Abstract
The maintenance of healthy colonic crypts is dependent on the integrity of the adult epithelial stem cells located within them. Perturbations in stem cell dynamics are generally believed to represent the first step towards colorectal tumorigenesis. Experimental manipulation of intestinal stem cells has greatly increased our understanding of them, but further progress has been slowed due to the absence of a reliable stem cell biomarker. In this review we discuss the candidate colonic stem cell biomarkers which have been proposed. Furthermore, we investigate the putative biomarkers for so-called colorectal cancer stem cells, a highly aggressive subpopulation of cells considered to drive tumour development.
Keywords: adult stem cells, cancer stem cells, colon, stem cell biomarkers
Introduction
The intestinal tract is characterized by the presence of numerous small inpocketings, or crypts, created by invagination of the surface epithelium. Each crypt unit is maintained by the adult stem cells located within it (Booth & Potten, 2000; Marshman et al. 2002). It is generally understood that the stem cells of the colon are positioned at the bottom of the crypt (Potten et al. 1997; Marshman et al. 2002). This is different from the small intestine in which the stem cells are thought to reside in an oscillating annulus approximately four cell diameters up from the base of the crypt, as the very bottom of the crypt is occupied by Paneth cells (Potten et al. 1997; Marshman et al. 2002). To date, the topographical location of intestinal stem cells has been inferred from indirect experiments, rather than definitively proved, primarily due to the absence of a reliable biomarker(s). Consequently, identifying a biomarker for intestinal stem cells is a priority. The fact that no definitive biomarker has yet been found suggests that this is no easy task.
For the most part, work has concentrated on identifying biomarkers of stem cells in the small intestine. However, we consider it pertinent to highlight the progress made in the search for a biomarker of colonic stem cells, considering the susceptibility of the colon to carcinogenesis. Credible candidates have been proposed, including Musashi-1 (Msi-1) (Nishimura et al. 2003), β1 integrin subunit (Fujimoto et al. 2002), EphB receptors (Holmberg et al. 2006) and, most recently, Lgr5 (Barker et al. 2007). Interestingly, Lgr5 is the only candidate which appears to have demonstrable specificity for colonic stem cells to the exclusion of all other progenitors. Consequently, it has emerged as one of the strongest candidates to date.
Our understanding of tumour development is constantly evolving. Whereas the stochastic model assumes that every tumour cell is capable of initiating tumour growth, the cancer stem cell (CSC) model suggests that only a small proportion of tumour cells are enriched for cancer-initiating activity and that these drive tumour development (Wang & Dick, 2005; Dalerba et al. 2007a). The isolation of CSCs has been reported in both haematological malignancies and solid tumours, based on the expression of cell surface biomarkers. At this point, prospective CSCs have been identified in acute myeloid leukaemia (Lapidot et al. 1994; Bonnet & Dick, 1997), breast cancer (Al-Hajj et al. 2003), brain tumours (Singh et al. 2004), prostate cancer (Collins et al. 2005), head and neck squamous cell carcinoma (Prince et al. 2007), pancreatic cancer (Li et al. 2007), liver cancer (Ma et al. 2007) and now colorectal cancer (CRC) (Dalerba et al. 2007b; O’Brien et al. 2007; Ricci-Vitiani et al. 2007). Identification of CSCs opens new avenues for targeted drug development to reduce tumour recurrence and, as such, may deliver far-reaching clinical benefits (Dalerba et al. 2007a). Notably, CRC is second in importance as a cause of cancer-related death in the Western world (Jemal et al. 2006; Ferlay et al. 2007). Consequently, the recent discovery of putative biomarkers for colorectal cancer stem cells (CR-CSCs) is likely to represent a highly significant breakthrough in tackling this disease.
