Abstract
Organisms that use the standard genetic code recognize UAA, UAG, and UGA as stop codons, while variant code species frequently alter this pattern of stop codon recognition. We previously demonstrated that a hybrid eRF1 carrying the Euplotes octocarinatus domain 1 fused to Saccharomyces cerevisiae domains 2 and 3 (Eo/Sc eRF1) recognized UAA and UAG, but not UGA, as stop codons. In the current study, we identified mutations in Eo/Sc eRF1 that restore UGA recognition and define distinct roles for the TASNIKS and YxCxxxF motifs in eRF1 function. Mutations in or near the YxCxxxF motif support the cavity model for stop codon recognition by eRF1. Mutations in the TASNIKS motif eliminated the eRF3 requirement for peptide release at UAA and UAG codons, but not UGA codons. These results suggest that the TASNIKS motif and eRF3 function together to trigger eRF1 conformational changes that couple stop codon recognition and peptide release during eukaryotic translation termination.
INTRODUCTION
Translation termination occurs when one of three stop codons, UAA, UAG or UGA, enters the ribosomal A site. Two classes of release factors mediate this process in both prokaryotes and eukaryotes, although the mechanisms are quite distinct. Prokaryotes have two class I release factors, RF1 and RF2. RF1 recognizes UAA and UAG stop codons through the action of the linear tri-peptide anticodon Pro-Ala-Thr (PAT), while RF2 recognizes UAA and UGA codons using the tri-peptide anticodon Ser-Pro-Phe (SPF) (Ito et al., 2000; Nakamura and Ito, 2002). In these critical stop codon recognition sequences, the first and third amino acids are thought to discriminate the second and third purine bases, respectively. The prokaryotic class II release factor, RF3, uses GTP hydrolysis to recycle the class I factors after polypeptide chain release (Zavialov et al., 2001). Eukaryotes have only one class I release factor, eRF1, which recognizes all three stop codons (Kisselev et al., 2003). The mechanism used by eRF1 to recognize the three stop codons is still unknown. The eukaryotic class II release factor, eRF3, facilitates eRF1 stop codon recognition and carries out GTP hydrolysis prior to polypeptide chain release (Alkalaeva et al., 2006; Salas-Marco and Bedwell, 2004).
eRF1 proteins from eukaryotic species share significant homology at the primary amino acid sequence level and are composed of three distinct functional domains (Song et al., 2000). Domain 1 is thought to recognize stop codons in the ribosomal A site, and contains the highly conserved TASNIKS and YxCxxxF motifs (Bertram et al., 2000; Frolova et al., 2002; Inagaki et al., 2002; Song et al., 2000). Domain 2, which contains the universally conserved GGQ motif, interacts with the peptidyl transferase center of the ribosome to trigger peptidyl-tRNA hydrolysis (Frolova et al., 1999; Seit-Nebi et al., 2001; Song et al., 2000). Domain 3 of eRF1 mediates eRF3 binding (Eurwilaichitr et al., 1999; Ito et al., 1998).
Organisms that deviate from the standard genetic code are called variant code organisms. Among these organisms, many ciliate species have reassigned one or more stop codons. For example, Tetrahymena species reassigned both UAA and UAG to glutamine codons and retain only UGA as a stop codon, while Euplotes species reassigned UGA to a cysteine codon and retain UAA and UAG as stop codons. Ciliate eRF1 proteins retain substantial overall amino acid sequence homology with eRF1s from standard code organisms. It has been suggested that highly conserved amino acid motifs in standard code organisms that are more degenerate in variant code organisms may represent key residues that mediate stop codon recognition (Kim et al., 2005; Knight et al., 2000; Lozupone et al., 2001; Song et al., 2000). The TASNIKS and YxCxxxF motifs (amino acids 55–61 and 122–128 of S. cerevisiae eRF1, respectively) have been implicated in stop codon recognition by this approach. Subsequent in vitro studies have shown that various mutations in these elements alter the efficiency of polypeptide chain release (Frolova et al., 2002; Seit-Nebi et al., 2002). However, definitive evidence that these sequence elements are responsible for stop codon recognition has remained elusive.
In a recent study, domain 1 from Euplotes octocarinatus eRF1 was fused to domains 2 and 3 of S. cerevisiae eRF1 and the resulting hybrid protein (referred to hereafter as Eo/Sc eRF1) was tested for the ability to complement a knockout of the essential SUP45 gene that encodes eRF1 (Salas-Marco et al., 2006). This Eo/Sc eRF1 could not support viability as the sole source of eRF1 in yeast cells, and was subsequently shown to recognize UAA and UAG stop codons, but not the UGA stop codon. These results demonstrated that domain 1 of Euplotes eRF1 was sufficient to recapitulate variant stop codon recognition. In the current study, we identified several mutations in Eo/Sc eRF1 that restore UGA recognition and define distinct yet cooperative roles for the TASNIKS and YxCxxxF motifs in eRF1 function. In particular, mutations in the Eo/Sc eRF1 TASNIKS motif eliminated the eRF3 requirement for peptide release at UAA and UAG codons, but not UGA codons. These results suggest that the TASNIKS motif and eRF3 function together to modulate the conformation of eRF1 during eukaryotic translation termination.
RESULTS
Identification of intragenic suppressor mutations in a hybrid Eo/Sc eRF1
We previously found that a hybrid Eo/Sc eRF1 was unable to support growth when provided as the only source of eRF1 in yeast cells (Salas-Marco et al., 2006). Dual luciferase readthrough assays (Keeling et al., 2004; Salas-Marco and Bedwell, 2005) revealed that Eo/Sc eRF1 efficiently recognized UAA and UAG as stop codons, but not the UGA codon. These findings led us to hypothesize that intragenic suppressor mutations that restore UGA recognition should also restore the growth of cells expressing Eo/Sc eRF1, thus identifying functional determinants of UGA recognition.
A centromeric plasmid carrying the SUP45 gene encoding the Eo/Sc hybrid eRF1 under SUP45 promoter control was subjected to random mutagenesis and a pooled mutant library was transformed into a sup45Δ strain that also carried a plasmid encoding the wild type SUP45 under GAL1 promoter control (Figure 1A). Transformants were selected at 30°C on plates containing glucose to shut off expression of the wild type SUP45 gene. Three independent suppressors that restored growth carried the same Cys to Ser substitution at codon 124 (C124S; S. cerevisiae eRF1 numbering) in the highly conserved YxCxxxF motif.
Figure 1.
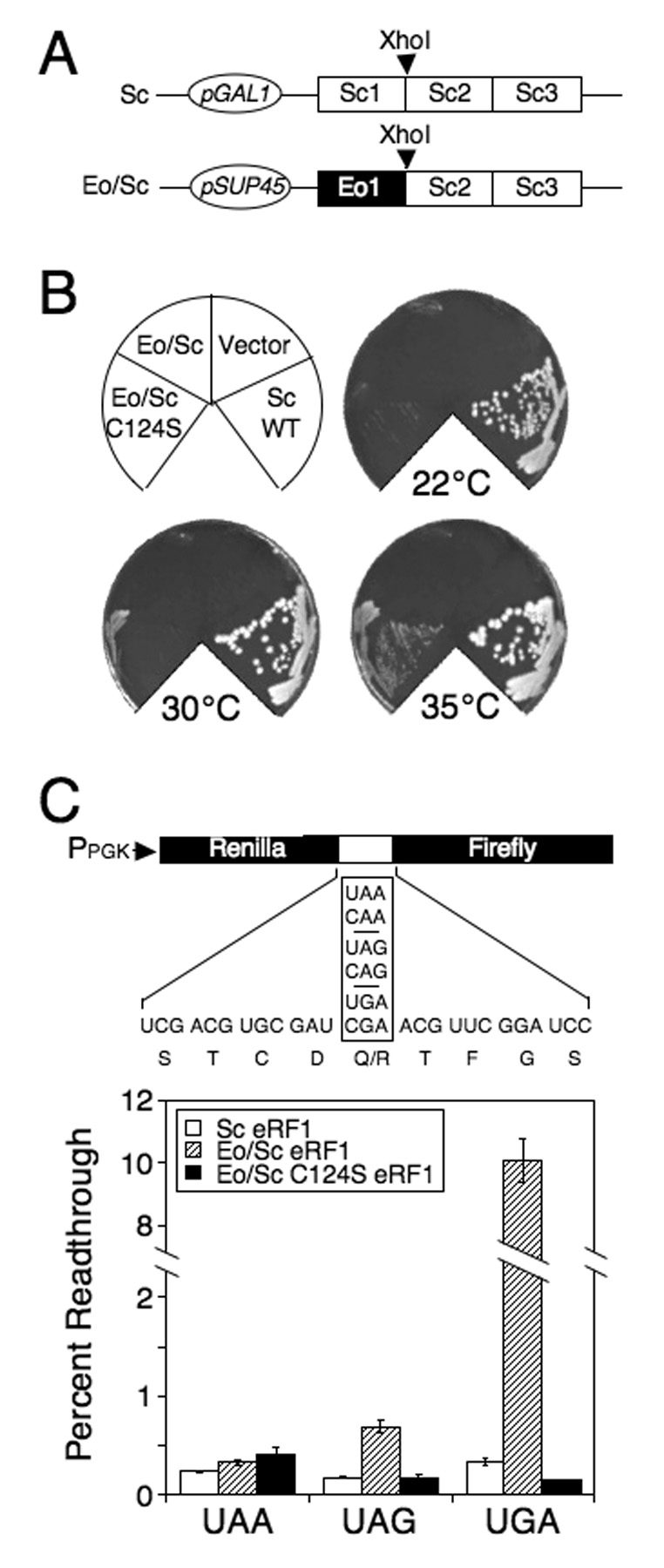
Mutagenesis of Eo/Sc eRF1. (A) The Eo/Sc eRF1 plasmid used for random mutagenesis and the Sc eRF1 plasmid under GAL1 promoter control used to support growth of the sup45Δ strain while screening for suppressors of the hybrid Eo/Sc eRF1. (B) Growth of strain carrying Eo/Sc C124S eRF1. Plates were incubated at 22°C, 30°C and 35°C for 5 days. (C) Readthrough levels of wild type Sc eRF1, Eo/Sc eRF1 and Eo/Sc C124S eRF1 at 35°C. Readthrough in strains carrying the Eo/Sc eRF1 that cannot support cell growth as the sole source of eRF1 was carried out using a galactose to glucose shift protocol as described in the Materials and Methods. Readthrough values are represented as mean ± SD.
