Abstract
Studies into posttranslational modifications of histones, notably acetylation, have yielded important insights into the dynamic nature of chromatin structure and its fundamental role in gene expression. The roles of other covalent histone modifications remain poorly understood. To gain further insight into histone methylation, we investigated its occurrence and pattern of site utilization in Tetrahymena, yeast, and human HeLa cells. In Tetrahymena, transcriptionally active macronuclei, but not transcriptionally inert micronuclei, contain a robust histone methyltransferase activity that is highly selective for H3. Microsequence analyses of H3 from Tetrahymena, yeast, and HeLa cells indicate that lysine 4 is a highly conserved site of methylation, which to date, is the major site detected in Tetrahymena and yeast. These data document a nonrandom pattern of H3 methylation that does not overlap with known acetylation sites in this histone. In as much as H3 methylation at lysine 4 appears to be specific to macronuclei in Tetrahymena, we suggest that this modification pattern plays a facilitatory role in the transcription process in a manner that remains to be determined. Consistent with this possibility, H3 methylation in yeast occurs preferentially in a subpopulation of H3 that is preferentially acetylated.
In eukaryotes, DNA is complexed with histone proteins to form nucleosomes, which are, in turn, folded into a series of less well-understood higher-order structures (1, 2). Such compaction implies a severe restriction to DNA by proteins seeking access to DNA as substrate in DNA-templated processes such as transcription or replication. Thus, multiple biochemical mechanisms exist to dynamically control the basic architecture of chromatin to facilitate these processes.
Posttranslational modifications of histone amino-termini have long been thought to play a central role in the control of chromatin structure and function (1, 2). Well documented are a large number of covalent modifications—acetylation, phosphorylation, methylation, ubiquitination, and ADP ribosylation—that take place on the “tail” domains of histones (1–4). Such diversity in the types of modifications and the remarkable specificity for residues undergoing these modifications suggest a complex hierarchy of order and combinatorial function that remains unclear. Of the covalent modifications known to take place on histone amino-termini, acetylation is perhaps the best studied and appreciated. Recent studies have identified previously characterized coactivators and corepressors that acetylate or deacetylate, respectively, specific lysine residues in histones in response to their recruitment to target promoters in chromatin (reviewed in refs. 2–8). These studies provide compelling evidence that chromatin remodeling plays a fundamental role in the regulation of transcription from nucleosomal templates.
Histone methylation, first described in 1964 (9), is one of the least-understood posttranslational modifications affecting histones. Early work suggests that H3 and H4 are the primary histones modified by methylation (reviewed in refs. 1, 10, and 11), and sequencing studies, using bulk histones, have shown that several lysines (e.g., 9 and 27 of H3 and 20 of H4) are often preferred sites of methylation, although species-specific differences appear to exist (1, 10). Interestingly, each modified lysine has the capacity to be mono-, di-, or trimethylated, adding yet another level of variation to this posttranslational “mark” (1, 11). The role(s) of histone methylation have remained elusive although several reports suggest that this modification may play a functional role, albeit undefined, in gene transcription (11–14).
To date, little information exists on the major enzyme systems responsible for bringing about the steady-state balance of histone methylation. In this report, we continue to exploit unique biological features of the ciliated protozoan Tetrahymena (15) to begin to characterize a macronuclear-specific histone methyltransferase (HMT). The utility of this model is underscored by the fact that the first nuclear (type A) histone acetyltransferase (HAT) was identified by using macronuclei as an enriched starting source of this transcription-related activity (16). We also show that lysine 4 of H3 is a major site of methylation in ciliates, yeast, and human HeLa cells, implying a highly conserved function for this site that currently is undefined.
Materials and Methods
Cell Culture and Preparation of Nuclei and Nuclear Extracts.
HeLa cells were grown at 37°C in DMEM containing 10% FBS under 95% air-5% CO2 and their nuclei were isolated as described and stored at −80°C (17). Tetrahymena thermophila (strains CU 427 or CU 428) was grown in enriched 1% proteose peptone as described (18). Macro- and micronuclei were isolated from vegetatively growing Tetrahymena as described by Gorovsky et al. (18) and purified by sedimentation at unit gravity according to Allis and Dennison (19). Macronuclear DNase I extracts were prepared as described by Ohba et al. (20). The wild-type yeast strain MX4–22A was grown in rich yeast extract/peptone/dextrose medium followed by standard nuclei and histone isolation (21).
