Abstract
The specific binding of HIV-1 nucleocapsid (NC) to the hinge region of the kissing-loop (KL) dimer formed by stemloop 1 (SL1) can have significant consequences on its ability to isomerize into the corresponding extended duplex (ED) form. The binding determinants and the effects on the isomerization process were investigated in vitro by a concerted strategy involving ad hoc RNA mutants and electrospray ionization-Fourier transform ion cyclotron resonance (ESI-FTICR) mass spectrometry, which enabled us to characterize the stoichiometry and conformational state of all possible protein-RNA and RNA-RNA assemblies present simultaneously in solution. For the first time, NC-hinge interactions were observed in constructs including at least one unpaired guanine at the 5′-end of the loop-loop duplex, whereas no interactions were detected when the unpaired guanine was placed at its 3′-end. This binding mode is supported by the presence of a grip-like motif described by recent crystal structures, which is formed by the 5′-purines of both hairpins held together by mutual stacking interactions. Using tandem mass spectrometry, hinge interactions were clearly shown to reduce the efficiency of KL/ED isomerization without inducing its complete block. This outcome is consistent with the partial stabilization of the extra-helical grip by the bound protein, which can hamper the purine components from parting ways and initiate the strand exchange process. These findings confirm that the broad binding and chaperone activities of NC induce unique effects that are clearly dependent on the structural context of the cognate nucleic acid substrate. For this reason, the presence of multiple binding sites on the different forms assumed by SL1 can produce seemingly contrasting effects that contribute to a fine modulation of the two-step process of RNA dimerization and isomerization.
Keywords: Genome dimerization and isomerization, RNA conformation, NC chaperone activity, ESI-FTICR mass spectrometry, tandem mass spectrometry
Introduction
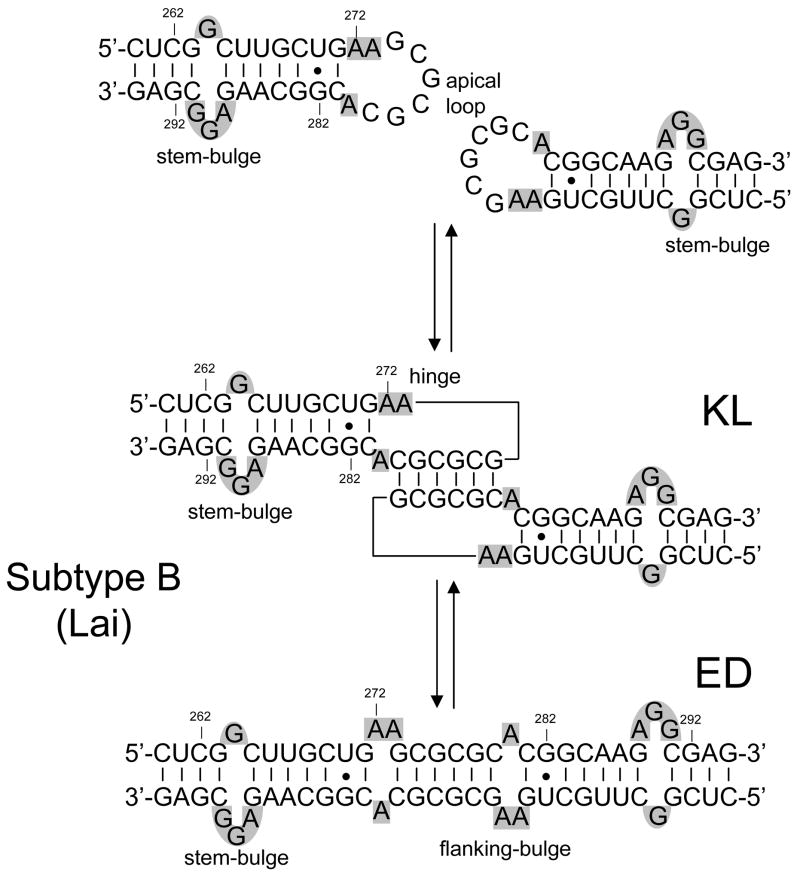
The dimerization of genomic RNA in human immunodeficiency virus type-1 (HIV-1) is mediated by a self-complementary sequence located in the apical loop of the stemloop 1 (SL1) domain of the packaging signal (Scheme 1).1–4 Mutations of this highly conserved region of the 5′-leader reduce the viral infectivity by disrupting both genome dimerization and packaging in vivo,4–7 thus making SL1 a potential target for the development of new therapeutic strategies. The mechanism of SL1 dimerization proceeds in vitro through the annealing of the palindromic loops of homologous copies to form a metastable kissing-loop (KL) dimer.1, 2, 4, 8–10 Subsequent steps involve melting of the double-stranded stems and formation of an extended duplex (ED) that is stabilized by a larger number of interstrand base pairs (Scheme 1).11–14 The rearrangement is facilitated in vitro by the activity of the nucleocapsid (NC) domain of the viral Gag polyprotein,13, 15–18 which presents broad nucleic acids binding and chaperone properties.19, 20 The observation that the genomic RNA of HIV-1 and Moloney murine leukemia virus experience a significant increase of their thermal stability upon protease-dependent viral maturation suggests that an analogous isomerization process may take place in vivo, as well.21–23 While complete 3D structures have been obtained for monomeric24–26 and dimeric27–35 forms of SL1, no high-resolution data are currently available for the corresponding NC assemblies. This fact has thus far hampered the elucidation of the functions performed by NC in the process of RNA dimerization and isomerization.
Scheme 1.
Two-step model of SL1 dimerization and isomerization. KL stands for kissing loop and ED for extended duplex. The sequence shown here corresponds to the subtype B (Lai variant) of HIV-1, but the nucleotides are numbered according to the subtype A sequence (Mal variant) for the sake of consistency. Stem- and flanking-bulges are highlighted in gray.