Topography and morphogenesis of the colonic crypt
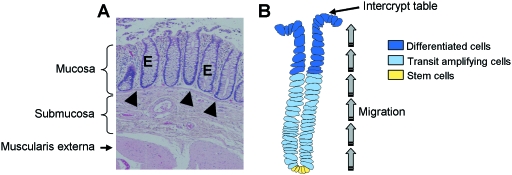
The colon is radially organized into four histologically distinct layers (Fig. 1A). At the luminal surface, simple columnar epithelium is folded into thousands of pit-like structures, termed crypts, which are supported by the lamina propria and a layer of smooth muscle cells, known as the muscularis mucosae. Collectively these form the mucosal lining. Underneath this lies the submucosa, which comprises connective tissue and contains the neural plexus (Meissner's plexus) that innervates the epithelium. Two bands of smooth muscle, forming the muscularis externa, are situated beneath the submucosa. An outer serous coat completely invests the colon, except for a few segments (Bannister, 1995).
Fig. 1.
Organization of the colonic epithelium. (A) Haematoxylin & eosin (H&E) stained section of healthy colon showing the epithelium, E, which is invaginated to form crypts which are embedded in the mucosa and underlined by the muscularis mucosae (arrowheads). (B) Topography of the colonic crypt. Stem cells reside at the bottom of the crypt and give rise to proliferative progenitors which migrate upwards, eventually differentiating into functionally competent cell types which are concentrated in the upper part of the crypt. Upon reaching the intercrypt table cells are targeted for programmed cell death and are shed into the lumen.
The crypts themselves are dynamic structures which are constantly self-renewing (Radtke & Clevers, 2005). The differentiated cells, which are mainly concentrated in the top third of the crypt, are turned over at a high rate and continually extruded into the lumen. They are replenished through a process of unidirectional transit amplification and lineage-specific differentiation of multipotent stem cells located in the crypt base (Fig. 1B). Together, the stem cell compartment and transit-amplifying region occupy the lower two-thirds of the crypt (Potten et al. 1997; Booth & Potten, 2000; Marshman et al. 2002; Radtke & Clevers, 2005). The transition from proliferation to differentiation constitutes the crypt–villus axis which is maintained by the Wnt signal (van de Wetering et al. 2002; Pinto & Clevers, 2005). There are three colonic epithelial lineages into which cells mainly mature: mucus-secreting goblet cell, absorptive enterocyte (also termed colonocyte) and less abundant enteroendocrine cell. These cell types function to lubricate the passage of waste material, absorb water and salts, and secrete hormones, respectively. Paneth cells are completely absent from colonic crypts and are found only in the small intestine (Potten et al. 1997; Marshman et al. 2002).
Characteristics of colonic epithelial stem cells
Stem cells are defined by their functional properties. They are relatively undifferentiated cells, capable of proliferation, which balance self-maintenance with the production of offspring that can differentiate into various functionally competent cell types. In addition, intestinal stem cells are able to respond to changes within the crypt to regulate stem cell number and crypt volume and have the capacity to regenerate the epithelium after injury (Potten & Loeffler, 1990).
The lower cryptal region of both the colon and small intestine harbours the multipotent stem cells. Unique environmental determinants, as well as intrinsic elements, are believed to maintain the stem cell phenotype in these cells, thereby creating a stem cell niche and preserving the integrity of the stem cell population (Spradling et al. 2001; McDonald et al. 2006). Functional responses to indirect experimental approaches have so far been used to predict stem cell position, number and life cycle.
Stem cell location
Stem cell location has been extrapolated from cell kinetic and radiation regeneration assays. By comparing the migration velocities of labelled cells at different positions in the crypt, the origin of cell migration, and by implication the position of the stem cells, has been pinpointed to the base of the crypt in the colon (Qiu et al. 1994) and about four cells up from the crypt base in the small intestine (Kaur & Potten, 1986). It has been observed that the proliferative regenerative response to small intestinal crypt insults is initiated in cells occupying the position attributed to stem cells (Potten et al. 1990). The bottom and lower regions of the crypt appear more sensitive to cytotoxic damage compared to the mid and upper crypt cells. Smaller doses of radiation that do not appear to affect cells in the mid and upper portions of the crypt, still render the crypt sterile, indicating that the cells in the mid and upper sections are not capable of repopulating the crypt. Accordingly the regenerative potential and consequently the stem cells must reside towards the base of the crypt (Hendry et al. 1989).