Methyl- and Acetyltransferase Activity Assays.
For labeling experiments involving Tetrahymena nuclei, 0.5 × 106 macronuclei or 15 × 106 micronuclei were incubated in methyltransferase (MTase) buffer (final concentration being 50 mM Tris, pH 8.0, 1 mM PMSF and 0.5 mM DTT) along with either 1.92 μCi of S-adenosyl-l-[methyl-3H]methionine (3H-AdoMet; 72 Ci/mmol; NEN Life Science Products) or 0.33 μCi of [3H]acetyl-CoA (3.2 Ci/mmol; Amersham Pharmacia) (each cofactor at 1.0 μM final) for 1 hr at 30°C in a total volume of 25 μl. Nuclei were pelleted, washed once with MTase buffer, and then resuspended in sample buffer before loading onto 12% SDS/PAGE gels. For labeling studies involving nuclei for microsequencing, 5 × 106 macronuclei, 1.5 × 107 HeLa nuclei, or yeast nuclei from 1 × 108 cells were labeled as described above, except that the reaction conditions were scaled up 10-fold. For MTase assays involving macronuclear extracts, 20 μg of extract was incubated with either 5 μg of chicken core histones or mononucleosomes (22) along with 0.55 μCi of 3H-AdoMet in MTase buffer for 1 hr at 30°C in a final volume of 50 μl. Typically, 10 μl of the reaction was spotted on Whatman P-81 for liquid scintillation counting while the remainder was precipitated with 20% trichloroacetic acid, collected by centrifugation, and then analyzed by gel electrophoresis followed by staining and fluorography. For peptide analyses, 5 μg of macronuclear extract was incubated with 5 μg of the appropriate peptide along with 0.55 μCi of 3H-AdoMet in MTase buffer for 1 hr at 30°C in a final volume of 25 μl that was spotted on Whatman P-81 and then used for scintillation counting. HAT assays with these peptides were performed as described (22), except that the amount of extract and final reaction volume were identical to those used in the HMT reactions. For the binding experiments involving Bio-Rex 70, 1 ml (≈1 mg) of macronuclear DNase I extract was incubated with 6 mg of Bio-Rex 70 for 2 h at 4°C. The supernatant (unbound fraction) was collected and bound proteins were eluted with 0.8 M NaCl in MTase buffer in a final volume equal to that of the input.
Gel Filtration Chromatography.
Typically, 0.5-ml aliquots of DNase I macronuclear extract were filtered and then concentrated to ≤50 μl before loading on a Superose 6 or 12 PC 3.2/30 column attached to a Smart System (Amersham Pharmacia) equilibrated with 20 mM Hepes, pH 7.9, 100 mM NaCl, 10% glycerol, 0.2 mM EDTA, and 1 mM DTT at 10°C. Fractions of 50 μl were collected at a flow rate of 0.4 ml/min and assayed for HAT and HMT activity as described above. The chromatographic system was calibrated by using the same buffer and the following standard proteins: thyroglobulin (669 kDa), apoferritin (440 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), ovalbumin (45 kDa), and carbonic anhydrase (31 kDa).
Electrophoresis and Western Blotting.
SDS/PAGE was performed as described by Laemmli (23). Acid urea gel electrophoresis was performed according to procedures described by Lennox and Cohen (24). Western blot analyses were performed by using secondary reagents and procedures from Amersham Life Sciences. Rabbit anti-hv1 was used at a dilution of 1:10,000 and rabbit anti-α was used at a dilution of 1:500 (25, 26).
Protein Microsequencing.
Methylated and unmethylated Tetrahymena, yeast, or HeLa histones were purified from sulfuric acid extracts (27) of 3H-AdoMet-labeled nuclei by RP-HPLC using a C8 column. Gradient conditions used for histone isolation were adapted from conditions previously described for isolation of HeLa histones (22). RP-HPLC-purified H3 or H4 was sequenced from their amino termini in an Applied Biosystems model 477A Protein Sequencer with an in-line 120A PTH-Analyzer (Applied Biosystems) using optimized cycles. After conversion, 50% of the sample was transferred to the RP-HPLC for phenylthiohydantoin (PTH)-amino acid identification and the other 50% was collected for determination of radioactivity by scintillation counting. Identification of mono- and dimethyllysine was determined by RP-HPLC analysis of PTH-derivatized mono- and dimethyllysine standards (Sigma). The N terminus of H4 was deblocked before sequencing as described (27).