The binding modes of NC with the different forms assumed in vitro by SL1 have been investigated using electrospray ionization (ESI)36, 37 and Fourier transform ion cyclotron resonance (FTICR)38, 39 mass spectrometry. Ever since its ability of handling intact oligonucleotide duplexes was first described in 1993,40, 41 this soft ionization technique has been successfully employed for the characterization of noncovalent complexes comprising nucleic acid substrates and ligands of very diverse nature and size (reviewed in ref.s 42–44). The range of information accessible by ESI-based approaches is not limited to the composition and stoichiometry revealed directly by the observed molecular masses, but includes also relative45–49 and absolute50–55 binding affinities calculated from the abundance of free and bound species at equilibrium in solution.56 Additional information can be obtained by inducing the gas-phase dissociation of the initial nucleic acid complex of interest, which can facilitate the characterization of ligand binding sites,57, 58 provide insights into the organization of the assembled components,59–62 and determine the kinetic stability of the interactions between bound species.63–69
Taking advantage of the unique capabilities afforded by ESI-FTICR mass spectrometry, we have recently characterized the RNA-RNA and protein-RNA complexes formed by selected SL1 mutants (subtype B, Lai isolate) in the absence and presence of NC.70 RNA constructs lacking salient structural motifs were used to unambiguously identify binding sites located in single-stranded regions of the two conformers (Scheme 1), in agreement with the preference of NC for unpaired nucleotides.71–74 This work demonstrated that specific interactions with the apical loop or the stem-bulge motif can have contrasting effects on RNA dimerization. In fact, loop binding was shown to hamper the palindrome’s participation in intermolecular base-pairing, in direct competition with the initial step of RNA dimerization. In contrast, stem-bulge interactions destabilize the contiguous intramolecular base pairs that define the hairpin secondary structure, thus facilitating the rearrangement of the KL conformer into the more stable ED dimer. Specific binding was also demonstrated between NC and the unpaired adenines that flank the annealed palindromes (Scheme 1). The fact that this type of interaction was observed only when such bases formed bulged motifs in the ED structure, but not when they were included in the junction region of the KL complex, could foreshadow a possible mechanism for NC recognition of the different conformers.70
In the current study, we focused on the mechanism of dimer isomerization mediated in vitro by NC. Considering the possible role of palindrome-flanking nucleotides in the initial steps of the structural rearrangement, we have explored the binding determinants using constructs in which the junction bases were appropriately varied. The effects of these mutations on the formation of stable NC complexes were evaluated by direct ESI-FTICR analysis. Conformer-specific constructs that are incapable of structural rearrangement were employed to investigate separately the binding and isomerization events. The functions of hinge nucleotides in RNA isomerization were then probed using transition-competent mutants, which required the implementation of sustained off-resonance irradiation collision induced dissociation (SORI-CID)75 to differentiate unambiguously between conformers according to their gas-phase kinetic stabilities. The results of the binding experiments were correlated with the ability of each KL assembly to convert to the corresponding ED isomer in the presence and absence of NC-junction interactions. Wild-type and mutants of the subtype A (Mal isolate) were also included in the study to investigate the effects of intermolecular base-pairing on the interplay between dimer association, protein binding, and structural rearrangement. The findings of the isomerization experiments were discussed in the context of the accepted two-step model of RNA dimerization and stabilization in vitro. A better understanding of the mechanism of NC participation in these processes is crucial for the development of new antiviral strategies that could provide valid alternatives against rapidly emerging drug-resistant strains.
Results and Discussion
Determinants of NC-hinge binding
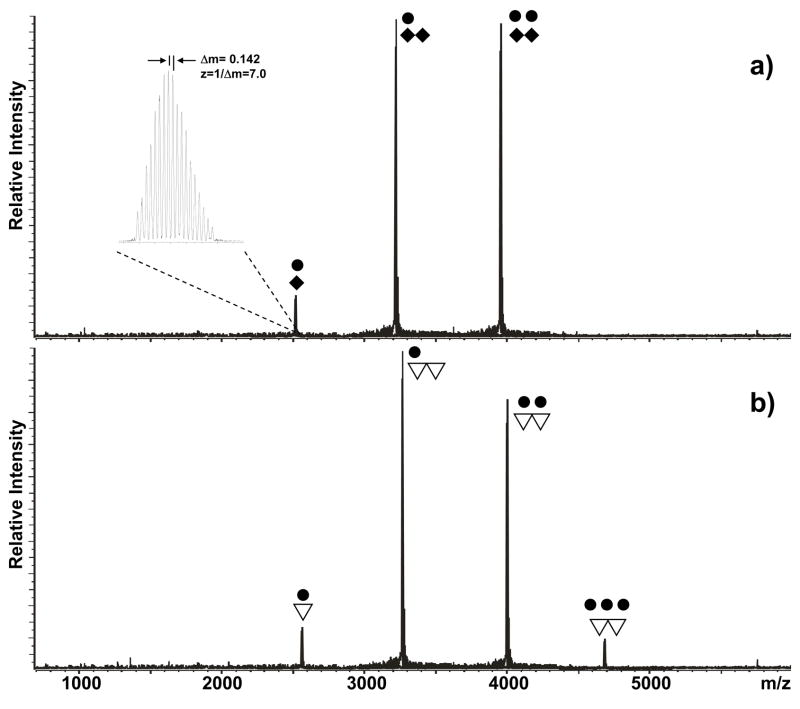
The SL1 domain of subtype B (SL1B, Lai isolate) presents adenine nucleotides in position 272, 273, and 280, which do not participate in canonic Watson-Crick pairing in either the KL or the ED conformer (Scheme 1). In previous work, a truncated duplex lacking the stem-bulge motifs was used to demonstrate that these unpaired nucleotides were capable of sustaining effective NC binding in vitro when they formed the small bulge motifs flanking the palindromes of the ED structure (Scheme 1).70 In contrast, no stable binding could be observed when such nucleotides were part of the hinge region between the kissing hairpins, as shown by the ESI-FTICR analysis of a mixture obtained by titrating a wild-type construct that was heat-denatured and rapidly cooled to obtain predominantly the kinetic KL product (Figure 1a). In this example, the formation of assemblies with a maximum of two protein units per RNA dimer is ascribable to the combination of the tight binding of NC to the hairpins’ stem-bulges and the absence of significant interactions with the hinging bases under the selected experimental conditions (see Materials and Methods).
Figure 1.
Nanospray-FTICR mass spectra of samples containing 30 μM NC (●) and 10 μM of either a) wild-type SL1B (◆), or b) SL1B-A272G (▽) in 150 mM ammonium acetate (pH 7.0) at room temperature. The signal corresponding to the −7 ion of the NC-SL1B complex is enlarged in the inset to show how the charge state of each species is unambiguously determined from the respective isotopic spacing. The corresponding molecular mass is then readily obtained from the charge state and the position of the peak on the m/z scale. Note that a maximum 2:2 protein to RNA ratio was observed for the KL assembly of wild-type SL1B, whereas a single base substitution in the junction region allowed for the binding of a third equivalent of NC.