Stem cell number
The regenerative capacity of the crypt after irradiation has been exploited to determine the number of actual stem cells in the colon and small intestine, as well as the number of clonogenic or potential stem cells. Potential stem cells are capable of functioning as stem cells and, therefore, regenerating the crypt in the event that the stem cell population is ablated (Potten & Loeffler, 1990). The clonogenic content of the colon was extrapolated from a modified radiation regeneration assay in mouse. A small population of actual stem cells was predicted, i.e. between 5 and 10, with an extra 16–36 potential stem cells anticipated (Cai et al. 1997). By comparison, in the small intestine there are an estimated 4–6 actual stem cells per crypt and a further 28–42 potential stem cells available, which are less radiosensitive the further up the crypt they are positioned (Potten et al. 1997).
Stem cell life cycle
Studies on small intestinal crypts indicate that actual stem cells divide more slowly compared to cells higher up the crypt, perhaps to allow for error-free DNA synthesis or more time for repair (Potten, 1986; Potten et al. 1997). This functional difference is illustrated by labelling all proliferating cells with tritiated thymidine (Potten, 1986). Although putative stem cells may be dividing more slowly, it is still possible to label them. Over time, the label in faster cycling, transit-amplifying cells disappears more quickly, thereby distinguishing slower cycling stem cells from other cells by retention of the label.
The long-term label-retaining property of stem cells has been exploited to isolate physically putative stem cells from rat colon. Kim et al. (2004) administered the DNA-labelling dye bromodeoxyuridine (BrdU) to male Fisher rats in drinking water for 2, 4 and 8 weeks, followed by a delabelling period of 2–8 weeks after which animals were sacrificed. Immunohistochemical analysis of whole colonic crypts after 2 weeks’ delabelling confirmed retention of the BrdU label at the bottom of the crypt (the putative stem cell location). After 8 weeks’ delabelling, fluorescence-activated cell sorting (FACS) analysis of cells in the proliferative zone detected BrdU-positivity in 3.8% of nuclei. These BrdU-positive cells are synonymous with long-term label-retaining cells (LRCs) and are therefore considered to represent an enriched stem cell population in the colon (Kim et al. 2004).
Putative biomarkers of colonic epithelial stem cells
Stem cells must be defined by their function, as they do not possess any distinguishing morphological features. Ultimately the number and position of intestinal stem cells can only be deduced, rather than definitively proved, from functional responses to experimental manipulation which may actually alter stem cell properties (Potten & Loeffler, 1990). Furthermore, the very existence of clonogenic cells may be contributing to the varied estimates in stem cell number (Potten et al. 1997). Consequently, a method of exclusively demarcating the intestinal stem cell compartment is of paramount importance and the best way to achieve this is by identifying a definitive biomarker.
Msi-1, an RNA-binding protein (Nakamura et al. 1994; Sakakibara et al. 1996), and Hes1, a transcriptional repressor (Sasai et al. 1992) transactivated by Msi-1 (Imai et al. 2001), have been assigned key roles in preserving the phenotype of neural stem cells and early neuronal progenitors (Nakamura et al. 2000; Okano et al. 2005). Similarly, these two proteins have been considered as prime candidates for investigation as putative biomarkers of stem cells and early lineage precursors of the intestinal epithelium (Kayahara et al. 2003; Nishimura et al. 2003; Potten et al. 2003).