Results
Histone Methylation in Tetrahymena Occurs Only in Transcriptionally Active Macronuclei.
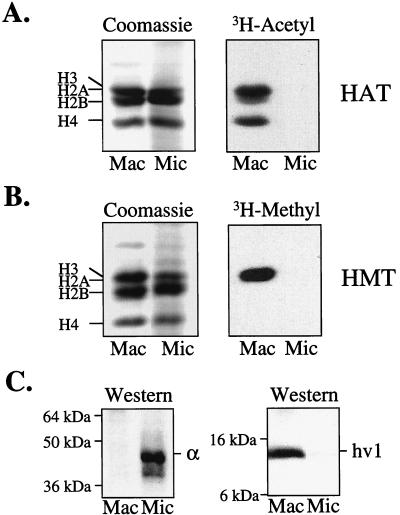
To gain insight into the possible function(s) of histone methylation, we examined the ability of highly purified populations of macro- and micronuclei to methylate endogenous histone substrates (for purity see Fig. 1C). As an internal comparison, macro- and micronuclei first were inspected for their ability to catalyze “in nucleo” histone acetylation. As expected from previous studies (28), macronuclei, but not micronuclei, contained potent endogenous HAT activities specific to all of the core histones (Fig. 1A). To determine the extent of endogenous HMT activity, macro- or micronuclei were incubated in the presence of 3H-AdoMet, and histones were analyzed as described above. As shown in Fig. 1B, a robust HMT activity specific for H3 (confirmed by RP-HPLC; not shown) was detected in macronuclei.
Figure 1.
HAT and HMT activities are associated exclusively with macronuclei in Tetrahymena. (A) Unit gravity-purified macro- or micronuclei, isolated from logarithmically growing Tetrahymena, were labeled in the presence of 3H-acetylCoA, and total proteins were resolved on a 12% SDS/PAGE gel and examined by Coomassie staining (Left) or fluorography (Right). (B) Same as in A except that nuclei were labeled in the presence of 3H-AdoMet. (C) Acid-soluble polypeptides from either macro- or micronuclei were resolved on a 12% SDS/PAGE gel, transferred to a membrane support, and probed with antibody to the micronuclear-specific protein α (Left) or with antibody to the macronuclear-specific protein hv1 (Right).
Like histone acetylation (28), HMT activity was not detected in micronuclei (Fig. 1B). Control experiments, using γ-labeled ATP, showed that both types of nuclei contained significant kinase activity toward histones, and using macronuclear extracts as a source of HMT (see Fig. 2), micronuclear H3 was methylated to a similar extent as chicken H3 (data not shown). These data demonstrate that micronuclei are enzymatically active with respect to at least some types of activities, and that the inability to detect HMT activity in the micronucleus is not because micronuclear H3 is a poor substrate. Finally, extracts derived from both types of nuclei showed that only macronuclei contained extractable HMT activity (data not shown). These results demonstrate that HMT activity, like histone acetylation, is restricted spatially to transcriptionally active macronuclei in Tetrahymena.
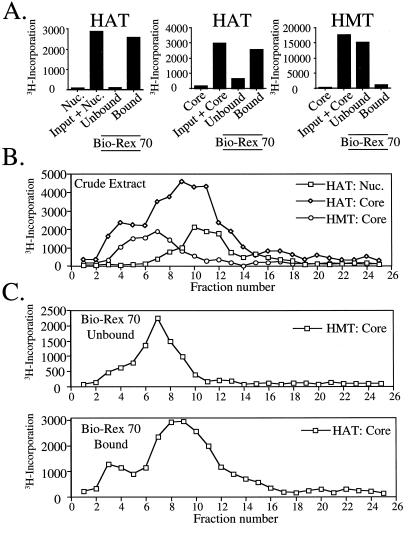
Figure 2.
Macronuclear HAT and HMT activities are physically separable. (A) DNase I extracts prepared from Tetrahymena macronuclei were incubated with the resin Bio-Rex 70 and bound proteins were eluted with 0.8 M NaCl before analysis of either HAT or HMT activity of the unbound or bound fractions by filter-binding assays using chicken core histones (Core) or nucleosomes (Nuc.) as substrate. The first two bars in each panel depict control experiments showing substrate only or substrate plus macronuclear extract (input) before the binding analysis. (B) Macronuclear extracts were fractionated by using a Superose 12 size exclusion column and collected fractions were analyzed for HMT and HAT activities by filter-binding assays as in A. (C) Bound- and unbound-Bio-Rex 70 fractions from DNase I extracts (see A) were fractionated by using Superose 12 and analyzed for free histone HAT and HMT activity. As only background levels of HAT and HMT activity were detected in the unbound- and bound-Bio-Rex 70 fractions, respectively, these data were omitted for clarity.