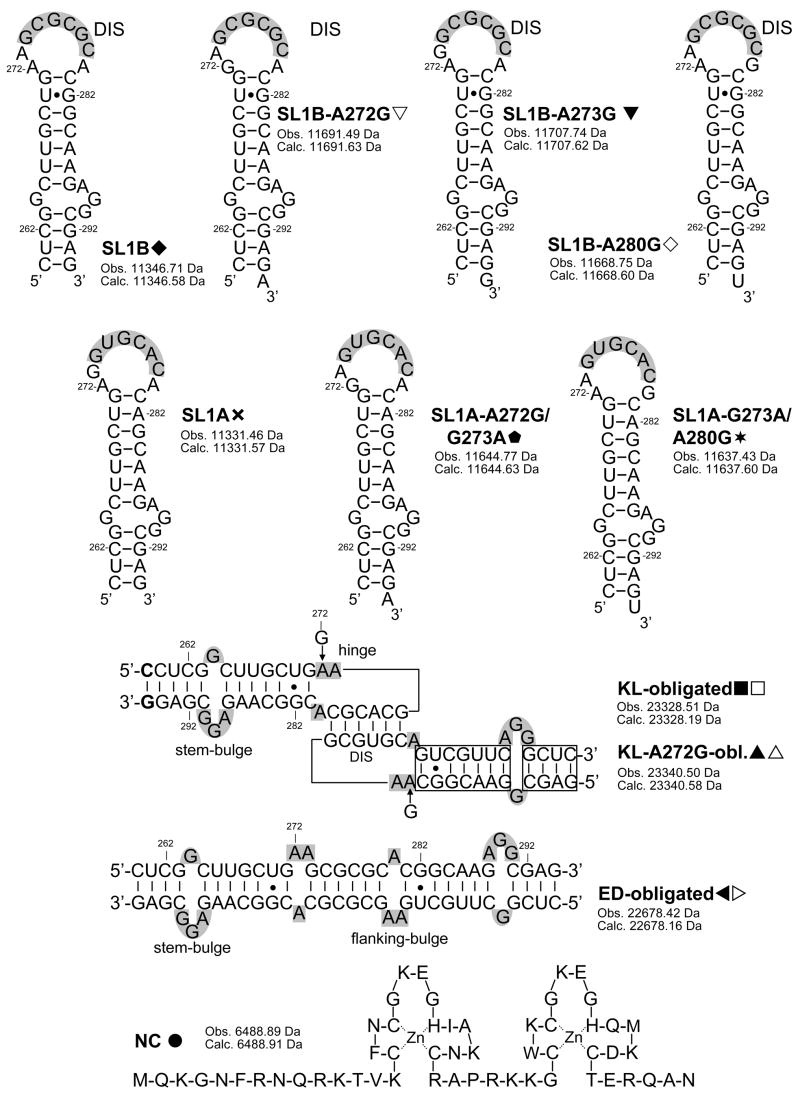
In light of the preference manifested by NC for exposed guanine bases,71–74 we tested the hypothesis that substituting any of the unpaired adenines with G may provide detectable interactions with the junction region in vitro (Scheme 2). Indeed, a maximum of three NC units were found to bind the SL1B-A272G mutant refolded according to the KL-inducing protocol, which was consistent with the introduction of a new viable site in addition to those located on each stem-bulge motif (Figure 1b). This stoichiometry was matched by the SL1B-A273G construct obtained by placing G on the immediate 5′side of the palindrome, but not by the SL1-A280G mutant with the substitution located on the 3′ side (Scheme 2). The latter displayed the 2:2 stoichiometry typical of the wild-type RNA, thus suggesting a close correlation between binding capabilities and structural context (Table 1).
Scheme 2.
Sequences and secondary structures of the constructs included in the study. Nucleotides are numbered according to the subtype A sequence (Mal variant) of HIV-1. The self-complementary sequences are highlighted in gray. Stems and flanking bulges of dimeric species are also highlighted in gray. For each species, the monoisotopic masses observed experimentally and calculated from sequence are included.
Table 1.
Summary of the maximum binding stoichiometries obtained by titrating SL1 constructs with NC.
| Construct | Maximum Binding to KL Form (Protein:RNA) |
|---|---|
|
| |
| B WT | 2:2 |
| A272G | 3:2 |
| A273G | 3:2 |
| A280G | 2:2 |
|
| |
| A WT | 3:2 |
| A272G/G273A | 3:2 |
| G273A/A280G | 2:2 |
The ability of junction nucleotides to direct specific protein interactions was further investigated in the subtype A (Mal isolate), which contains G in position 273 (SL1A, Scheme 2). As expected from the homology between this junction and that of SL1B-A273G, wild-type SL1A allowed for a maximum binding of three NC units per dimer under the same experimental conditions (Table 1), which indicated effective hinge interactions. In agreement with the binding modes exhibited by the corresponding subtype B constructs, the double-mutant SL1A-A272G/G273A containing G on the 5′ side of the palindrome allowed for hinge binding, whereas SL1A-G273A/A280G with G on the other side did not (Table 1).
The high degree of conformational polymorphism exhibited by the junction region offers an excellent explanation for the results afforded by these binding experiments. High-resolution structures obtained by nuclear magnetic resonance (NMR) have shown that purine 272 and 273 rest in different tucked-in positions that increase the overall stability of the kissing dimer.27, 32 In contrast, structures provided by X-ray diffraction place the same nucleobases in bulged-out conformations that may be stabilized by dimer-dimer interactions in the crystal packing.31, 33 In a dynamic situation in which both positions may be allowed in solution with different frequency, NC could readily recognize the exposed guanines and trap them in the extra-helical conformation. In the crystal structure, the bulged-out purines are held together by stacking interactions that confer a compact morphology to the motif.33 The rather small surface presented by this grip-like formation on the side of KL provides an excellent rationale for the detection of only one NC unit bound to the junction region. In this direction, control experiments performed with increasing protein concentrations did not produce higher order assemblies, but induced instead dimer dissociation and melting of the hairpin structures, consistent with earlier studies (see for example 53, 70, 73 and references therein). No additional binding was observed at lower ionic strength (i. e., 10 mM ammonium acetate), which is expected to strengthen protein-nucleic acids interactions. The fact that mutating the purine in position 280 did not have any detectable effect on protein binding is consistent with the steric situation of this nucleobase, which has been invariably described as intra-helical by both NMR and X-ray crystallography.
Conformational state and kinetic stability of NC-SL1 assemblies
In the case of KL-obligated mutants, the formation of assemblies with a 3:2 protein to RNA ratio constitutes an unambiguous indicator of the ability of hinge nucleotides to participate in specific protein interactions. In constructs capable of isomerization, however, the same stoichiometry and molecular mass could be shared by corresponding KL and ED assemblies, the latter upon partial saturation of the stem-bulge and flanking-bulge sites (Scheme 1).70 Considering the possible ambiguities arising from the simultaneous presence of both conformers during isomerization, we have tested tandem mass spectrometry76, 77 as a possible alternative to distinguish between dimeric forms, which would not rely on the stoichiometry of the assemblies observed in solution. This method involves isolating the complex of interest in the FTICR cell and then activating it by sustained off-resonance irradiation collision-induced dissociation (SORI-CID)75 to obtain products that are characteristic of the precursor ion structure. Based on the mounting evidence in support of the gas-phase preservation of hydrogen bonds and stacking interactions in nucleic acid structures,64–67, 78, 79 we explored the possibility that the widely different number of intermolecular base pairs in the two conformers may translate into readily recognizable dissociation patterns.