Nishimura et al. (2003) immunohistochemically analysed the expression pattern of Msi-1 in 155 normal human colonic crypts and found that 19.0 ± 7.53 (mean ± SD) cells stained positively for Msi-1 in each crypt. The majority of cells (14.6 ± 5.77, mean ± SD) were located in the lower region of the crypt which corresponds to the expected position of the colonic stem cells. Similarly, Msi-1 expression in adult BDF1 mouse colon was concentrated in cells at the bottom of the crypt, further associating Msi-1 activity with the anticipated location of the colonic stem cell niche (Potten et al. 2003). However, Nishimura et al. (2003) also observed that most Msi-1-positive cells were situated in cell positions 1–10, implying that a proportion of the lineage precursor cells retained Msi-1 expression.
In mouse small intestine, a similar situation was reported when Msi-1 positive cells were scored along 50 longitudinal crypt sections (Potten et al. 2003). The frequency of Msi-1-positive cells was highest in lower crypt cells; however, immunoreactivity was also observed above the position attributed to stem cells. This suggested that the immediate proliferating progenitor cells in the small intestine were also identified by the presence of Msi-1. The only other explanations for the high frequency of Msi-1-positive cells are that either stem cell number has been grossly underestimated, or the half-life of Msi-1 is longer than the cycling time of the stem cells (Potten et al. 2003). However, it is more likely that Msi-1 continues to be expressed in the first one or two tiers of transit-amplifying cells (Potten et al. 2003), which retain the potential to revert to a full stem cell phenotype in the event of injury to the crypt (Potten & Loeffler, 1990).
Hes1 expression has been investigated in mouse small intestinal epithelium (Kayahara et al. 2003). Hes1 was found co-expressed in all Msi-1-positive cells that lie just above the Paneth cells, and in the crypt base columnar cells that intercalate the Paneth cells. No Msi-1 or Hes1 expression was found in fully differentiated cells. Hes1 expression was, however, found to be expressed in mid to lower crypt cell nuclei and therefore stained a broader range of cells than Msi-1. Consequently, Hes1/Msi-1 co-expression may delineate small intestinal stem cells, and Hes1 single expression may distinguish lineage precursor cells (Kayahara et al. 2003). No comparable data are available for Hes1 expression in the colon.
It is worth noting that the Hes1 single and Hes1/Msi-1 double-positive cells also expressed the proliferation marker Ki67, adding further weight to the argument that Hes1 single-positive cells are transit-amplifying cells (Kayahara et al. 2003). However, the indicated high proliferation index of double-positive cells casts doubt on the suitability of Hes1 and Msi-1 as biomarkers, considering the slower cell cycle of the stem cells. Consequently, further studies need to be undertaken to clarify their reproducibility as biomarkers of colonic and small intestinal stem cells.
An in vitro culture system has been developed to propagate proliferative progenitor cells of the colon (Whitehead et al. 1999). Accordingly, it is considered to be a method by which stem cells can be isolated, although this has been difficult to confirm because it is not possible to identify retrospectively the position within the crypt that any colony-forming cell originally occupied (Whitehead et al. 1999). This so-called clonogenic assay has been used as a tool to evaluate the potential of employing integrin expression as a biomarker of colonic stem cells (Fujimoto et al. 2002). The integrin superfamily of transmembrane glycoproteins are important mediators of cell adhesion, as well as recognized modulators of signalling cascades which influence cell growth, proliferation, differentiation, migration and apoptosis (Howe et al. 1998). Initially, fluorescence analysis of frozen sections of colonic mucosa established that both α2 and β1 integrin subunits localize to the lower portion of the crypt and, most interestingly, β1 integrin expression was restricted to the bottom third of the crypt (Fujimoto et al. 2002). Furthermore, enrichment of colony-forming cells was achieved by separating crypt cells based on their expression of β1 integrin and culturing them using the clonogenic assay (Whitehead et al. 1999; Fujimoto et al. 2002). Nevertheless, further experiments are required to confirm the specificity of β1 integrin expression for colonic stem cells.