Macronuclear HMT Activity Is Not Physically Associated with Known HAT Activities.
Based on our current understanding, incorporation of 3H-acetate into macronuclear histones (Fig. 1A) reflects a combination of multiple HAT activities that on extraction, prefer free (non-nucleosomal; e.g., Gcn5) or nucleosomal (e.g., Esa1-like) histone substrates (see ref. 20 for details). We wondered whether the macronuclear HMT activity has a preference for histones in either of these forms and whether the HMT interacts, physically or functionally, with any known HAT activities. To address the first issue, macronuclear extracts were incubated with either chicken nucleosomes or free core histones in the presence of 3H-AdoMet or 3H-acetyl-CoA (Fig. 2A). As expected, HATs present in this extract acetylate either free or nucleosomal histones well. In contrast, the HMT activity displayed clear preference for free histones and failed to label nucleosomal substrates (data not shown). This result is consistent with other reports demonstrating that HMTs from other cellular sources, when extracted from chromatin, prefer free histone substrates (11).
We next sought to determine whether a direct physical association might exist between HATs and HMT(s) in these extracts. As shown in Fig. 2A, essentially all detectable HAT activities, capable of acetylating either nucleosomal (Left) or free (Center) histones, bind reversibly to Bio-Rex 70. In contrast, essentially all of the HMT activity (Fig. 2A, Right) is not bound by Bio-Rex 70 under identical chromatographic conditions, suggesting that these activities are not tightly associated. In support, extracts were fractionated over a Superose 12 size exclusion column wherein HMT activity was well resolved from the nucleosomal HAT activity (Fig. 2B). Because partial overlap between the HMT activity and the free histone-specific HAT activity was observed, isolated Bio-Rex 70 activities (bound vs. unbound) were further fractionated on Superose 12 (Fig. 2C). Results showed that these two separated activities yielded similar distributions as observed with crude extract, suggesting that each activity is physically distinct (Fig. 2C). Using protein standards for calibration on either Superose 6 or 12 (see Materials and Methods), we estimate the apparent size of the macronuclear HMT activity at ≈400 kDa (Fig. 2 B and C). Sizes predicted for the free histone-specific HAT activity (≈200 kDa) and nucleosomal HAT activity (≈150 kDa) agree well with previously reported values from Tetrahymena (20, 29).
Macronuclear HAT and HMT Activities Are Not Functionally Linked.
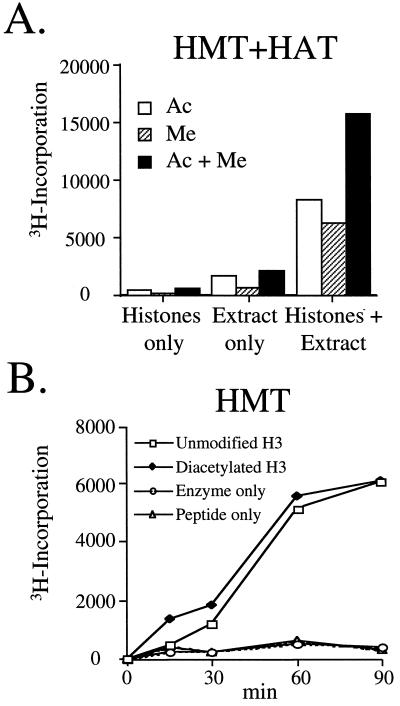
To test whether functional synergism or antagonism exists between macronuclear HAT and HMT activities, macronuclear extracts were assayed with free histones or model peptide substrates in the presence of 3H-AdoMet and/or 3H-acetyl-CoA. As shown in Fig. 3A, 3H-incorporation observed with chicken core histones on the addition of both radiolabeled cofactors (i.e., 3H-acetyl-CoA and 3H-AdoMet) was approximately the sum of that observed when each was used individually, demonstrating that significant synergism or antagonism between these two types of activities was not apparent. To further explore this idea, we compared the ability of the HMT activity to methylate H3 NH2-terminal peptides (residues 1–20) that were either unmodified or diacetylated at lysines 9 and 14 (lysine 14 is a highly preferred site of acetylation in vitro of the Gcn5 family of HATs; refs. 30 and 31). As shown in Fig. 3B, incubation of macronuclear extracts with either peptide substrate resulted in similar amounts of 3H-methyl incorporation, suggesting that acetylation at lysines 9 and 14 in H3 does not influence the ability of this activity to methylate the H3 N terminus. Furthermore, these data suggest that lysines 4 and/or 18, the only available lysines in our diacetylated peptide that are not acetylated, may be preferred site(s) of methylation by the macronuclear HMT activity (see below).