Mutations at C124 of the YxCxxxF motif restore UGA recognition by Eo/Sc hybrid eRF1
The Eo/Sc C124S eRF1 mutant restored cell viability, but was associated with a slow growth and cold-sensitive phenotype (Figure 1B and Table 1). To determine the efficiency of stop codon recognition by Eo/Sc C124S eRF1, a dual luciferase-based readthrough assay was carried out (Grentzmann et al., 1998; Howard et al., 2000; Keeling et al., 2004; Salas-Marco and Bedwell, 2004, 2005; Salas-Marco et al., 2006). This assay monitors the efficiency of stop codon recognition, since efficient release factor binding should preclude suppression of the stop codon by a near-cognate tRNA. As controls, stop codon recognition was also measured in strains expressing either wild type Sc eRF1 or the original Eo/Sc hybrid eRF1 (Figure 1C). Stop codon recognition was efficient (<0.4% readthrough) at all three stop codons in the strain expressing wild type Sc eRF1. Since the Eo/Sc eRF1 (expressed under SUP45 promoter control) was unable to support cell viability, growth of the strain expressing this construct was maintained by a plasmid expressing wild type Sc eRF1 under GAL1 promoter control. This strain was grown in SM galactose medium, and then shifted to SM glucose to shut off wild type eRF1 expression. After growth for 6 more generations, the residual level of Sc eRF1 was <10% of normal (Salas-Marco et al., 2006), which allowed us to estimate the stop codon recognition of Eo/Sc eRF1. We found that Eo/Sc eRF1 exhibited low readthrough (<0.7%) at UAA and UAG stop codons, but >10% readthrough at a UGA codon. These results confirmed our previous finding that Eo/Sc eRF1 efficiently recognizes UAA and UAG codons, but is severely compromised in UGA recognition.
Table 1.
Summary of readthrough and growth data for eRF1 variants used in this study
| Growth¥ |
Percent readthrough† |
eRF3 dependence for release activity |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type of eRF1 | Mutations | 22°C | 30°C | 35°C | UAA | UAG | UGA | UAA | UAG | UGA |
| Eo/Sc eRF1 | WT | − | − | − | 0.32±0.03 | 0.69±0.07 | 10.70±0.69 | YES | YES | nr |
| Eo/Sc eRF1 | C124S | − | +/− | + | 0.40±0.06 | 0.17±0.04 | 0.14±0.004 | YES | YES | YES |
| Eo/Sc eRF1 | C124N | − | + | + | 0.32±0.04 | 0.20±0.03 | 0.12±0.02 | nd | nd | nd |
| Eo/Sc eRF1 | A75S | − | + | ++ | 0.13±0.02 | 0.17±0.02 | 0.86±0.13 | nd | nd | nd |
| Eo/Sc eRF2 | A75S/C124S | +/− | + | ++ | nd | nd | nd | nd | nd | nd |
| Eo/Sc eRF1 | A75S/C124N | − | ++ | +++ | 0.22±0.05 | 0.29±0.04 | 0.11±0.02 | nd | nd | nd |
| Eo/Sc eRF1 | E57S | − | − | − | nd | nd | nd | nd | nd | nd |
| Eo/Sc eRF1 | S58N | − | − | − | nd | nd | nd | nd | nd | nd |
| Eo/Sc eRF1 | E57S/S58N | − | +/− | ++ | 0.24±0.07 | 0.20±0.04 | 0.71±0.11 | NO | NO | YES |
| Eo/Sc eRF1 | E57S/C124S | − | + | +++ | nd | nd | nd | nd | nd | nd |
| Eo/Sc eRF1 | S58N/C124S | − | ++ | +++ | nd | nd | nd | nd | nd | nd |
| Eo/Sc eRF1 | E57S/S58N/C124S | ++ | +++ | +++ | 0.15±0.01 | 0.15±0.01 | 0.18±0.02 | nd | nd | nd |
| Eo/Sc eRF1 | E57S/S58N/A75S | + | ++ | +++ | nd | nd | nd | nd | nd | nd |
| Eo/Sc eRF1 | E57S/S58N/CΔ19 | − | +/− − | +/− | 0.14±0.01 | 0.16±0.02 | 6.9±0.51 | NO | NO | YES |
| Sc eRF1 | WT | +++ | ++++ | ++++ | 0.26±0.02 | 0.14±0.01 | 0.39±0.06 | YES | YES | YES |
| Sc eRF1 | C124S | ++ | +++ | +++ | 0.34±0.04 | 0.22±0.03 | 0.17±0.03 | nd | nd | nd |
| Sc eRF1 | S57E | + | ++ | ++ | 0.19±0.02 | 0.22±0.02 | 1.04±0.04 | nd | nd | nd |
| Sc eRF1 | N58S | + | ++ | ++ | 0.34±0.02 | 0.19±0.01 | 0.93±0.11 | nd | nd | nd |
| Sc eRF1 | S57E/N58S | +/− | + | ++ | 0.18±0.01 | 0.20±0.02 | 0.80±0.05 | nd | nd | nd |
| Sc eRF1 | depleted | − | − | − | 5.27±0.54 | 4.68±0.08 | 18.36±2.10 | nd | nd | nd |
| Sc eRF1 | S57E/C124S | − | − | − | 0.58±0.07 | 1.13±0.05 | 0.62±0.04 | nd | nd | nd |
| Sc eRF1 | N58S/C124S | − | − | − | 5.07±0.57 | 4.78±0.58 | 0.58±0.10 | nd | nd | nd |
| Sc eRF1 | S57E/N58S/C124S | − | − | − | 4.01±0.30 | 3.89±0.06 | 2.58±0.27 | nd | nd | nd |
| Sc eRF1 | CΔ19 | − | +/− − | + | 1.4±0.20 | 1.1±0.10 | 6.6±0.30 | YES | YES | YES |
Symbols used to denote growth on YPD plates: (−) no growth; (+/− −) very weak growth; (+/−) weak growth; (+) modest growth; (++)moderate growth; (+++) good growth; (++++) optimal wild type growth as seen at 30°C. nd, not determined. nr, not recognized.
Readthrough values are represented as mean ± SD.
We found that the strain expressing Eo/Sc C124S eRF1 exhibited <0.4% readthrough at UAA and UAG stop codons, indicating that efficient recognition was retained at these stop codons (Figure 1C). Remarkably, only ~0.15% readthrough was measured at the UGA stop codon, which was less than the readthrough observed with wild type Sc eRF1. These results indicate that the C124S mutation successfully restored efficient recognition of the UGA stop codon without significantly compromising recognition of the UAA or UAG stop codons.
The cysteine residue of the YxCxxxF motif in domain 1 is universally conserved among known eukaryotic eRF1 proteins (including variant code ciliate species) (Kim et al., 2005; Kisselev et al., 2003). To determine whether other amino acid substitutions at this position also restore UGA recognition by Eo/Sc eRF1, we tested each of the other 18 amino acids at this position. We found that only C124N supported cell viability, and it resulted in a cold-sensitive phenotype like the C124S mutation (Table 1). Readthrough assays carried out with strains expressing Eo/Sc C124N eRF1 revealed very low readthrough (~0.1%) at the UGA stop codon, and 0.2 to 0.4% readthrough at the UAA and UAG codons (Figure 2B). These results indicate that the C124N mutation, like C124S, is able to efficiently restore UGA stop codon recognition without compromising UAA and UAG recognition.