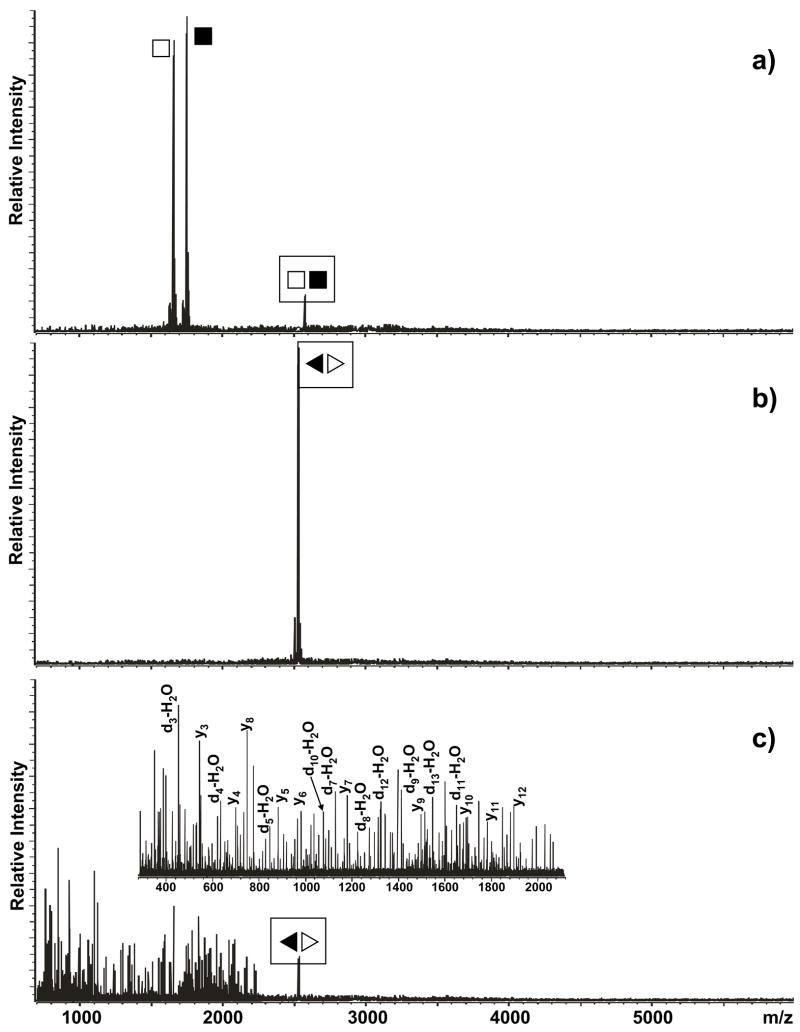
The hypothesis was initially tested using conformer-specific constructs that are capable of folding exclusively into either a KL or an ED structure (Scheme 2).70 Submitted to SORI-CID, the former readily dissociated in the individual hairpin components, whereas the latter remained intact at the same activation regime (Figure 2a and b, see Materials and Methods) Further, the ED-obligated dimer required significantly harsher activation conditions to provide detectable products, which did not include the intact unzipped strands, but consisted instead of fragments produced by cleavage of the oligonucleotides’ covalent backbones (Figure 2c).80, 81 As described in detail for DNA duplexes, the process of strand dissociation in the gas phase follows a multistep mechanism that is largely defined by kinetics considerations.79 In this context, the overall activation barrier and reaction time required to completely unzip the 28 interstrand pairs of the ED structure are expected to exceed those necessary to dissociate the 6 intermolecular pairs of the KL counterpart (Scheme 1), thus accounting for the ability of the former to withstand strand dissociation under the selected experimental conditions. The species observed upon harsher activation of the ED dimer were produced by a process involving partial dissociation of the interstrand base pairs and covalent fragmentation of the exposed regions.79
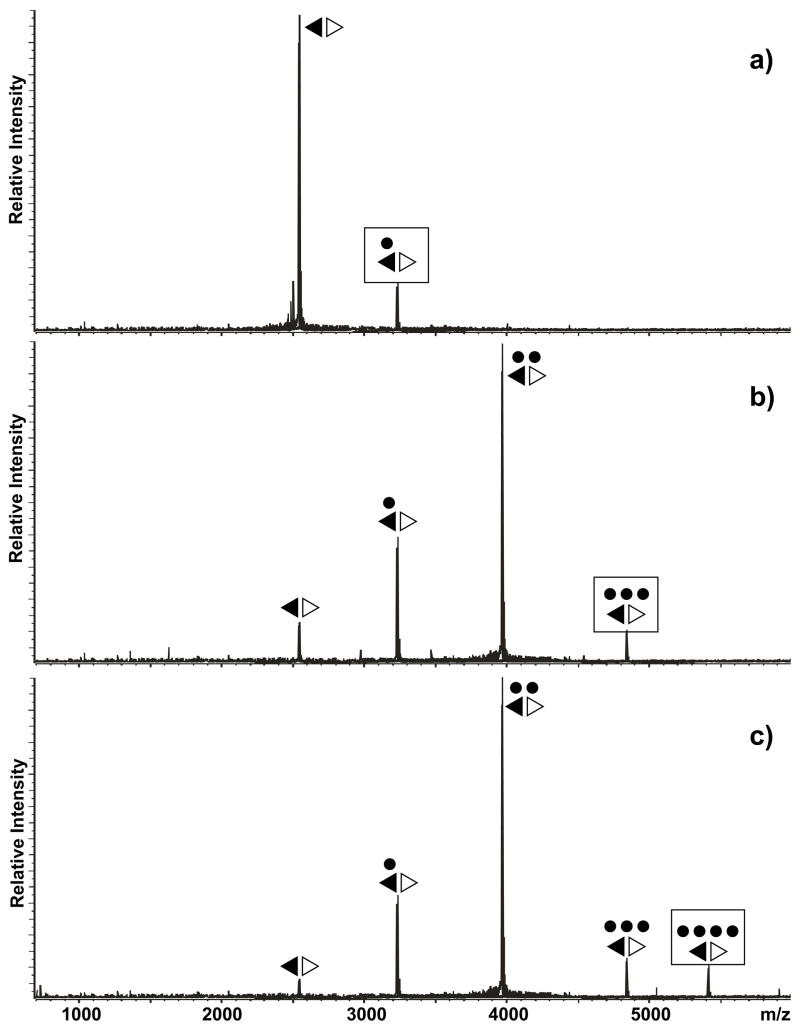
Figure 2.
Product ion spectra obtained by submitting a) the KL-obligated dimer (■□) and b) the ED-obligated dimer (◀▷) to mild SORI-CID (see Materials and Methods). Panel c) was obtained by submitting the ED-obligated dimer to harsher activation conditions. Boxed symbols identify each precursor ion. Note that the KL dimer readily dissociated into its monomeric components, whereas the ED conformer remained intact under identical activation conditions. At more energetic regimes, the latter provided typical d-H2O and y series, which were produced by covalent fragmentation of the RNA backbone.
The dissociation patterns exhibited by the ribonucleoprotein assemblies were found to mirror very closely those of the corresponding RNA substrates in the absence of NC. Consistent with the greater number of intramolecular base pairs, NC complexes of the ED-obligated construct did not undergo strand dissociation under mild activation conditions, but displayed a progressive loss of protein units (Figure 3). In contrast, KL-obligated assemblies underwent facile cleavage of interstrand hydrogen bonds, which allowed for the detection of monomeric components in either free or NC-bound form (Figure 4). Further disruptions of the specific protein-RNA interactions were responsible for the formation of the remaining products.