Figure 3.
Macronuclear HAT and HMT activities are functionally independent. (A) DNase I extracts prepared from Tetrahymena macronuclei were incubated with chicken core histones and 3H-acetyl-CoA (Ac) and/or 3H-AdoMet (Me) before filter binding and liquid scintillation counting. (B) A peptide containing N-terminal sequences 1–20 of H3 or a similar H3 peptide that was di-acetylated at lysines 9 and 14 was incubated with or without macronuclear extract and 3H-AdoMet before filter binding and liquid scintillation counting.
Lysine 4 of H3 Is the Preferred Methylation Site in Tetrahymena.
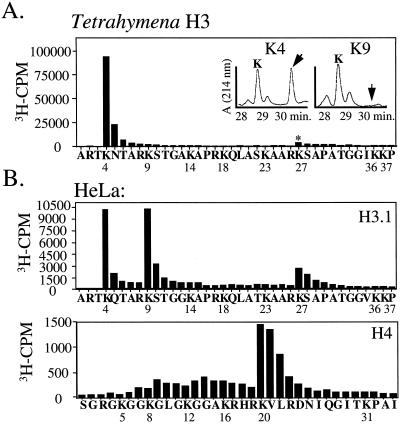
As an initial step toward identifying the site(s) of H3 methylation, several unmodified H3 peptides were assayed for their ability to be methylated by the macronuclear HMT activity. While a 1–20 residue H3 peptide was found to be methylated approximately 7-fold over “extract alone” controls, a shorter H3 peptide, representing residues 7–20, was not methylated (data not shown), suggesting that the preferred methylation site in H3 resides between sequences 1 and 7 of H3. To more definitively determine which residue(s) in H3 are methylated, H3 from 3H-AdoMet-labeled macronuclei was microsequenced and 3H-incorporation associated with each cycle was determined. The results clearly show that lysine 4 is the major site of methylation in macronuclear H3 under these conditions (Fig. 4A). Lysine 27 also appears to be methylated, albeit to a lesser extent (even after considering repetitive yield). Essentially all of the methyllysine eluting at position 4 was monomethylated (see Fig. 4A Inset and Table 1). Furthermore, comparison of the mass ratio between mono- or unmethylated lysine 4 revealed that approximately 47% of the total lysine at position 4 from macronuclei is methylated (Table 1). Finally, RP-HPLC amino acid analysis of lysine 27 did not reveal the presence of methyllysine at this position, suggesting that the inability to methylate lysine 27 well is not a consequence of pre-existing methylation at this site (see Table 1).
Figure 4.
Lysine 4 is a major site of active H3 methylation in Tetrahymena and HeLa cells. (A) RP-HPLC-purified H3 isolated from 3H-AdoMet-labeled macronuclei was subjected to N-terminal automated sequencing and 3H radioactivity eluted from each cycle was counted. Amino acids identified at each cycle of microsequencing are listed; numbers correspond to the known positions of lysine residues. * denotes the position of a second, relatively minor peak of methylation occurring at lysine 27. The inset corresponds to RP-HPLC analysis of lysines 4 and 9 from the above sequencing run showing the positions of mono- or unmethylated lysine. (B) RP-HPLC purified H3.1 and H4 isolated from 3H-AdoMet-labeled HeLa cell nuclei was subjected to N-terminal automated sequencing and 3H radioactivity eluted from each cycle was counted. Analysis was as in A.
Table 1.
Methyllysine/lysine ratio in H3
| H3 | Site of methylation
|
||
|---|---|---|---|
| 4 % Methylated | 9 % Methylated | 27 % Methylated | |
| Tetrahymena | 47 | None detected | None detected |
| Yeast | 34* | None detected | Not determined |
| HeLa | 7 | 33 | Not determined |
Levels of unmodified and mono- or dimethyllysine were determined by N-terminal sequencing followed by RP-HPLC analysis. Peaks representing either lysine or methyllysine were integrated and converted to % methyllysine at the residue indicated. Unless otherwise stated, values represent monomethyllysine.