Figure 2.
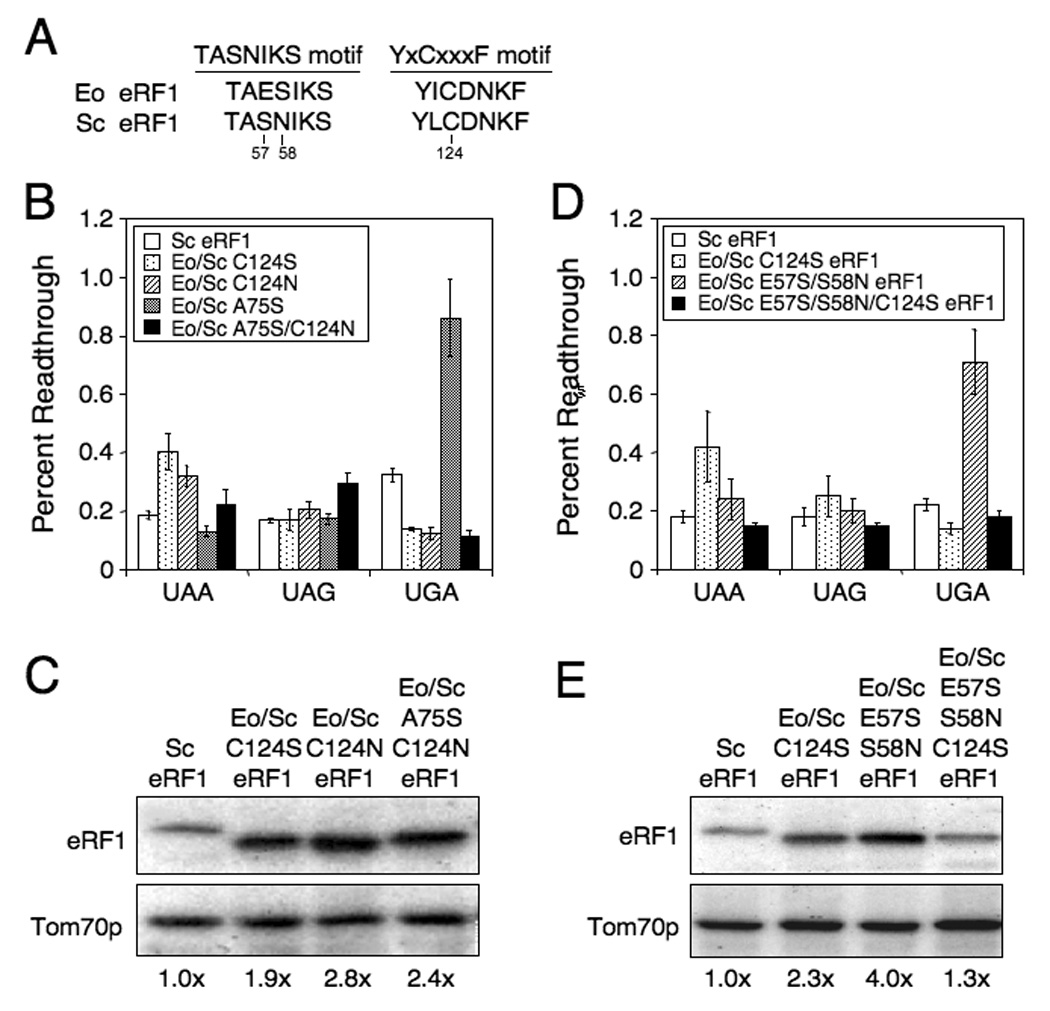
Efficiency of stop codon recognition mediated by Eo/Sc eRF1 suppressor mutants. (A) Alignment of TASNIKS and YxCxxxF motifs from Saccharomyces eRF1 and Euplotes eRF1. Numbering from Saccharomyces eRF1 is used. (B) Readthrough of stop codons measured in strains expressing wild type Sc eRF1 or Eo/Sc C124S, C124N, A75S or A75S/C124N eRF1. (C) Western blot quantitation of eRF1 levels from strains in (B). (D) Readthrough of stop codons measured in strains expressing wild type Sc eRF1 or Eo/Sc C124S, E57S/S58N, or E57S/S58N/C124S eRF1. (E) Western blot quantitation of eRF1 levels from strains in (D). All strains were grown in SM glucose medium at 35°C. Readthrough values are represented as mean ± SD.
While characterizing the C124N mutation, we found a fast growing colony that had spontaneously acquired a second mutation, A75S (S. cerevisiae eRF1 numbering; corresponds to T75 in Sc eRF1). Interestingly, this mutation is located near C124 in the three-dimensional structure of eRF1. Strains expressing Eo/Sc eRF1 proteins with a combination of either the A75S/C124N or A75S/C124S mutations grew better at 30°C and 35°C than a strain expressing an Eo/Sc eRF1 with either C124 mutation alone (Table 1). Furthermore, the Eo/Sc A75S eRF1 supported cell growth as the sole source of eRF1. Readthrough assays carried out on strains expressing Eo/Sc A75S eRF1 showed very low readthrough (0.2%) at UAA and UAG codons, but 0.9% readthrough at UGA (Figure 2B). Thus, the A75S mutation restored UGA recognition less efficiently than the C124S or C124N mutations. However, the combination of the A75S and C124N mutations resulted in less than 0.2% readthrough at the UGA codon, and 0.2% to 0.3% readthrough at UAA and UAG codons. Given that growth is better when A75S is combined with either C124N or C124S, it appears that these mutations together optimize an aspect of eRF1 function not reflected by the readthrough assay.
Since changes in the steady-state level of the Eo/Sc eRF1 suppressors might influence the overall termination efficiency, we next examined the relative abundance of each Eo/Sc eRF1 by western blot analysis. We found that the steady-state level of eRF1 was not reduced by any of the suppressor mutations. In fact, the steady-state level of the hybrid eRF1 suppressors was elevated 2 to 3-fold in the strains that had regained UGA recognition (Figure 2C). These results are consistent with our recent finding that a novel regulatory mechanism increases steady-state eRF1 protein levels when translation termination is compromised in Saccharomyces cerevisiae (Kallmeyer and Bedwell, unpublished results).
TASNIKS mutations also restore UGA recognition and function cooperatively with C124S in the Eo/Sc hybrid eRF1
Previous studies have implicated the NIKS motif (residues 58 to 61 of S. cerevisiae eRF1) in stop codon recognition (Frolova et al., 2002; Seit-Nebi et al., 2002). This motif is highly conserved in eRF1 proteins from standard code organisms, and crosslinking studies suggest that it is in close proximity to stop codons in the A site (Chavatte et al., 2002). Furthermore, this motif is frequently altered in variant code organisms, suggesting that divergence from the consensus TASNIKS sequence may contribute to changes in stop codon recognition (Inagaki and Doolittle, 2001; Knight et al., 2000). In Euplotes octocarinatus eRF1a (used in this study), this motif has the sequence TAESIKS, which differs from the consensus TASNIKS by the presence of a glutamic acid (E) at residue 57 and serine (S) at residue 58 (Figure 2A). To determine the importance of these variations on stop codon recognition, we introduced single amino acid changes in the Eo/Sc hybrid eRF1 (E57S or S58N) or a double mutant (E57S/S58N) that restored the standard TASNIKS motif. Viability assays indicated that the single mutations did not allow the growth of cells expressing these mutants as the sole source of eRF1. In contrast, the Eo/Sc eRF1 containing both mutations (Eo/Sc E57S/S58N eRF1) restored viability, but growth was again cold sensitive (Table 1).
We next asked whether combining mutations in the TASNIKS and YxCxxxF motifs could further improve growth of the strain expressing Eo/Sc eRF1. We found that strains expressing Eo/Sc eRF1 proteins containing either single TASNIKS mutation, E57S or S58N, in conjunction with the YxCxxxF mutation C124S mutation were viable and had better growth than a strain expressing the Eo/Sc C124S eRF1 alone (Table 1). The triple mutant, Eo/Sc E57S/S58N/C124S eRF1, exhibited the best growth. These results indicated that changes in the TASNIKS motif and C124 are cooperative with respect to cell growth. We also examined the affects of combining the E57S/S58N mutations with A75S in Eo/Sc eRF1. Viability assays showed that this combination of mutations also improved growth as compared to either of the two TASNIKS mutations or the A75S mutation separately. The E57S/S58N/A75S mutant grew less well than the E57S/S58N/C124S mutant, but better than the A75S/C124S mutant.