EphB receptors are important regulators of cell positioning and migration in the intestinal epithelium (Batlle et al. 2002). They are expressed in a gradient manner, where activity is highest at the bottom of colonic and small intestinal crypts and lowest at the crypt–villus junction, where differentiation is initiated (Batlle et al. 2002; van de Wetering et al. 2002). Consequently, EphB receptors have been considered candidate stem cell biomarkers worth exploring.
Recently, both EphB2 and EphB3 receptor expression was reported in the lower cryptal region of mouse colon, which incorporates the putative stem cell population (Holmberg et al. 2006). Inhibition of EphB2/EphB3 signalling was shown to reduce the number of crypt cells expressing proliferation indices, whereas constitutive overactivation of EphB2 receptor signalling correlated with an increase in the number of proliferating cells compared with wild-type crypts. It was established that these receptors function to regulate proliferation positively by promoting cell cycle re-entry. However, taking into account the strong relationship between EphB signalling and proliferation and the additional finding by Holmberg et al. (2006) that stem cell number remained unchanged in the absence of EphB2/EphB3 signalling in the small intestine, the evidence suggests that EphB receptor expression is unlikely to be an independent biomarker of colonic stem cells.
Most recently, Lgr5 has been proposed as a biomarker of colonic stem cells (Barker et al. 2007). Also known as Gpr49, Lgr5 is a Wnt response element whose expression has been observed in a handful of cells at the base of both colonic and small intestinal crypts (Barker et al. 2007). In the colon, the Lgr5 signal is restricted to a small number of cells situated in the bottom of the crypts which corresponds to the predicted location of the stem cells. Importantly, Lgr5-positive colon cells appear to fulfil the major criteria which define stem cells in that they are both self-maintaining and multipotent. Indeed, it has been demonstrated that Lgr5-expressing cells differentiate into the expected functional lineages of the colonic epithelium (Barker et al. 2007). In short, Lgr5 is the first biomarker to be proposed which appears to identify distinctly colonic stem cells and as such it is one of the most promising candidates to be revealed to date.
Cancer stem cells
Colorectal cancer is a prevalent disease in the Western world. In Europe, there were an estimated 412 900 incident cases in 2006 and approximately 207 400 deaths occurred, constituting 12.2% of all cancer deaths (Ferlay et al. 2007). A similar pattern is recorded for the USA, where more than 140 000 new diagnoses are expected each year and more than 55 000 people die from the disease, accounting for 10% of total cancer deaths (Jemal et al. 2006). Accordingly, CRC is routinely listed as the second most common cause of cancer-related death in the developed world, behind lung cancer (Toms, 2004; Jemal et al. 2006; Ferlay et al. 2007). Consequently, understanding the principles of colorectal tumour initiation, growth and metastasis is of paramount importance if this disease is to be decisively combated and the mortality rate reduced.
The long-held view of tumour development, described in the stochastic model, is that every tumour cell is equally capable of initiating neoplastic growth. This theory is now challenged by the cancer stem cell theory, which suggests that only a small proportion of cells within a tumour actually possess cancer-initiating potential and that these so-called CSCs are responsible for tumour development and sustain tumour growth (Wang & Dick, 2005; Dalerba et al. 2007a). CSCs are proposed to possess stem-cell-like properties, including the ability to self-renew and differentiate. Consequently, they can give rise to heterogeneous tumours. Yet by virtue of self-maintenance, their numbers and accordingly their aggressive properties are consistently retained within a tumour, facilitating tumour expansion and spread (Dalerba et al. 2007a).