*Both mono- and dimethyllysine were detected (19% and 15%, respectively).
Conservation of H3 Methylation Sites in HeLa Cells.
To our knowledge, lysine 4 methylation in H3 has not been reported in higher organisms. To determine to what extent this pattern of histone methylation is conserved, HeLa nuclei were incubated in the presence of 3H-AdoMet and analyzed as described above for Tetrahymena. In agreement with earlier results (11), each of the major human H3 isoforms (H3.1 and H3.2/.3), as well as H4, are methylated (data not shown). To determine the pattern of site utilization, H3.1 and H4 from 3H-AdoMet-labeled nuclei were microsequenced and analyzed as in Fig. 4A. The results indicate that lysine 4 is a major site of methylation in H3.1 (Fig. 4B), and in addition, several other previously characterized methylation sites (1) were detected by this labeling approach: lysines 9 and 27 of H3 and lysine 20 of H4 (Fig. 4B).
Histone Methylation in Yeast Occurs Preferentially on a Subpopulation of H3 That Is Also Acetylated.
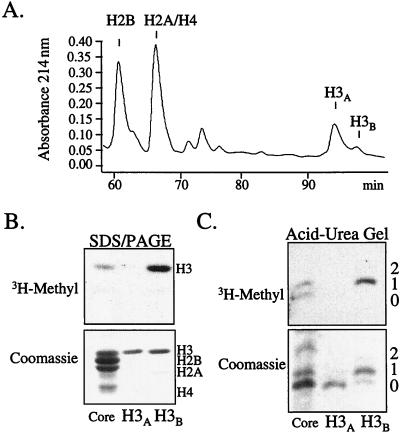
Although nuclei from Tetrahymena and HeLa cells readily incorporate 3H-methyl (and 3H-acetate) into histones, yeast nuclei incorporate 3H-methyl much less efficiently, precluding our ability to identify active methylation sites by a microsequencing approach. However, RP-HPLC amino acid analysis from these microsequencing attempts revealed that 34% of lysine 4 was both mono- and dimethylated (19% and 15% respectively; see Table 1). This result is in agreement with one report that lysine 4 in yeast H3 may be a potential site of methylation (39). In the course of performing these studies, we found that yeast H3 was consistently resolved into two peaks by RP-HPLC; the major peak (labeled H3A in Fig. 5A) was followed by a minor peak (labeled H3B in Fig. 5A). The basis for this separation is unknown. Surprisingly, SDS/PAGE analysis, followed by staining and fluorography, showed that H3B was preferentially methylated relative to H3A (Fig. 5B). No other histones were found to be significantly methylated under these conditions (data not shown).
Figure 5.
H3 methylation in yeast occurs preferentially on acetylated isoforms. (A) Nuclei isolated from logarithmically growing yeast were labeled with 3H-AdoMet and acid-soluble histones were purified by RP-HPLC. The RP-HPLC profile shows the presence of a major and minor H3 peak designated as H3A and H3B, respectively. To date, we have not observed this second peak with Tetrahymena or HeLa H3. (B) RP-HPLC-purified H3A and H3B were analyzed on a 12% SDS/PAGE gel followed by examination by Coomassie staining (Lower) or fluorography (Upper). For comparison, the amount of H3B was normalized with H3A. (C) RP-HPLC-purified H3A and H3B was subjected to acid-urea gel analysis before examination by Coomassie staining (Lower) or fluorography (Upper). For comparison, H3B was normalized with H3A. Numbers to the right indicate the positions of shifted H3 isoforms.
We then sought to determine whether the H3A and H3B peaks differed with respect to acetylated species of H3. As shown in Fig. 5C, acid-urea gel analysis showed that the preferentially methylated H3B fraction is enriched in acetylated (slower migrating) forms of H3 (Fig. 5C Upper), a result confirmed by immunoblotting with antibodies directed to acetylated H3 (data not shown). In contrast, H3A, the major form of H3 in yeast, appears to be mostly unmodified (i.e., unmethylated and unacetylated).