Readthrough assays carried out with a strain expressing Eo/Sc E57S/S58N eRF1 showed 0.2% readthrough at UAA and UAG, but 0.7% readthrough at UGA (Figure 2D). These results indicated that the E57S/S58N mutations were less efficient in restoring UGA recognition than the C124S mutation alone. However, the E57S/S58N/C124S triple mutant exhibited less than 0.2% readthrough at all three stop codons. This level of readthrough was lower than the Eo/Sc hybrid eRF1 carrying the two TASNIKS mutations at all three stop codons, and less than the hybrid eRF1 carrying the C124S mutation at the UAA and UAG codons. Again, these results suggest that the C124S and E57S/S58N mutations act cooperatively to fine-tune overall eRF1 function. Consistent with this premise, western blot analysis of the steady-state levels of these eRF1 proteins revealed a 2.3-fold excess of Eo/Sc C124S eRF1 and a 4-fold excess of Eo/Sc E57S/S58N eRF1, while the Eo/Sc E57S/S58N/C124S eRF1 level was reduced to a level only 1.3-fold above normal (Figure 2E). Since increased eRF1 levels correlate with a defect in translation termination, these results suggest that overall eRF1 function was better when all three mutations were present.
TASNIKS and C124 mutations reduce stop codon recognition by S. cerevisiae eRF1
Our results indicate that both the TASNIKS motif and C124 are important for UGA recognition by Eo/Sc eRF1. To determine the role of the corresponding residues in an eRF1 from a standard code organism, we introduced the C124S mutation and altered the TASNIKS motif to the TAESIKS element found in Euplotes eRF1 by introducing single (S57E or N58S) or double mutations (S57E/N58S) into domain 1 of Sc eRF1. Viability assays showed that Sc eRF1s with the single mutations, C124S, S57E or N58S could support cell viability, but all reduced the growth rate relative to a strain expressing wild type Sc eRF1 (Table 1). The double TASNIKS mutant (S57E/N58S) resulted in a more severe growth defect. Readthrough assays carried out with a strain expressing Sc C124S eRF1 showed 0.35% and 0.2% readthrough at UAA and UAG respectively, similar to wild type Sc eRF1 (Figure 3A). Readthrough at the UGA codon was less than 0.2%, which was about two fold less than the readthrough observed with wild type Sc eRF1. These results indicate that the C124S mutation makes UGA recognition more efficient in a standard code eRF1, as it does in the variant code Eo/Sc eRF1.
Figure 3.
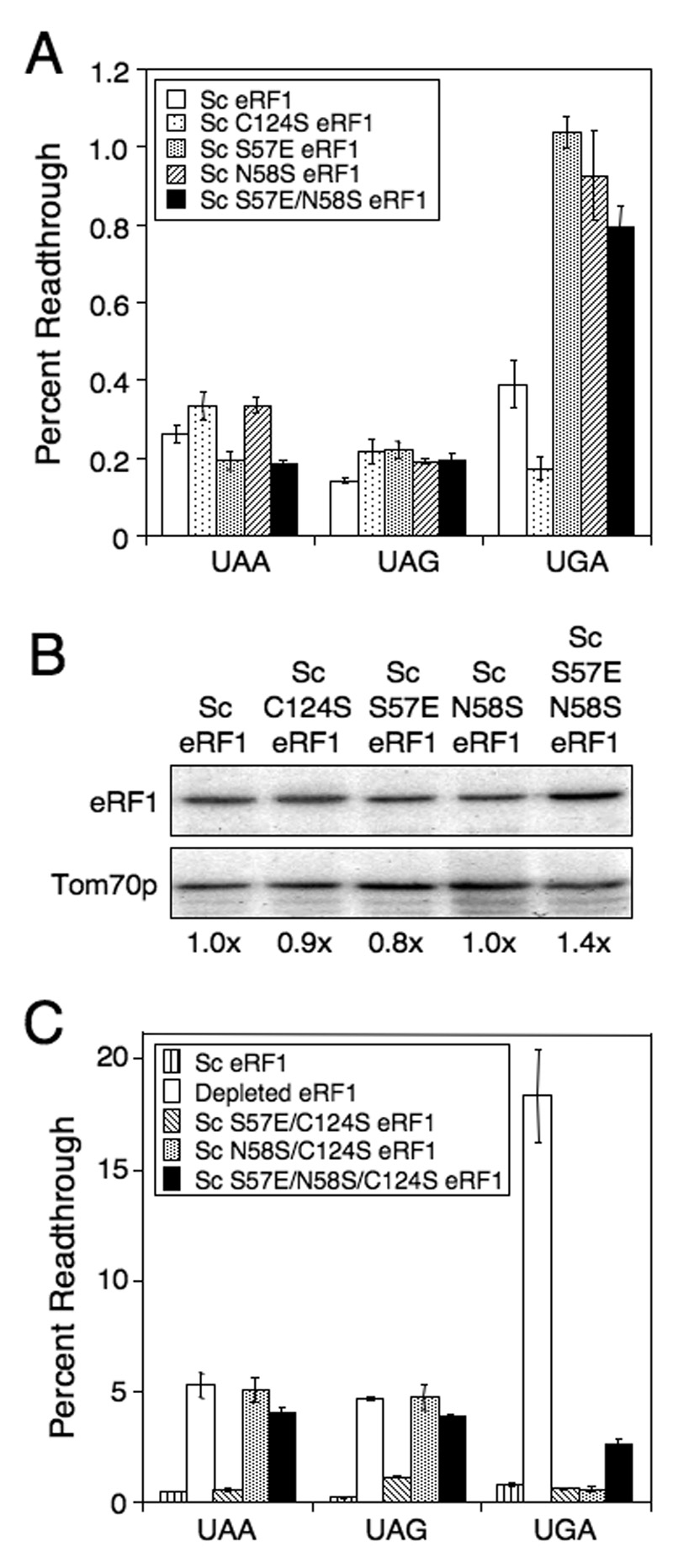
Efficiency of stop codon recognition mediated by Sc eRF1 carrying C124S or TASNIKS mutations. (A) Readthrough of stop codons measured in strains expressing Sc eRF1 with C124S or viable TASNIKS mutations. (B) Western blot quantitation of eRF1 levels from strains in (A). (C) Readthrough in strains carrying Sc eRF1, depleted of eRF1, or carrying combinations of Sc eRF1 mutations that result in an inability to support growth as the sole source of eRF1. For mutant eRF1 proteins unable to support growth, readthrough was measured using a galactose to glucose shift protocol as described in the Materials and Methods. All strains were grown in SM glucose medium at 35°C. Readthrough values are represented as mean ± SD.
Readthrough assays of Sc eRF1 proteins carrying the S57E, N58S and S57E/N58S mutations showed a low level of readthrough (0.2 to 0.35%) at the UAA and UAG stop codons. However, 0.8% to 1.0% readthrough was observed at the UGA stop codon in strains expressing these mutant proteins, a ≥2-fold increase in readthrough compared to wild type Sc eRF1 (Figure 3A). This suggests that these changes in the TASNIKS motif of Sc eRF1 diminished the efficiency of UGA recognition, consistent with the observation that the reciprocal mutations that improved the TASNIKS homology in Eo/Sc eRF1 increased the efficiency of UGA recognition. The protein levels of all of these Sc eRF1 mutants were essentially normal (Figure 3B).
When we combined the S57E or N58S TASNIKS mutations with C124S in Sc eRF1, we found that none of the strains expressing pairwise combinations or the triple mutant were viable (Table 1). To determine the effect of the mutations on stop codon recognition, we carried out readthrough assays on strains expressing these mutant Sc eRF1 proteins in yeast cells after wild type Sc eRF1 had been depleted by growth in the absence of wild type Sc eRF1 for six generations. A strain constitutively expressing wild type Sc eRF1 was used as a positive control, while a strain depleted of eRF1 was used as a negative control (Figure 3C). The wild type strain exhibited 0.5% readthrough at the UAA stop codon in this series of experiments, while 5.3% readthrough was observed in the eRF1-depleted strain. The strain expressing Sc S57E/C124S eRF1 exhibited a wild type level of readthrough (0.6%) at the UAA codon. In contrast, the strains expressing Sc N58S/C124S eRF1 and S57E/N58S/C124S eRF1 resulted in 5.1% and 4.0% readthrough, respectively, at the UAA stop codon (8 to 10-fold higher than wild type eRF1). This level was similar to the readthrough observed following eRF1 depletion.
At the UAG stop codon, readthrough in the strain expressing wild type eRF1 was 0.2%, while readthrough in the eRF1-depleted strain was 4.7%. Readthrough in the strain expressing Sc S57E/C124S eRF1 was 1.1% (5 fold higher than wild type eRF1). Readthrough in strains expressing Sc N58S/C124S eRF1 or Sc S57E/N58S/C124S eRF1 was 4.8% and 3.9% respectively, again near the level observed following eRF1 depletion. At the UGA stop codon, 0.8% readthrough was observed in the strain expressing wild type Sc eRF1, while 18.4% readthrough was measured following eRF1 depletion. Readthrough at the UGA codon in strains expressing Sc eRF1 carrying a single TASNIKS mutation and C124S (S57E/C124S or N58S/C124S) was normal (0.6%), while readthrough in a strain expressing Sc eRF1 with both TASNIKS mutations and C124S (S57E/N58S/C124S) was 2.6%, or 4.3-fold higher than the strain expressing wild type Sc eRF1.