The potency of CSCs is demonstrated using xenograft models. The CSC population is refined by FACS sorting of tumour cells according to the expression of ‘signature’ cell surface biomarkers which enrich subfractionated populations for cancer-initiating activity. Proposed biomarkers include CD133 (Singh et al. 2004; Collins et al. 2005; Ma et al. 2007), CD44 (Al-Hajj et al. 2003; Collins et al. 2005; Li et al. 2007; Prince et al. 2007), CD34 (Lapidot et al. 1994; Bonnet & Dick, 1997), CD24 (Al-Hajj et al. 2003; Li et al. 2007) and epithelial-specific antigen (ESA) (Al-Hajj et al. 2003; Li et al. 2007). Injection of CSC-enriched populations into immunocompromised mice at low concentrations results in the formation of tumours with equivalent histology and phenotypic heterogeneity to the original neoplasm, whereas injection of non-cancer stem cells, even at high concentrations, results in the growth of few or no tumours (Al-Hajj et al. 2003; Singh et al. 2004; Li et al. 2007; Prince et al. 2007). Research so far suggests that the molecular ‘signature’ which specifically identifies CSCs is likely to constitute a combination of cell surface proteins that are co-ordinately expressed or repressed. This appears to be the case for pancreatic cancer, in which the CD44+ CD24+ ESA+ phenotype possessed the greatest tumorigenic potential (Li et al. 2007), and for breast cancer, in which rare CD44+ CD24− ESA+ cells were found to propagate the disease (Al-Hajj et al. 2003).
Identification of novel biomarkers of CR-CSCs
Recent work suggests that the tumorigenic cell population of CRC can be isolated according to the positive expression of specific cell surface biomarkers, namely CD133 (O’Brien et al. 2007; Ricci-Vitiani et al. 2007), CD44, CD166 and epithelial cell adhesion molecule (EpCAM), otherwise described as ESA (Dalerba et al. 2007b). In addition, the presence of aldehyde dehydrogenase was proposed as a useful indicator of tumour initiation potential, but only in a subset of CRC xenograft samples (Dalerba et al. 2007b). The functionality of these biomarkers was evaluated using a combination of flow cytometry to sort the disaggregated tumour cell population and xenograft modelling to determine the tumour initiation potential of the cellular fraction in question (Dalerba et al. 2007b; O’Brien et al. 2007; Ricci-Vitiani et al. 2007).
Using limiting dilution analysis, O’Brien et al. (2007) calculated that the frequency of CR-CSCs in an unfractionated population of tumour cells is 1 in 5.7 × 104. However, a CD133+ population of cells was > 200-fold enriched for CRC-initiating activity, with an estimated 1 in every 262 cells being a CR-CSC. CD133+ cells readily developed into tumours upon transplantation at low concentrations into non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice, whereas CD133− cells and non-segregated populations did not induce tumour formation (O’Brien et al. 2007; Ricci-Vitiani et al. 2007). Importantly, the xenografts displayed equivalent morphologic features to the parental tumour which were reproducibly maintained upon serial transplantation of tumours into secondary and tertiary recipients. The molecular heterogeneity seen in the original tumour was recapitulated, as demonstrated by the presence of CD133+ and CD133− cells at similar ratios to the original tumour, indicating that CD133+ cells have stem-cell-like properties of self-renewal and differentiation and are responsible for generating differentiated, yet neoplastic CD133− progeny.