Discussion
Using the ciliate model, we demonstrate that Tetrahymena macronuclei contain a robust endogenous HMT activity that is missing from micronuclei during vegetative growth. During each vegetative cell cycle, micronuclei undergo DNA replication followed by mitosis. Our results then suggest that HMT activity is not correlated closely with either DNA replication or mitosis, although asynchronous cells have been used for our analyses. However, micronuclei, isolated from 5-hr conjugating Tetrahymena, the sexual phase of the life cycle in which micronuclei undergo a rapid series of meiotic and mitotic divisions, did not display HMT activity (data not shown), suggesting that histone methylation is not coupled to meiosis or mitosis in this system. In contrast, macronuclei isolated from this stage display a potent H3-specific HMT activity (data not shown). Thus, histone methylation, like transcription-associated histone acetylation (28), appears to be localized specifically to macronuclei in all stages of the life cycle, suggesting a potential link to the transcription process. The observation that H3 methylation in yeast (this report, Fig. 5), HeLa cells (12), and chicken erythrocytes (14) is enriched in subpopulations of acetylated H3 provides additional correlative evidence of a role of histone methylation in transcriptional regulation.
Direct evidence linking histone methylation and transcription recently has been provided by Stallcup and coworkers (32), who identified an arginine-specific HMT as a two-hybrid partner with the steroid hormone receptor coactivator family member GRIP-1. This activity (referred to as CARM1 for coactivator associated arginine methyltransferase 1), like the ciliate HMT activity described here, is highly selective for H3, although the site(s) of this arginine-based methylation has yet to be identified. Interestingly, GRIP-1/nuclear receptor-mediated transcriptional activation is enhanced by cotransfection of this HMT. Based on the apparent difference in amino acid specificity (arginine vs. lysine) between CARM1 and the Tetrahymena macronuclear HMT activity reported here, it seems unlikely that these activities are caused by the same catalytic component. Nevertheless, these data raise the intriguing possibility that coactivator complexes, containing distinct histone-modifying activities, such as HMTs with both arginine and lysine specificity, are recruited to specific promoters to modify chromatin structure as part of the transcription process.
In this report, we have used a radiolabeling/microsequencing approach to identify sites of methylation in histones after incubation of isolated nuclei in the presence of 3H-AdoMet. Methylation of H3 at lysine 4 has not been widely described (e.g., see table 4–13 of ref. 1). Earlier comparisons of the N-terminal amino acid sequences of H3 from calf thymus and human spleen failed to reveal methylation at lysine 4 (33, 34), leaving open the possibility that methylation at this site in HeLa cells is a property unique to cultured and/or transformed cells. However, recent amino acid analysis of pig thymus H3 shows the presence of monomethyllysine at position 4, indicating that this site normally is methylated in at least this mammalian tissue (C. Crane-Robinson, personal communication). Consistent with our Tetrahymena and yeast results, methylation at lysine 4 also has been reported in other ciliates, yeast, and plants from bulk sequencing analyses (35–39). These results collectively suggest that lysine 4 methylation in H3 is a highly conserved eukaryotic modification.
Modification of lysine 4 in H3, a residue normally not found to be acetylated in any organism, suggests a nonrandom pattern of histone modification. Similarly, we note that methylation of lysine 20 in H4 (see Fig. 4B and ref. 1), an assembly partner with H3 whose amino-terminal tail is functionally redundant with that of H3 (2–4), does not overlap with any of the known acetylation sites in this core histone (lysines at 5, 8, 12 and 16). Together, these data reinforce an emerging concept that different lysines, even those embedded in the same histone tail, have distinct covalent modification “marks” that presumably direct distinct functional roles that remain to be determined. To what extent specific sequential or combinatorial patterns of histone modification (e.g., methylation and acetylation) work together to facilitate specific processes (see refs. 32 and 40) remains an important issue for future studies.
Acknowledgments
We thank C. A. Mizzen for providing histone and nucleosomal substrates as well as for his critical review of the manuscript. Also, we thank all current laboratory members for their helpful discussions and technical advice and C. Crane-Robinson for kindly sharing unpublished data. This research was supported by grants from the National Institutes of Health to C.D.A. (GM53512) and B.D.S. (GM20039).