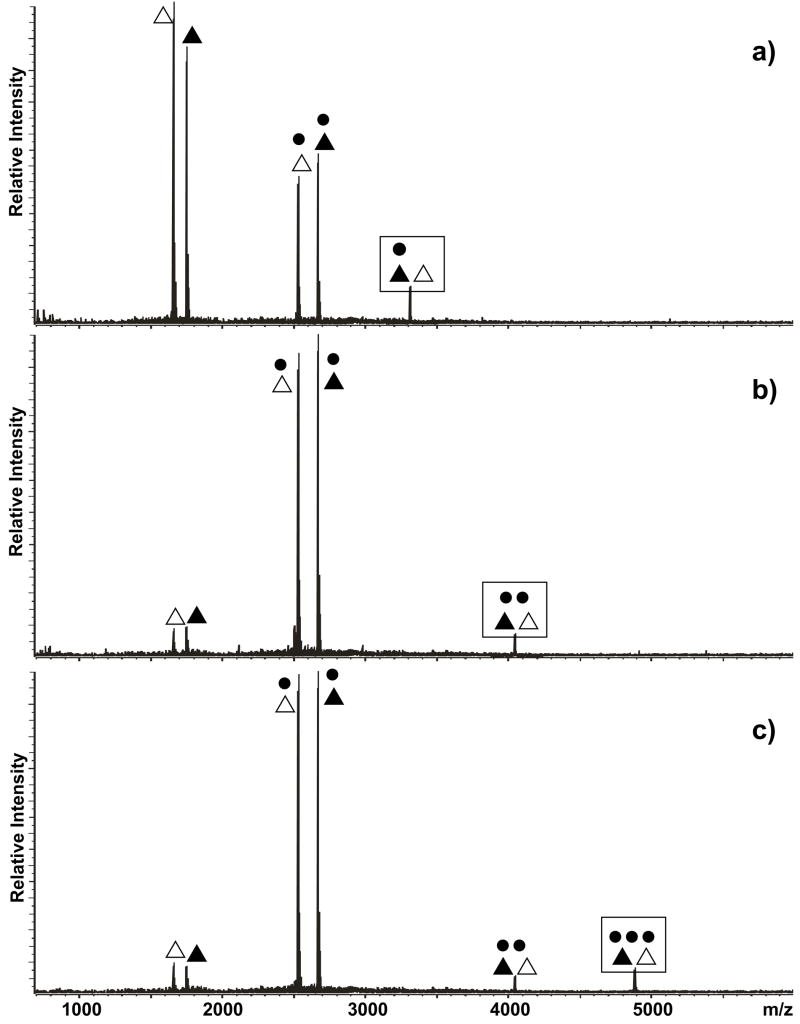
Figure 3.
Product ion spectra obtained by submitting the a) 1:2, b) 3:2, and c) 4:2 complexes of NC (●) and ED-obligated dimer (◀▷) to mild SORI-CID (see Materials and Methods). Boxed symbols identify each precursor ion. Note that the sequential loss of NC units constitutes a common thread among these spectra, whereas no dimer dissociation was observed. The fact that the 2:2 product was the predominant species obtained from the higher order precursors provides a measure of the intrinsic kinetic stability of this assembly.
Figure 4.
Product ion spectra obtained by submitting the a) 1:2, b) 2:2, and c) 3:2 complexes of NC (●) and KL-obligated dimer (▲△) to mild SORI-CID (see Materials and Methods). Boxed symbols identify each precursor ion. Note that both NC loss and dimer dissociation were observed in these spectra.
The ability of gas-phase techniques to probe the kinetic stability of protein-nucleic acid complexes has been described for intact E. coli ribosomes, which showed an excellent agreement between the order by which proteins dissociated from a target subunit and the extent of their interactions in the complex.60 Although it may not be always possible to draw direct correlations between gas-phase and solution behavior of noncovalent complexes,82 the binding properties observed in solution for the various ribonucleoprotein assemblies were found to be strikingly consistent with those revealed by tandem mass spectrometry in the absence of solvent. In the case of ED-obligated assemblies, the predominant 2:2 intermediate formed by stepwise NC losses from higher order assemblies (e. g., Figure 3b and c) clearly corresponds to the stable 2:2 species formed in solution upon binding to the high affinity sites on the stem-bulge motifs.70 In the case of KL-obligated complexes, the fact that NC was readily lost in the gas phase by the 3:2 assembly but not by the more stable 2:2 counterpart (Figure 4c and b, respectively) agrees with the relatively weak binding affinity afforded in solution by the junction site (Figure 1b).
NC-hinge interactions and isomerization efficiency
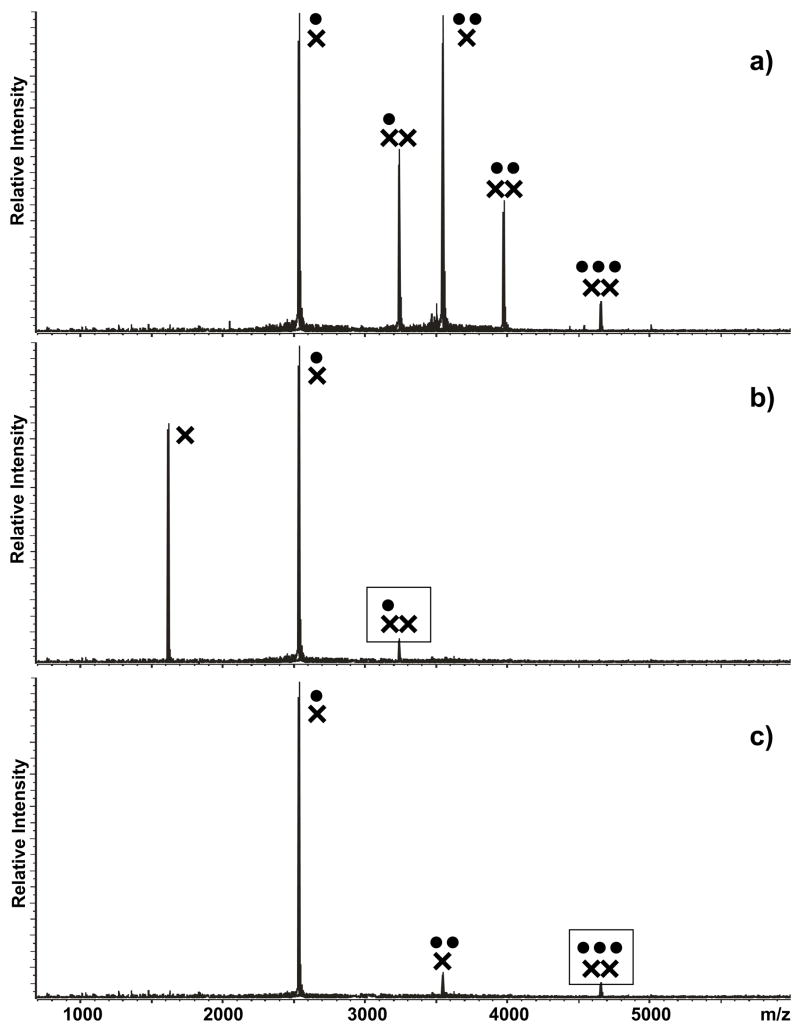
The possible consequences of hinge binding on the isomerization process were investigated by allowing the initial assemblies to undergo KL-ED conversion and by using tandem mass spectrometry to assess the conformational state of the products obtained in solution. For this purpose, each substrate was initially refolded according to the KL-inducing protocol and then mixed with a predetermined amount of NC. Based on experiments in which the process was completed with increasing protein concentrations, we determined that a 3 fold excess of NC over total RNA provided the best results for exemplification purposes (see Materials and Methods). As a direct consequence of simultaneous RNA-RNA and protein-RNA binding equilibria, a range of monomeric and dimeric complexes with multiple stoichiometries were readily observed in solution by ESI-FTICR. In the case of wild-type SL1A, the monomeric assemblies contained a maximum of two NC units per RNA, which is consistent with the presence of two high-affinity sites on the apical loop and stem-bulge regions of the hairpin structure (Figure 5a).70 The dimeric assemblies included up to three protein units, as expected from having G in the junction region. Submitted to mild SORI-CID, these dimers provided dissociation patterns replicating those observed for the KL-obligated mutant (Figure 5b and c), thus confirming the initial conformation of the RNA substrate.