With 1 in 262 CD133+ cells estimated to possess cancer-initiating potential (O'Brien et al. 2007), it is clear, however, that not all CD133+ cells are CR-CSCs. Consequently, additional biomarkers might be predicted to further enrich the CR-CSC population. Furthermore, CD133+ has limitations as a CR-CSC biomarker because it appears not to predict tumour initiation potential in Dukes' stage A tumours (Ricci-Vitiani et al. 2007). To this end, Dalerba et al. (2007b) have identified an entirely different three-molecule ‘signature’ for CR-CSCs. They observed that CD133 expression varied in tumours, with some tumours being completely negative. They found concurrence between CD133 and CD44 positivity and observed that the number of CD133+ cells was usually greater than the number of CD44+ cells, which therefore must represent a subset of the CD133+ population. The group focused on the tumour initiation potential of CD44+/EpCAMHIGH CRC cells. These antigens have been identified already as biomarkers for human breast (Al-Hajj et al. 2003) and pancreatic (Li et al. 2007) CSCs. Several colorectal tumours were enriched for CD44+/EpCAMHIGH. Subcutaneous injection of purified CD44+/EpCAMHIGH cells into NOD/SCID mice resulted in a high frequency of tumours with equivalent phenotypic and morphologic characteristics to the original lesion, whereas CD44−/EpCAMLOW cells appeared to lack tumour-initiating activity. Further subfractionation of the CD44+/EpCAMHIGH cell population using the mesenchymal stem cell marker CD166 more accurately specified the ‘signature’ of CR-CSCs, as demonstrated by successful xenografting of CD44+/EpCAMHIGH/CD166+ cells from two independent primary tumours (Dalerba et al. 2007b).
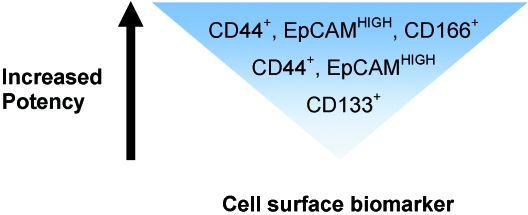
Taken as a whole, current research (Dalerba et al. 2007b; O'Brien et al. 2007; Ricci-Vitiani et al. 2007) implies that the CR-CSC population can be sequentially enriched based on the activity of a repertoire of four cell surface biomarkers, namely, CD133, CD44, EpCAM and CD166. CD133+ cells reportedly represent a larger proportion of a colorectal tumour, compared to CD44+ cells (Dalerba et al. 2007b). Therefore it can be deduced that isolation of CR-CSCs based upon CD133 expression alone would correlate with lowest tumour initiation potency in comparison with enrichment using the three-molecule ‘signature’, CD44+/EpCAMHIGH/CD166+, which would be expected to correlate with greatest tumour initiation potency (Fig. 2).
Fig. 2.
The comparative tumour initiation potency of enriched CR-CSC populations prospectively isolated according to different combinations of cell surface biomarkers, extrapolated from work completed by Dalerba et al. (2007b), O'Brien et al. (2007) and Ricci-Vitiani et al. (2007). CR-CSCs can be separated from the tumour bulk by flow cytometry based on the expression of a repertoire of cell surface proteins. The resulting cellular fractions appear to have different tumour initiation potencies, depending on the combination of cell surface biomarkers expressed.
Conclusion
Molecular biomarkers are hugely beneficial because they enable the definitive identification of particular cell types and cell populations. The adult stem cells of the colon are of particular interest because they both sustain the perpetual self-renewal of healthy colonic epithelium and are the target cell for cancer-initiating mutations (Potten et al. 1997; McDonald et al. 2006). The predisposition of the colon to tumorigenesis compared to the small intestine suggests that identifying a biomarker for colonic stem cells should be a priority. Definitive proof of their putative location at the bottom of the crypt is essential if further understanding of the homeostasis of normal colonic epithelium and the mechanisms of colorectal tumour development is to be achieved. Biomarkers for colonic stem cells and early progenitor cells have been proposed, but there is general uncertainty as to whether they exclusively demarcate the stem cell population.
Identification of biomarkers for CR-CSCs will enable greater understanding of how colorectal tumours grow and progress. To this end, a repertoire of cell surface biomarkers has been suggested to distinguish phenotypically the CSC subset from the non-tumorigenic cells forming the bulk of the tumour. The ability prospectively to isolate the cancer-initiating population and maintain these cells in culture without any significant phenotypic alterations (Ricci-Vitiani et al. 2007) offers a potential avenue for evaluating the efficacy of currently used drugs and for pre-clinical testing of new therapeutic approaches.