These results indicated that Sc S57E/C124S eRF1 exhibits only a partial defect in UAG recognition. In contrast, the strain expressing Sc N58S/C124S eRF1 had a severe defect in both UAA and UAG recognition, but retained efficient UGA recognition. This demonstrated that these two mutations were sufficient to convert Sc eRF1 to a UGA-only pattern of recognition. The Sc S57E/N58S/C124S eRF1 had a severe defect in UAA and UAG recognition, and also exhibited a partial defect in UGA recognition. This indicated that the S57 and N58 residues in the TASNIKS motif are important for UAA and UAG recognition, but are less important for UGA recognition. In contrast, the C124S mutation exerted a negative effect on UAA and UAG stop codon recognition when combined with the N58S TASNIKS mutation, indicating that C124 influences stop codon recognition in conjunction with this TASNIKS residue. Only when all three mutations are combined did UGA recognition deteriorate. When taken together, these results demonstrate that the TASNIKS and YxCxxxF motifs function cooperatively to mediate efficient stop codon recognition in Sc eRF1.
In vitro peptide release activity of Eo/Sc eRF1 suppressor mutants at UAA, UAG and UGA stop codons
We next examined the relative efficiency of the Eo/Sc eRF1 suppressors in mediating polypeptide release at different stop codons using a recently described in vitro peptide release assay (Alkalaeva et al., 2006). In this assay, mammalian pre-termination complexes (pre-TCs) containing peptidyl-tRNA in the P site and a stop codon in the A site were assembled on Met-Val-His-Cys (MVHC-STOP) mRNAs (encoding a MVHC tetrapeptide followed by either a UAA, UAG or UGA stop codon) using 40S and 60S ribosomal subunits, purified initiation and elongation factors, and aminoacylated tRNAs. Pre-TCs were purified by sucrose gradient centrifugation, and peptide release was initiated by the addition of eRFs. Since previous studies demonstrated that GTP hydrolysis by eRF3 is a prerequisite for efficient stop codon recognition and peptide release (Alkalaeva et al., 2006; Salas-Marco and Bedwell, 2004), peptide release induced by Eo/Sc eRF1 suppressors was investigated in the absence and in the presence of eRF3. Release of M-V-H-[35S]C tetrapeptide as a function of time was monitored by scintillation counting of supernatants after TCA precipitation of reaction mixtures.
We first verified the ability of yeast eRFs (Sc eRF1 and Sc eRF3) to promote peptide release on mammalian pre-TCs. Incubation of pre-TCs for 20 minutes with saturating amounts of Sc eRF1 alone or in combination with Sc eRF3 in the presence of GTP resulted in nearly complete peptide release, just as with human (Hs) eRFs (Figure 4A). Release mediated by the combination of Sc eRF1 and Sc eRF3 was inhibited by the non-hydrolyzable GTP analog GMPPNP, again as with Hs eRFs (Alkalaeva et al., 2006). To investigate the kinetics of peptide release, the concentrations of Sc eRFs were reduced, but release factors continued to be in excess over pre-TCs so that their recycling was not required. Like Hs eRF1, Sc eRF1 alone promoted slow peptide release, which was strongly increased by Sc eRF3 in the presence of GTP (Figure 4B, closed and open circles). These results indicated that the behavior of yeast and human eRFs was identical in this in vitro system, which justified the use of mammalian pre-TCs to study the activities of yeast release factors.
Figure 4.
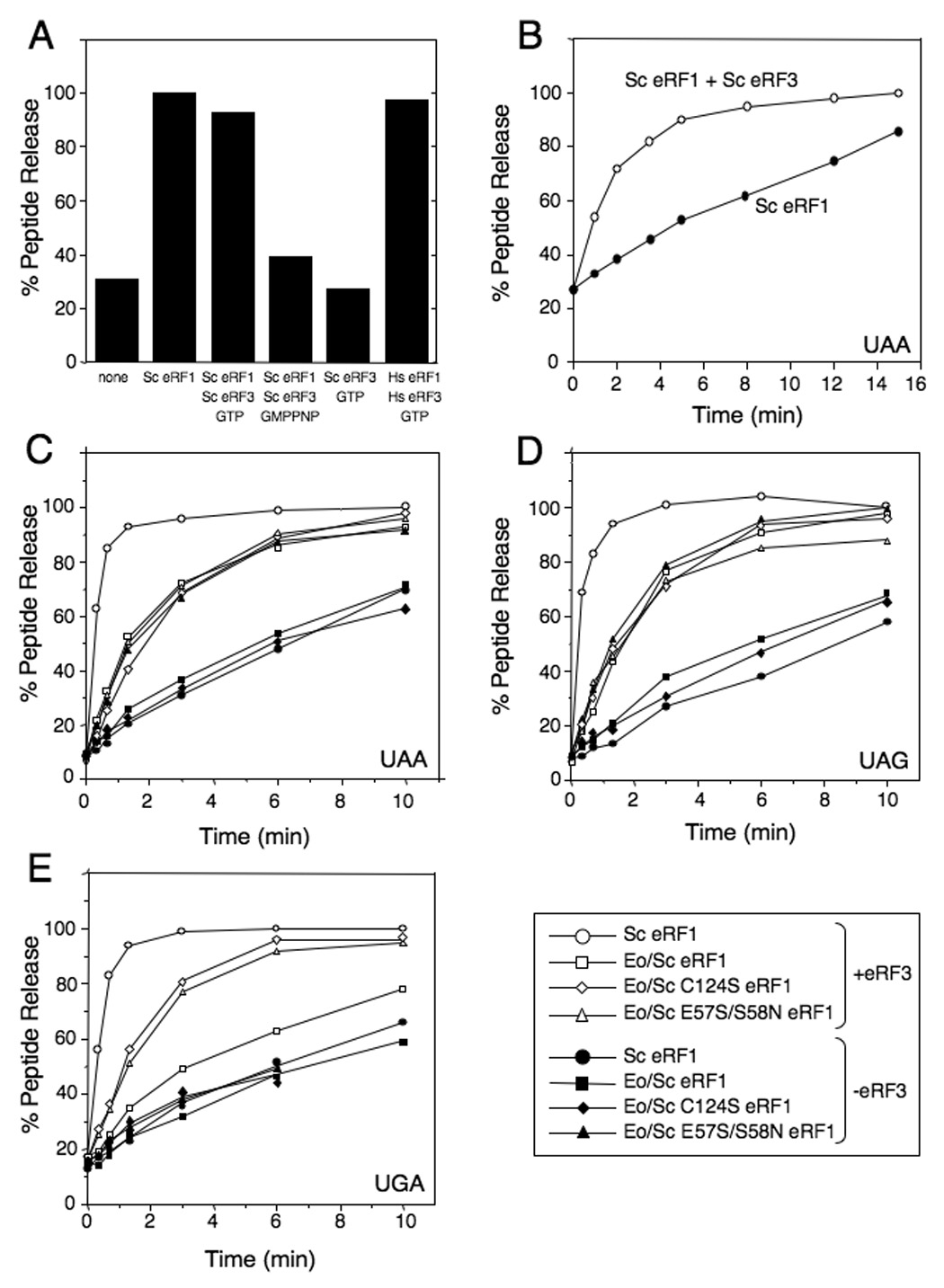
Eo/Sc eRF1 mutants restore peptide release at the UGA stop codon. A) Endpoint peptide release assay with excess Sc eRF1 and Sc eRF3. B) Kinetics of peptide release with Sc eRF1 (+/− Sc eRF3). C) Kinetics of peptide release with Eo/Sc eRF1 and mutant derivatives (+/− Sc eRF3) at UAA stop codon. D) Kinetics of peptide release with Eo/Sc eRF1 and mutant derivatives (+/− Sc eRF3) at UAG stop codon. E) Kinetics of peptide release with Eo/Sc eRF1 and mutant derivatives (+/− Sc eRF3) at UGA stop codon. Representative data for each experiment is shown. Reactions with eRF1s are denoted by closed symbols, while reactions with eRF1s and Sc eRF3 are denoted by open symbols.