Figure 5.
a) Nanospray-FTICR mass spectrum of a sample containing 30 μM NC (●) and 10 μM wild-type SL1A (×) in 150 mM ammonium acetate (pH 7.0) at room temperature. Panel b) and c) include the product ion spectra obtained by submitting the 1:2 and 3:2 complexes to mild SORI-CID (see Materials and Methods). Boxed symbols identify each precursor ion. Note that the 3:2 complex represented the maximum stoichiometry observed in this experiment and that activation of the different precursor ions resulted in both NC loss and dimer dissociation.
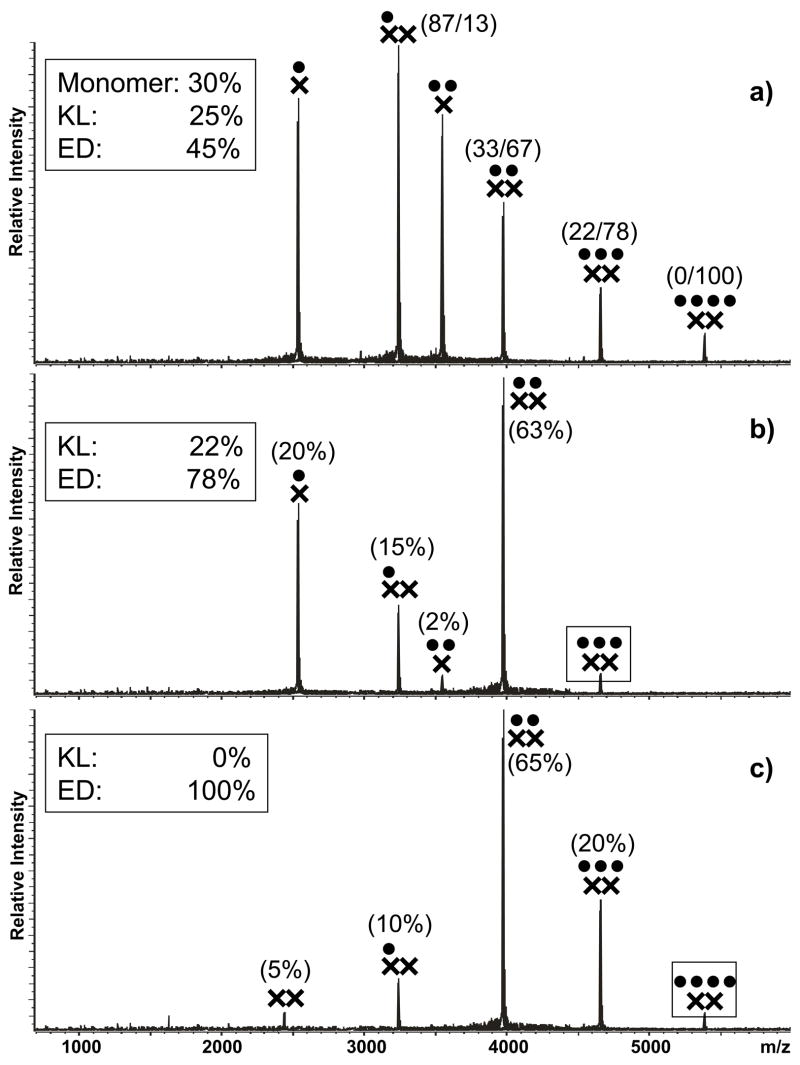
Dimer isomerization was subsequently performed by incubating the initial mixtures at 37°C for at least 3 h to allow for the processes taking place in solution to reach steady-state.16–18 At the end, the ESI-FTICR spectrum obtained from wild-type SL1A contained familiar monomeric and dimeric species, together with a previously absent 4:2 complex (Figure 6a). The conformational state of all dimeric assemblies in the sample was again interrogated by mild SORI-CID, which revealed very distinctive dissociation patterns. In particular, the 4:2 assembly displayed exclusively the stepwise loss of NC characteristic of the ED form (Figure 6c), whereas the lower order complexes afforded species originating from protein loss, as well as dimer dissociation. For example, the 3:2 assembly provided products with 2:2, 1:2, and 0:2 stoichiometries that were indicative of an ED conformer (Figure 6b). However, the concomitant detection of a 1:1 species could be explained exclusively by the dissociation of a KL structure, thus suggesting that a combination of both RNA conformers contributed to the lower order complexes observed in solution. Based on the relative intensities of the recorded signals and taking in account that a small portion of the 2:2 product could be ascribed to a KL dimer (see for example Figure 4c), we estimated that the 3:2 population corresponding to this precursor ion contained approximately 22% of KL versus ~78% of ED dimer (see Materials and Methods for details). This treatment was carried out for each individual assembly detected in solution (Figure 6a) and the results were combined to estimate that approximately 25% of the total RNA in the sample was present in the KL conformation, ~45% in the ED conformation, and ~30% in monomeric form.
Figure 6.
a) Nanospray-FTICR mass spectrum of a sample containing 30 μM NC (●) and 10 μM wild-type SL1A (×) in 150 mM ammonium acetate (pH 7.0) after 3 hour incubation at 37°C. Panel b) and c) include the product ion spectra obtained by submitting the 3:2 and 4:2 complexes to mild SORI-CID (see Materials and Methods). Boxed symbols identify each precursor ion. In panel b) and c), the percentages in parentheses indicate the normalized intensity of the corresponding signal compared to the total intensity of the product species, as described in Materials and Methods. Dimeric products were assigned to the ED conformer, whereas monomeric products were assigned to KL. The percentages were then summed to determine the partitioning between the two conformers within each precursor ion population, as reported in the box. In panel a), the KL/ED proportions within each ion signal were used to obtain the overall proportions of monomeric, KL, and ED forms in solution (summarized in Table 2).
Completed for all the constructs in the study, this analysis afforded a direct comparison of the KL/ED partitioning before and after incubation (Table 2), which enabled us to draw interesting correlations between the ability to sustain hinge interactions and the propensity to undergo isomerization under the selected experimental conditions. In particular, structures containing A in position 272 and 273, which did not allow for detectable protein binding, provided the higher yields of ED formation within each subtype series. This was clearly shown for subtype A by the SL1A-G273A/A280G mutant, which exhibited ~66% of total RNA in the ED form. In the subtype B series, the wild-type and SL1B-A280G constructs yielded ~89% and ~87% of total RNA, respectively. It should be noted that the presence of NC was strictly necessary to achieve isomerization under these conditions, as demonstrated by control samples that did not undergo detectable conversion in the absence of protein even after extended incubation periods. In this direction, it has been demonstrated that binding of NC to the stem-bulge motifs is necessary to destabilize the hairpins’ double-stranded stems for strand exchange and isomerization to take place.13, 70
Table 2.