Acknowledgments
The authors wish to thank S. F. Rahman-Casañs (Durham) for providing the H&E micrograph of colon histology. We are grateful to AICR, Department of Health R & D fund and EU FP6 for their financial support.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LH. Alimentary system. In: Williams PL, editor. Gray's Anatomy. 38. London: Churchill Livingstone; 1995. pp. 1782–1785. [Google Scholar]
- Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Booth C, Potten CS. Gut instincts: thoughts on intestinal epithelial stem cells. J Clin Invest. 2000;105:1493–1499. doi: 10.1172/JCI10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WB, Roberts SA, Potten CS. The number of clonogenic cells in crypts in three regions of murine large intestine. Int J Radiat Biol. 1997;71:573–579. doi: 10.1080/095530097143905. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007a;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007b;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
- Hendry JH, Potten CS, Ghafoor A, Moore JV, Roberts SA, Williams PC. The response of murine intestinal crypts to short-range promethium-147 beta irradiation: deductions concerning clonogenic cell numbers and positions. Radiat Res. 1989;118:364–374. [PubMed] [Google Scholar]
- Holmberg J, Genander M, Halford MM, et al. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Howe A, Aplin AE, Alahari SK, Juliano RL. Integrin signaling and cell growth control. Curr Opin Cell Biol. 1998;10:220–231. doi: 10.1016/s0955-0674(98)80144-0. [DOI] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- Kaur P, Potten CS. Cell migration velocities in the crypts of the small intestine after cytotoxic insult are not dependent on mitotic activity. Cell Tissue Kinet. 1986;19:601–610. doi: 10.1111/j.1365-2184.1986.tb00761.x. [DOI] [PubMed] [Google Scholar]
- Kayahara T, Sawada M, Takaishi S, et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535:131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Cheung S, Hellerstein MK. Isolation of nuclei from label-retaining cells and measurement of their turnover rates in rat colon. Am J Physiol Cell Physiol. 2004;286:C1464–1473. doi: 10.1152/ajpcell.00139.2003. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. Bioessays. 2002;24:91–98. doi: 10.1002/bies.10028. [DOI] [PubMed] [Google Scholar]
- McDonald SA, Preston SL, Lovell MJ, Wright NA, Jankowski JA. Mechanisms of disease: from stem cells to colorectal cancer. Nat Clin Pract Gastroenterol Hepatol. 2006;3:267–274. doi: 10.1038/ncpgasthep0473. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sakakibara S, Miyata T, et al. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Wakabayashi N, Toyoda K, Kashima K, Mitsufuji S. Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig Dis Sci. 2003;48:1523–1529. doi: 10.1023/a:1024763723240. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Potten CS. Cell cycles in cell hierarchies. Int J Radiat Biol Relat Stud Phys Chem Med. 1986;49:257–278. doi: 10.1080/09553008514552541. [DOI] [PubMed] [Google Scholar]
- Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- Potten CS, Owen G, Roberts SA. The temporal and spatial changes in cell proliferation within the irradiated crypts of the murine small intestine. Int J Radiat Biol. 1990;57:185–199. doi: 10.1080/09553009014550431. [DOI] [PubMed] [Google Scholar]
- Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol. 1997;78:219–243. doi: 10.1046/j.1365-2613.1997.280362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Booth C, Tudor GL, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JM, Roberts SA, Potten CS. Cell migration in the small and large bowel shows a strong circadian rhythm. Epithelial Cell Biol. 1994;3:137–148. [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Imai T, Hamaguchi K, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- Toms JR. CancerStats Monograph 2004. London: Cancer Research UK; 2004. [Google Scholar]
- Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Whitehead RH, Demmler K, Rockman SP, Watson NK. Clonogenic growth of epithelial cells from normal colonic mucosa from both mice and humans. Gastroenterology. 1999;117:858–865. doi: 10.1016/s0016-5085(99)70344-6. [DOI] [PubMed] [Google Scholar]