Abbreviations
- HMT
histone methyltransferase
- HAT
histone acetyltransferase
- MTase
methyltransferase
- 3H-AdoMet
S-adenosyl-l-[methyl-3H]methionine
References
- 1.van Holde K E. In: Chromatin, Springer Series in Molecular Biology. Rich A, editor. New York: Springer; 1988. pp. 111–148. [Google Scholar]
- 2.Wolffe A P. Chromatin: Structure and Function. San Diego: Academic; 1998. pp. 97–108. [Google Scholar]
- 3.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 5.Davie J R. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 6.Wolffe A P, Hayes J J. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornberg R D, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 8.Berger S L. Curr Opin Genet Dev. 1999;11:336–341. [Google Scholar]
- 9.Murray K. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 10.von Holt C, Brandt W F, Greyling H J, Lindsey G G, Retief J D, Rodrigues J D, Schwager S, Sewell B T. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 11.Duerre J A, Buttz H R. In: Protein Methylation. Paik W K, Kim S, editors. Boca Raton, FL: CRC; 1990. pp. 125–138. [Google Scholar]
- 12.Annunziato A T, Eason M B, Perry C A. Biochemistry. 1995;34:2916–2924. doi: 10.1021/bi00009a023. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers R, Tanguay R M. Biochem Biophys Res Commun. 1985;133:823–829. doi: 10.1016/0006-291x(85)90978-7. [DOI] [PubMed] [Google Scholar]
- 14.Hendzel M J, Davie J R. J Biol Chem. 1989;264:19208–19214. [PubMed] [Google Scholar]
- 15.Gorovsky M A, Glover C, Johmann C A, Keevert J B, Mathis D J, Samuelson M. Cold Spring Harbor Symp Quant Biol. 1978;42:493–503. doi: 10.1101/sqb.1978.042.01.052. [DOI] [PubMed] [Google Scholar]
- 16.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 17.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorovsky M A, Yao M C, Keevert J B, Pleger G L. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 19.Allis C D, Dennison D K. Dev Biol. 1982;93:519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- 20.Ohba, R., Steger, D. J., Brownell, J. E., Mizzen, C. A., Cook, R. G., Côtê 90, J., Workman, J. L. & Allis C. D. (1999) Mol. Cell. Biol.19, 2061–2068. [DOI] [PMC free article] [PubMed]
- 21.Edmondson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 22.Mizzen C A, Brownell J E, Cook R G, Allis C D. Methods Enzymol. 1999;304:675–696. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lennox R W, Cohen L H. Methods Enzymol. 1989;170:532–549. doi: 10.1016/0076-6879(89)70063-x. [DOI] [PubMed] [Google Scholar]
- 25.Stargell L A, Bowen J, Dadd C A, Dedon P C, Davis M, Cook R G, Allis C D, Gorovsky M A. Genes Dev. 1993;7:2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- 26.Allis C D, Allen R L, Wiggins J C, Chicoine L G, Richman R. J Cell Biol. 1984;99:1669–1677. doi: 10.1083/jcb.99.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vavra K J, Allis C D, Gorovsky M A. J Biol Chem. 1982;257:2591–2598. [PubMed] [Google Scholar]
- 29.Brownell J E, Allis C D. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant P A, Eberharter A, John S, Cook R G, Turner B M, Workman J L. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 31.Kuo M-H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Nature (London) 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 32.Chen D, Ma H, Hong H, Koh S S, Huang S-M, Schurter B T, Aswad D W, Stallcup M R. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 33.DeLange R J, Hooper J A, Smith E L. Proc Natl Acad Sci USA. 1972;69:882–884. doi: 10.1073/pnas.69.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohe Y, Iwai K. J Biochem. 1981;90:1205–1211. doi: 10.1093/oxfordjournals.jbchem.a133573. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi T, Hayashi H, Fusauchi Y, Iwai K. J Biochem. 1984;95:1741–1749. doi: 10.1093/oxfordjournals.jbchem.a134788. [DOI] [PubMed] [Google Scholar]
- 36.Waterborg J B. J Biol Chem. 1990;265:17157–17161. [PubMed] [Google Scholar]
- 37.Waterborg J H, Robertson A J, Tatar D L, Borza C M, Davie J R. Plant Physiol. 1995;109:393–407. doi: 10.1104/pp.109.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandt W F, von Holt C. FEBS Lett. 1986;194:278–281. doi: 10.1016/0014-5793(86)80099-0. [DOI] [PubMed] [Google Scholar]
- 39.Brandt W F, von Holt C. FEBS Lett. 1976;65:386–390. doi: 10.1016/0014-5793(76)80153-6. [DOI] [PubMed] [Google Scholar]
- 40.Strahl, B. D. & Allis, C. D. (1999) Nature (London), in press.