Percent partitioning between monomeric, kissing-loop (KL), and 7extended duplex (ED) forms at different stages of the two-step process of dimerization and isomerization. The percentage of each species in solution was estimated as described in Materials and Methods. Each determination was repeated in triplicate and the final values carried on average a standard deviation of ±2.5%. The first column reports estimates obtained at room temperature in the absence of NC, the second after 3 h incubation at 37°C. The third column reports estimates obtained immediately after addition of three fold NC over total RNA at room temperature, the fourth after 3 h incubation at 37°C in the presence of NC.
| No NC | 3× NC | ||||
|---|---|---|---|---|---|
| t = 0 h | t = 3 h | t = 0 h | t = 3 h | ||
| B WT | Mono % | 11 | 12 | 5 | 1 |
| KL % | 89 | 88 | 95 | 10 | |
| ED % | 0 | 0 | 0 | 89 | |
| Total Dimer % | 89 | 88 | 95 | 99 | |
| A272G | Mono % | 13 | 13 | 5 | 2 |
| KL % | 87 | 87 | 95 | 22 | |
| ED % | 0 | 0 | 0 | 76 | |
| Total Dimer % | 87 | 85 | 95 | 98 | |
| A273G | Mono % | 11 | 15 | 4 | 0 |
| KL % | 89 | 85 | 96 | 22 | |
| ED % | 0 | 0 | 0 | 78 | |
| Total Dimer % | 89 | 84 | 96 | 100 | |
| A280G | Mono % | 9 | 11 | 5 | 1 |
| KL % | 91 | 89 | 95 | 12 | |
| ED % | 0 | 0 | 0 | 87 | |
| Total Dimer % | 91 | 89 | 95 | 99 | |
| A WT | Mono % | 33 | 34 | 50 | 30 |
| KL % | 67 | 66 | 50 | 25 | |
| ED % | 0 | 0 | 0 | 45 | |
| Total Dimer % | 67 | 65 | 50 | 70 | |
| A272G, G273A | Mono % | 33 | 31 | 47 | 25 |
| KL % | 67 | 69 | 53 | 23 | |
| ED % | 0 | 0 | 0 | 52 | |
| Total Dimer % | 67 | 68 | 53 | 75 | |
| G273A, A280G | Mono % | 36 | 38 | 47 | 20 |
| KL % | 64 | 62 | 53 | 14 | |
| ED % | 0 | 0 | 0 | 66 | |
| Total Dimer % | 64 | 62 | 53 | 80 | |
Isomerization and dimer association
A close examination of the data summarized in Table 2 provides further information about the effects of isomerization on the overall outcome of the two-step process. Before addition of NC, the two subtype series manifested intrinsic differences in the monomer/dimer partitioning under the same experimental conditions, as expected from the different base composition of their respective self-complementary sequences. In subtype B, the loop-loop interactions defining the kissing complex consist of six intermolecular pairs involving exclusively G and C nucleotides, whereas the subtype A series includes two A-U pairs that afford lower intermolecular stabilization. For this reason, it was not surprising that the former exhibited significantly higher percentages of dimeric species than the latter (Table 2). Considering that these estimations entailed an average standard deviation of ±2.5% (see Materials and Methods), the excellent agreement of monomer/dimer partitioning within each series confirmed that these base substitutions in the palindrome-flanking region did not have significant consequences on the stability of the respective dimers in the absence of NC.
The presence of NC produced immediate and contrasting effects on the degree of dimer association exhibited by the two subtypes, which were clearly independent of its activity in structural rearrangement. In fact, the mere addition of protein induced the partial dissociation of the subtype A kissing complexes before isomerization could take place, as revealed by a marked increase of the percentage of monomer from an average of ~34% to an average of ~48% of total RNA in solution (Table 2). Conversely, a decrease of the percentage of monomer from an average of ~11% to ~5% of total RNA highlighted a stronger association of the subtype B counterparts. As discussed above, the different base composition of the palindromic sequences can again provide the basis for the observed behavior. In a situation involving concomitant RNA-RNA and protein-RNA binding in solution, the single-stranded loop of a monomeric hairpin can be alternatively involved in intermolecular pairing or specific interactions with NC. In the subtype A series, protein binding is capable of competing more effectively with loop-loop annealing due to the relatively weaker stabilization afforded by the two A-U base pairs. This effect is counterbalanced in subtype B by the stronger association of G-C pairing and by the uncanny ability of NC to facilitate the annealing of G-rich oligonucleotides,74 the mechanism of which is still the object of intense investigation.20
The favorable effects of isomerization on dimer association are clearly demonstrated by the fact that all the constructs in the study yielded the maximum percentage of total dimer at the end of their incubation period in the presence of NC, regardless of their subtype and sequence (Table 2). This observation is not surprising in light of the thermodynamic drive provided by the formation of the additional interstrand base pairs defining the ED structure. In fact, stabilizing the final product of the two-step process is expected to shift both equilibria to the right (Scheme 1). This effect was more noticeable for constructs of the subtype A series, due to the less favorable monomer/dimer partitioning established at the beginning of the process. Consistent with the inverse correlation between junction binding and isomerization efficiency, the highest percentage of dimer in solution was achieved in this series by the SL1A-G273A/A280G mutant that did not sustain hinge interactions.
Implications of NC-hinge interactions on the isomerization mechanism
The leading models proposed for the isomerization mechanism agree on the fact that base pairing between annealed palindromes is conserved throughout the process.83, 84 In a concerted multi-step progression, the loop-loop helix of the KL complex is extended in both directions by gradually dissociating the stems’ intramolecular pairs and by reconstituting them in intermolecular fashion to complete the ED structure.17, 26, 85 In the absence of NC, one of the initial steps requires mobilizing the flanking purines from their relatively stable tucked-in conformations,27, 32 which can be mediated for example by base protonation.86 In the same direction, NC binding can prevent the participation of these nucleotides in junction stabilization by constraining them into the bulged-out conformation observed in the crystal structures of the kissing conformer (Figure 7a).31, 33 However, this intrinsically destabilizing effect is countered by the fact that the extra-helical grip consists of purines contributed by both hairpins, which are held together by stacking interactions and may receive further stabilization by NC binding. Considering that such nucleotides are expected to form distinct flanking-bulges in the final ED product (Figure 7d),28–30, 32, 35 the grip structure must necessarily dissolve and the same-strand purines must part ways for strand exchange to take place (see for example the cartoons in Figure 7b–c). In this context, specific NC-junction interactions may hamper the grip melting with unfavorable consequences on the initial steps of helix extension and strand exchange.
Figure 7.