We next compared the rate of peptide release induced by Sc eRF1, Eo/Sc eRF1, Eo/Sc E57S/S58N eRF1, and Eo/Sc C124S eRF1 in the presence or absence of Sc eRF3. As in the case of Sc eRF1, slow peptide release at UAA and UAG stop codons promoted by the original Eo/Sc eRF1 hybrid alone was strongly stimulated by Sc eRF3•GTP (Figures 4C and 4D, closed and open squares). However, peptide release promoted by Eo/Sc eRF1 and Sc eRF3•GTP was slower than peptide release promoted by Sc eRF1 and Sc eRF3•GTP, which was most likely due to the heterogeneity of the system: even though mammalian pre-TCs could tolerate yeast release factors well, the use of the Eo/Sc eRF1 hybrid introduced an additional challenge. Consistent with the in vivo data, peptide release induced by Eo/Sc eRF1 on the UGA codon was slow even in the presence of Sc eRF3•GTP (Figure 4E, closed and open squares). Again, reflecting the in vivo situation, the E57S/S58N and C124S mutations in Eo/Sc eRF1 restored its activity at the UGA stop codon: the rates of peptide release on the UGA codon by Eo/Sc E57S/S58N eRF1 and Eo/Sc C124S eRF1 in the absence and in the presence of Sc eRF3•GTP (Figure 4E, closed and open triangles and diamonds, respectively) were similar to the rates of peptide release by Eo/Sc eRF1 on UAA and UAG codons (Figure 4C and 4D, closed and open squares). These in vitro data confirmed the in vivo results described above that the E57S/S58N and C124S mutations restore the peptide release activity of Eo/Sc eRF1 at the UGA stop codon.
The C124S mutation did not change the activity of Eo/Sc eRF1 at UAA and UAG codons, and as in the case of the original Eo/Sc eRF1 hybrid, the low activity of Eo/Sc C124S eRF1 alone was also strongly enhanced by Sc eRF3•GTP (Figures 4C and 4D, closed and open diamonds). However, the rate of peptide release on UAA and UAG codons by Eo/Sc E57S/S58N eRF1 alone was as high as the rates of peptide release on these codons by either Eo/Sc eRF1 or Eo/Sc C124S eRF1 in the presence of Sc eRF3•GTP, and was not stimulated further by Sc eRF3•GTP (Figure 4C and 4D, open and closed triangles). This suggested that in addition to restoring peptide release activity on the UGA stop codon, the E57S/S58N mutations also enabled Eo/Sc eRF1 to promote efficient peptide release in vitro at UAA and UAG codons independently of eRF3.
The Eo/Sc E57S/S58N eRF1 suppresses an eRF1 mutation that reduces eRF3 binding in vivo
After finding that the E57S/S58N mutations in Eo/Sc eRF1 enabled it to promote efficient peptide release at UAA and UAG codons independently of eRF3 in vitro, we next examined whether these TASNIKS mutations also altered the eRF3 requirement for efficient stop codon recognition by eRF1 in vivo. To test this, we used a modified form of Eo/Sc E57S/S58N eRF1, E57S/S58N/CΔ19, which carried a deletion of the last 19 amino acids (referred to as CΔ19). This deletion was previously shown to greatly reduce the ability of eRF1 to bind eRF3, resulting in a slow growth phenotype and increased readthrough at all three stop codons (Eurwilaichitr et al., 1999; Kallmeyer et al., 2006).
We found that a strain expressing CΔ19 eRF1 grew more slowly than strains expressing either wild type Sc eRF1 or Eo/Sc E57S/S58N eRF1, while a strain expressing Eo/Sc E57S/S58N/CΔ19 eRF1 had an even stronger growth defect (Table 1). When we examined the efficiency of stop codon recognition, we found that the Sc CΔ19 eRF1 strain exhibited elevated readthrough at all three stop codons (1.4% readthrough at UAA, 1.1% readthrough at UAG, and 6.6% readthrough at UGA stop codons) (Figure 5A). This general increase in readthrough caused by the CΔ19 mutation is consistent with a previous report (Eurwilaichitr et al., 1999). In contrast, the Eo/Sc E57S/S58N/CΔ19 eRF1 strain exhibited < 0.2% readthrough at UAA and UAG codons (5 to 7-fold lower than in the Sc CΔ19 eRF1 strain), but 6.9% readthrough at the UGA codon (similar to the Sc CΔ19 eRF1 strain). These results indicate that this mutant protein retains efficient stop codon recognition at UAA and UAG codons despite a reduced ability to associate with eRF3, suggesting that the TASNIKS mutations result in a reduced requirement for eRF3 during the termination process at these two codons in vivo. In contrast, recognition of the UGA codon was not restored to a comparable extent. Western blot analysis showed that the steady-state level of Sc CΔ19 eRF1 protein was elevated 11-fold relative to wild type eRF1, consistent with a strong defect in translation termination (Figure 5B). Interestingly, the Eo/Sc E57S/S58N/CΔ19 eRF1 level was 18-fold higher than wild type eRF1. This suggested that this mutant protein retained a severe defect in some aspect of the termination process.
Figure 5.
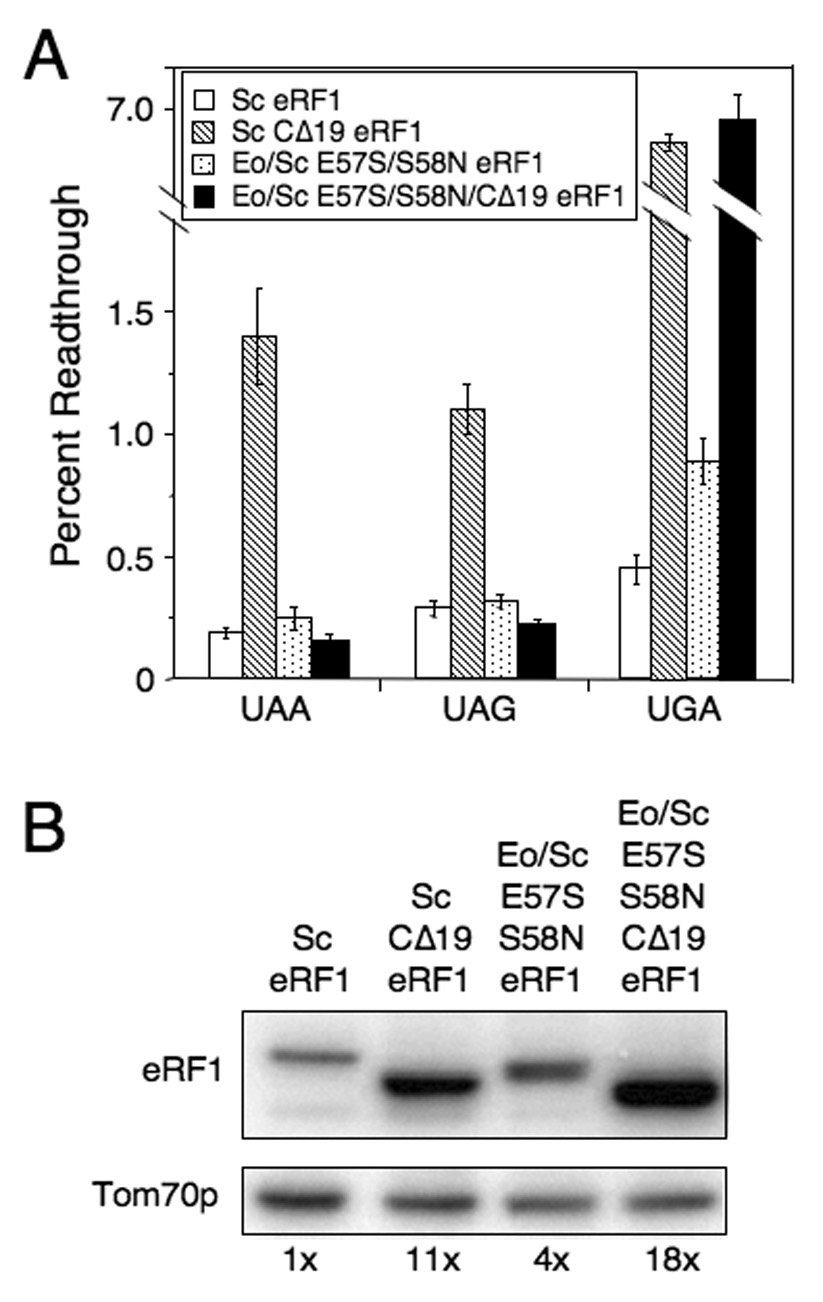
Effect of reducing eRF3 binding on Eo/Sc E57S/S58N eRF1 function in vivo. A) Readthrough in strains carrying Sc eRF1, Sc CΔ19 eRF1, Eo/Sc E57S/S58N eRF1, or Eo/Sc E57S/S58N/CΔ19 eRF1. B) Western blot quantitation of eRF1 levels in strains above. All strains were grown in SM glucose medium at 30°C. Readthrough values are represented as mean ± SD.
Although our results demonstrated that both the TASNIKS and YxCxxxF motifs play cooperative roles in stop codon recognition, they are spatially separated in the surface of eRF1 domain 1, making it unlikely that both are in direct contact with the stop codon during the recognition process. Taking this into consideration, the finding that the TASNIKS mutations alter the eRF3 requirement for efficient stop codon recognition by eRF1 in vivo and in vitro suggests a potential alternative important role for the TASNIKS motif that is distinct from direct interaction with stop codons.
