] [Google Scholar]
- Yoza B, Arakaki A, Matsunaga T. DNA extraction using bacterial magnetic particles modified with hyperbranched polyamidoamine dendrimer. J. Biotechnol. 2003b;101:219–228. doi: 10.1016/S0168-1656(02)00342-5. [DOI] [PubMed] [Google Scholar]