DISCUSSION
Mutations that restore standard stop codon recognition are consistent with the cavity model of stop codon recognition
Various models have been proposed to explain how eRF1 mediates stop codon recognition. First, phylogenetic comparisons of eRF1 proteins from diverse species found that the NIKS motif is highly conserved among standard code organisms, but frequently diverges in variant code organisms (Kim et al., 2005; Knight et al., 2000). By analogy to features of the peptide anticodon found in prokaryotes, Nakamura noted that residues T55/A56/S57 (the “TAS” of the TASNIKS motif; S. cerevisiae numbering) shares key features of the bacterial PAT and SPF motifs, and suggested that it may function as a linear peptide anticodon in eRF1 (Nakamura et al., 2000). In another model, it was proposed that the region at the end of α-helix 3 (which includes the TASNIKS motif) forms a flexible element that can assume a tight or relaxed conformation as a function of the stop codon recognized (Muramatsu et al., 2001). Specifically, it was suggested that amino acids G54 and T55 (S. cerevisiae numbering) interact with the second nucleotide of the stop codon, while S57 and N58 recognize the third nucleotide.
Subsequently, a genetic screen in yeast identified several mutations throughout domain 1 that increased readthrough of UAG or UGA stop codons (Bertram et al., 2000). Based on these results, a model was proposed in which the three nucleotides of the stop codon bind to three pockets (or cavities) located near many of the identified mutations (Figure 6A). In this model, cavities 1, 2, and 3 bound the first, second, and third nucleotides of the stop codon, respectively. Later, a study that considered these data in conjunction with evolutionary rates of amino acid changes in domain 1 agreed in principle with the cavity-binding model, but proposed that the orientation of the stop codon binding to the 3 pockets be reversed (Inagaki et al., 2002). Finally, Kisselev and colleagues probed the effect of mutations in residues of domain 1 that are conserved in standard code organisms, but have diverged in many variant-code organisms. They found that both the NIKS residues within the TASNIKS motif (S. cerevisiae residues 55–61) and the YxCxxxF motif (S. cerevisiae residues 122–128) influence polypeptide chain release, leading them to propose a non-linear model in which stop codon recognition is modulated by positive and negative determinants (Frolova et al., 2002; Seit-Nebi et al., 2002).
Figure 6.

Models of eukaryotic stop codon recognition and translation termination. A) The location of key determinants of eRF1 function on the three-dimensional structure of human eRF1. Only the relevant portion of domain 1 is shown. Residues identified by genetic screens in this study or by Stansfield and co-workers (Bertram et al., 2000) are indicated in yellow (residues identified in this study are in black type, while those identified by Stansfield and co-workers are in purple type). White circles indicate cavities 1, 2, and 3. The residue designations and numbering corresponds to human eRF1 (+3 relative to Sc eRF1). B) Model for eukaryotic translation termination at UAA/UAG vs. UGA stop codons.
In the current study, we identified mutations that restored UGA recognition to a yeast strain expressing a hybrid Eo/Sc eRF1 that initially recognized only UAA and UAG stop codons. This stringent selection demanded that UAA and UAG recognition be retained while gain-of-function mutations restored UGA recognition. We identified the C124S mutation in three independent suppressors, suggesting that this mutation was one of the strongest suppressors that could be obtained. The identification of mutations at C124 was unexpected, since this residue is conserved in all known standard and variant code eukaryotes. In the model of Inagaki et al., it was proposed that C124 is involved in the recognition of the first base of the stop codon (Inagaki et al., 2002). This cannot be correct, since we found that this residue is a key determinant of UGA recognition. Residue C124 (C127 in human eRF1) is positioned directly between cavities 2 and 3 in the model of Stansfield and co-workers (Bertram et al., 2000) (Figure 6A), which would put it in an ideal position to discriminate nucleotides in the second and third position of codons. Furthermore, the A75S mutation (which corresponds to V78 in human eRF1), which also restored UGA recognition (though to a lesser extent than C124S), is located in α-helix 3 directly adjacent to C124 (C127 in human eRF1) in the floor of cavity 3. The location of these new suppressor mutations, like the mutations identified by Stansfield and co-workers, is consistent with the cavity model of stop codon recognition.
The eRF1 TASNIKS motif facilitates eRF3-regulated conformational changes that couple stop codon recognition and peptide release
The eRF1 TASNIKS motif is highly conserved in organisms that utilize the standard genetic code, but is less conserved in variant code organisms (Kim et al., 2005; Knight et al., 2000). This observation led to models suggesting that the TASNIKS motif is directly involved in stop codon recognition (Muramatsu et al., 2001; Nakamura et al., 2000). Consistent with this premise, we found that the E57S/S58N mutations that converted the TAESIKS sequence of Euplotes eRF1a to the standard code consensus TASNIKS (S60/N61 in human eRF1; see Figure 6A) also restored UGA recognition. However, while this result could be interpreted to suggest that these residues play a direct role in stop codon recognition, other data presented here suggest an alternative/additional function for the TASNIKS motif, which is coupling stop codon recognition to polypeptide release in conjunction with eRF3.
We found that wild type eRF1 and most Eo/Sc eRF1 suppressor mutants required eRF3 for efficient peptide release in vitro at all three stop codons. However, Eo/Sc E57S/S58N eRF1 required eRF3 for peptide release at the UGA codon, but not UAA and UAG codons. Moreover, Eo/Sc E57S/S58N/CΔ19 eRF1 containing a deletion of the C-terminal 19 amino acids (which reduced the ability of eRF1 to interact with eRF3) suppressed the readthrough phenotype associated with Sc CΔ19 eRF1 at UAA and UAG codons, but not at the UGA codon. This demonstrated that the E57S/S58N mutations also make Eo/Sc eRF1 less dependent on eRF3 for termination at UAA and UAG codons in vivo, but do not alter the eRF3 requirement for termination at the UGA stop codon. When taken together, these results suggest that the TASNIKS motif facilitates eRF3-mediated conformational changes in eRF1 that couple UAA/UAG and UGA stop codon recognition to peptide release in distinct ways.
A conformational model of eRF1 function
Recent studies have provided significant new information regarding the overall mechanism of eukaryotic translation termination. eRF1 performs three distinct functions during translation termination: it recognizes stop codons, activates the GTPase activity of eRF3, and triggers peptide release. Peptide release is strongly stimulated by eRF3 in a GTP-dependent manner, although the mechanism of this stimulation is not yet understood. eRF3 could stimulate peptide release by enhancing eRF1’s association with pre-TCs, by increasing the catalytic rate of peptidyl-tRNA hydrolysis, or both. There is some evidence that eRF3 stimulates the initial binding of eRF1 to pre-TCs. Binding of the eRF1 (GGQ→AGQ) mutant (which is defective in peptide release) to pre-TCs in the absence of eRF3•GTP results in a 2 nucleotide forward shift of a corresponding ribosomal toe-print (Alkalaeva et al., 2006). However, this shift appears at much lower concentrations of eRF1 if eRF3•GTP is present (Pestova, unpublished results). If the solution structure of eRF1 is similar to its crystal structure (Song et al., 2000), then eRF1 must adopt a more closed conformation to fit into the ribosomal binding pocket since the distance between decoding and peptidyl transferase centers is smaller than the distance between the stop codon recognition cavities and the GGQ motif of crystallized eRF1. Binding to eRF3•GTP might theoretically close eRF1’s structure to meet the ribosomal constraints, thereby enhancing eRF1’s association with pre-TCs. However, pre-TCs that contain eRF1/eRF3•GTP remain inactive in peptide release because the GGQ loop of eRF1 is not properly positioned at the PTC, and eRF1 must induce ribosome-dependent hydrolysis of GTP by eRF3 in order to adopt the conformation and/or position required to trigger peptide release (Alkalaeva et al., 2006).
Importantly, eRF3 mutations that reduce the efficiency of GTP hydrolysis increase readthrough in a codon-dependent manner (Salas-Marco and Bedwell, 2004). Since readthrough is a measure of the efficiency of stop codon recognition by eRF1, this suggests that a reduced rate of GTP hydrolysis leads to less efficient recognition of those stop codons at which readthrough increased. Thus, in addition to the conformational changes required to activate eRF1 to induce peptide release at all stop codons, GTP hydrolysis also leads to changes that facilitate the recognition of a subset of stop codons. This suggests that stop codon recognition occurs in two stages: initial recognition prior to GTP hydrolysis, and tightening of the interaction once GTP is hydrolyzed.
The second stage might be particularly important for recognition of a subset of termination codons, which includes all four UGA-N stop signals (Salas-Marco and Bedwell, 2004). The initial recognition of a stop codon might in turn influence the rate of activation of GTP hydrolysis, making it codon-dependent. Efficient recognition of a UAA or UAG stop codon might activate rapid GTP hydrolysis by eRF3 and promote efficient peptide release, whereas eRF1 binding of a UGA codon might activate GTP hydrolysis less efficiently (possibly to allow discrimination between UGA and UGG codons). Peptide release would occur at the UGA codon as long as GTP hydrolysis took place before dissociation of the termination complex. This might partially account for the higher basal readthrough observed at the UGA stop codon (Bonetti et al., 1995).
Recent studies using a defined peptide release assay (Alkalaeva et al., 2006; this study) found that eRF1 alone promotes slow peptide release. To achieve a higher rate of peptide release, eRF1 probably must undergo similar conformational/positional rearrangements as when it is bound by eRF3 and GTP hydrolysis occurs. However, in the absence of eRF3 such changes are slow. Remarkably, we found that Eo/Sc E57S/S58N eRF1 did not require eRF3 for peptide release at UAA and UAG codons, but required eRF3 for release at the UGA codon. This suggests that Eo/Sc E57S/S58N eRF1 must be capable of efficiently and spontaneously achieving the conformation and position required for efficient peptide release on pre-TCs with UAA and UAG stop codons, but not with a UGA stop codon.
Based on the preceding considerations, we propose the following model for the codon-dependent stimulation of eRF1-induced peptide release by eRF3 (Figure 6B). eRF1 alone exhibits low affinity binding to pre-TCs (state 1) and may not bind to pre-TCs by itself unless eRF3 is unavailable. The eRF1/eRF3•GTP complex has a higher affinity for pre-TCs, in part probably due to conformational changes in eRF1 (state 2). eRF1/eRF3•GTP cannot facilitate peptide release until GTP is hydrolyzed. Upon binding to pre-TCs and stop codon recognition, eRF1 induces GTP hydrolysis by eRF3, which could be faster at UAA and UAG codons and slower at the UGA codon. GTP hydrolysis induces a rearrangement of the termination complex, which includes tightening the interaction between eRF1 and the stop codon, particularly in the case of the UGA codon, and proper positioning of the GGQ loop in the PTC. After GTP hydrolysis, the final positions/conformations of eRF1 on pre-TCs with UAA/UAG (state 3) and UGA (state 4) codons are probably somewhat different. Both states 3 and 4 can facilitate efficient polypeptide release, although the higher basal readthrough observed at UGA codons (Bonetti et al., 1995) suggests that peptide release in state 3 might be somewhat more efficient. These two conformations of active eRF1 may be necessary to allow eRF1 to readily recognize UAA and UAG codons in state 3, while allowing it to not only recognize UGA, but also discriminate against the UGG (tryptophan) codon in state 4. Our data suggest that Eo/Sc E57S/S58N eRF1 gained the ability to reach state 3, but not state 4, in the absence of eRF3.
Among species that use a variant genetic code, two types of stop codon reassignment are found. Some variant code species use only UAA and UAG as stop codons (e.g., Euplotes and Blepharisma species), while others use only UGA as a stop codon (e.g., Tetrahymena, Stylonychia and Loxodes species) (Inagaki et al., 2002; Kim et al., 2005). Our model could readily explain why these two specific patterns of stop codon usage have repeatedly arisen during the evolution of variant code species (Lozupone et al., 2001; Tourancheau et al., 1995). Since it is implicit in our model that peptide release at UAA and UAG stop codons requires a different eRF1 conformation than release at UGA codons, the observed stop codon specificity could arise if eRF1 lost the ability to acquire either state 3 or 4 (thus eliminating peptide release at UAA/UAG or UGA codons, respectively). Future studies will be needed to further define how eRF1 couples stop codon recognition and peptide release, and how eRF3 stimulates this process.
MATERIALS AND METHODS
Strains and plasmids
The strains and plasmids used are described in Table 2, Supplemental Materials.
Dual luciferase readthrough assays
Dual luciferase readthrough assays of viable eRF1 mutants were performed as described (Salas-Marco et al., 2006). Readthrough assays of non-viable mutants were measured following a galactose to glucose shift procedure. First, a sup45Δ strain (YDB447) that co-expressed both wild type Sc eRF1 under GAL1 promoter (pDB967) and the indicated Eo/Sc hybrid eRF1 under SUP45 promoter control was grown in SM galactose medium for several generations at 35°C. During mid-log growth the cells were harvested and re-suspended in SM glucose medium to a cell density of 0.01 A600 units/ml. After six generations, cells were harvested and assayed for readthrough levels using the dual luciferase assay.
Peptide release assay
Peptide release was performed as described (Alkalaeva et al., 2006). Pre-termination complexes (pre-TCs) were formed on MVHC-STOP mRNA (a derivative of the previously described MVHL-STOP mRNA) with the mammalian initiation and elongation factors, ribosomal subunits and aminoacyl tRNAs that included [35S]Cys-tRNACys, and purified by centrifugation through a 10–30% linear sucrose gradient for 95 min in an SW55 rotor at 4°C. For the initial characterization of peptide release mediated by Sc eRF1 and Sc eRF3 (Figure 4A), 0.02 pmol pre-TCs were incubated in 40 µl reaction mixtures with 10 pmol Sc eRF1, 20 pmol Sc eRF3, and 1 mM GTP (or 1 mM GMPPNP as indicated) for 20 minutes at 37°C. To determine the kinetics of peptide release, 0.5 pmol Sc eRF1, Eo/Sc eRF1, or mutant derivatives of Eo/Sc eRF1 and 1 pmol Sc eRF3 were used, while the amount of pre-TCs remained at 0.02 pmol. Release of M-V-H-[35S]C tetrapeptide was monitored by scintillation counting of supernatants after TCA precipitation of reaction mixtures.
Supplementary Material
SUPPLEMENTAL DATA Supplemental Materials include one table describing the strains and plasmids used in this study.
ACKNOWLEDGEMENTS
The authors thank Mark Walter for helpful discussions and Mick Tuite for providing pUKC802. This work was supported by NIH grants RO1 GM 68854 (DMB) and RO1 GM 80623 (TVP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In Vitro Reconstitution of Eukaryotic Translation Reveals Cooperativity between Release Factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eurwilaichitr L, Graves FM, Stansfield I, Tuite MF. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, Atkins JF. Sequence specificity of aminoglycoside-induced stop codon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol. 2000;48:164–169. [PubMed] [Google Scholar]
- Inagaki Y, Blouin C, Doolittle WF, Roger AJ. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res. 2002;30:532–544. doi: 10.1093/nar/30.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Doolittle WF. Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res. 2001;29:921–927. doi: 10.1093/nar/29.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Ebihara K, Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y. A tripeptide 'anticodon' deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- Kallmeyer AK, Keeling KM, Bedwell DM. Eukaryotic Release Factor 1 Phosphorylation by CK2 Protein Kinase Is Dynamic but Has Little Effect on the Efficiency of Translation Termination in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1378–1387. doi: 10.1128/EC.00073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OT, Yura K, Go N, Harumoto T. Newly sequenced eRF1s from ciliates: the diversity of stop codon usage and the molecular surfaces that are important for stop codon interactions. Gene. 2005;346:277–286. doi: 10.1016/j.gene.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. The early evolution of the genetic code. Cell. 2000;101:569–572. doi: 10.1016/s0092-8674(00)80866-1. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight RD, Landweber LF. The molecular basis of nuclear genetic code change in ciliates. Curr Biol. 2001;11:65–74. doi: 10.1016/s0960-9822(01)00028-8. [DOI] [PubMed] [Google Scholar]
- Muramatsu T, Heckmann K, Kitanaka C, Kuchino Y. Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett. 2001;488:105–109. doi: 10.1016/s0014-5793(00)02391-7. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K. A tripeptide discriminator for stop codon recognition. FEBS Lett. 2002;514:30–33. doi: 10.1016/s0014-5793(02)02330-x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K, Ehrenberg M. Mimicry grasps reality in translation termination. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM. GTP Hydrolysis by eRF3 Facilitates Stop Codon Decoding during Eukaryotic Translation Termination. Mol Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol. 2005;348:801–815. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Fan-Minogue H, Kallmeyer AK, Klobutcher LA, Farabaugh PJ, Bedwell DM. Distinct paths to stop codon reassignment by the variant-code organisms Tetrahymena and Euplotes. Mol Cell Biol. 2006;26:438–447. doi: 10.1128/MCB.26.2.438-447.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1-- mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Tourancheau AB, Tsao N, Klobutcher LA, Pearlman RE, Adoutte A. Genetic code deviations in the ciliates: evidence for multiple and independent events. EMBO J. 1995;14:3262–3267. doi: 10.1002/j.1460-2075.1995.tb07329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL DATA Supplemental Materials include one table describing the strains and plasmids used in this study.