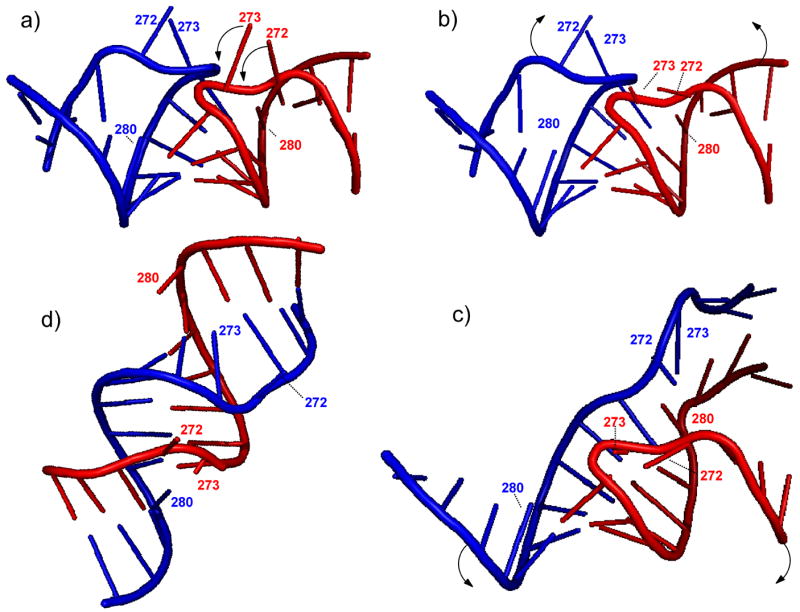
a) Loop-loop interactions in the KL dimer formed by wild type SL1A (PDB: 1XPF).33 The junction bases are labeled according to the subtype A sequence (Mal variant). The arrows give an idea of the putative rearrangements that may take place during the isomerization process. The graphical rendition in panel b) was created using Pymol91 to visualize the expected stacking between hinge and loop bases to extend the initial loop-loop helix. The arrows show the possible unzipping of the intramolecular base pairs of the KL dimer, which is necessary to enable strand exchange. The graphical rendition in panel c) depicts a more advanced stage of strand exchange, in which a greater number of intermolecular base pairs are being formed in concerted fashion. The final result of the isomerization process consists of a full-fledged duplex structure d), represented here by the palindromic region of the ED dimer of wild type SL1A (PDB: 1Y99).28
In the absence of high-resolution data for the corresponding ribonucleoprotein assemblies, the different binding behaviors afforded by the flanking G and A can offer some insights into the nature of the specific NC-junction interactions. The required participation of at least one G to provide detectable binding cannot be explained solely on the basis of the known affinity of NC for this type of nucleotide. Indeed, unpaired As can effectively sustain protein binding when they are included in the flanking-bulges of the ED structure, as demonstrated by the detection of 4:2 assemblies for constructs with A in position 272 and 273 (e. g., wild-type SL1B).70 The different behaviors could be perhaps explained by the fact that the crystal structure of the subtype A kissing dimer presents G273 in a syn conformation, whereas the corresponding A273 adopts an anti conformation in subtype B.33 In addition, it is possible that the different proton affinities afforded by the two purines may have very distinctive effects on junction dynamics and grip structure,86 which should be further investigated.
Conclusions
The direct mass spectrometric analysis of assemblies formed by simultaneous protein-RNA and RNA-RNA equilibria in solution has provided new valuable insights into the multifaceted activities performed by NC in the two-step process of SL1 dimerization and isomerization in vitro. The ability of tandem mass spectrometry to afford an unambiguous reading of the conformational state of the ribonucleoprotein assemblies of interest has added an additional dimension to the type of information attainable from complex sample mixtures without prior separation procedures. For this reason, the concerted application of ESI-FTICR and SORI-CID has enabled us to investigate not only the binding modes of NC, but also its chaperone activities in conformer conversion. Taking advantage of this approach, appropriate RNA mutants were designed to identify the structural determinants of the specific NC-SL1 interactions and investigate their role in the two-step model.
The results provided by these experiments have clearly shown that the effects induced by protein binding vary as a function of the specific site and that the outcome is highly dependent on the RNA structural context. In the case of monomeric SL1, NC can hamper the formation of the kissing complex in vitro by occluding the single-stranded palindrome and precluding loop-loop contacts between cognate hairpins. Alternatively, NC can bind with similar affinity to the stem-bulge motif, which results in the destabilization of the contiguous double-stranded structures. This event is inconsequential for monomeric RNA, but becomes an important factor in the context of the KL dimer for its role in facilitating the strand exchange necessary to complete isomerization. Upon structural rearrangement, the stem-bulge sites are recreated in the ED conformer with contributions from the opposite strands, but the local destabilizing effects induced by NC are not sufficient to produce observable effects on overall strand association. In constructs including a G on the 5′ side of the annealed palindromes, additional binding is provided by unpaired purines that assume a bulging extra-helical conformation in the junction of the KL conformer. This grip-like structure comprising nucleotides from both hairpins is stabilized by the binding of NC, which results in partial inhibition of strand exchange. Regardless of their identity, the junction purines are capable of sustaining protein interactions when they rearrange into the small bulge motifs of the ED structure, which have no observable consequences on strand association.
Finally, it should be noted that the 9-nt stretch including the palindrome and the flanking purines is highly conserved within each HIV-1 subtype.87 Further, an in vitro selection scheme designed to identify dimerization-competent constructs from randomized libraries has shown a very high degree of convergence toward only two autocomplementary sequences and very few variations of the flanking nucleotides.88 This study has also revealed a strong tendency of the central and flanking bases to covariate with a definite preference for the presence of AG and AA at the 5′ side of the subtype A and B palindromes, respectively, as shown by their wild-type sequences. Therefore, it is very remarkable that subtype A has evolved in nature to present simultaneously the more weakly associated palindrome and a junction that is less conducive to NC-mediated isomerization and stabilization, at least in vitro. The fact that isolated SL1 domains of the two subtypes exhibit clearly different modes of interaction with NC suggests the possible existence of distinct modulating mechanisms for the respective processes of structural rearrangement and genome compaction. The presence of the SL1 domain in the context of much larger RNA structures in vivo may also present significant topological constraints to the mechanism of structural rearrangement described here. The significance of these differential effects in the later stages of viral maturation will be the object of future investigations.
Materials and Methods
NC and RNA Constructs
Recombinant NC was expressed in E. coli BL21 (DE3)-pLysE, purified under non-denaturing conditions, and extensively desalted by ultrafiltration against 150 mM ammonium acetate with pH adjusted to 7.0.53, 70 The purity and integrity of the protein were confirmed directly by ESI-FTICR. All RNA constructs in the study (Scheme 2) were purchased from IDT (Coralville, IA), desalted by ultrafiltration, and quality-controlled by ESI-FTICR. Each stock concentration was determined by UV absorbance using molar extinction coefficients calculated from the respective sequences. The rationale for the design of the conformer-obligated constructs was discussed in detail in reference 70.
Binding and isomerization experiments
All experiments were performed using 5–10 μM solutions of each RNA substrate, which were obtained by appropriate dilution of the desired stock into 150 mM ammonium acetate buffer (pH 7.0). Before use, each sample was heated to 95°C for 5 minutes and then quickly cooled on ice to allow for proper folding of the RNA hairpins and obtain predominantly the KL conformer. The conformational state of each initial substrate was confirmed by SORI-CID, as described below. Binding experiments were completed by adding a predetermined volume of NC stock at room temperature. Although a wide range of protein-RNA ratios was explored, typical samples consisted of 30 μM and 10 μM final concentrations of NC and RNA, respectively. Isomerization experiments were carried out by incubating aliquots of the above samples in a water bath at 37°C for up to 3 h. More extensive incubation periods did not result in significant differences in the observed outcome.
Mass spectrometry
All analyses were performed on a Bruker Daltonics (Billerica, MA) Apex III FTICR mass spectrometer equipped with a 7T actively-shielded superconducting magnet and a nano-ESI source built in house.70 Desolvation temperature, skimmer voltage, and other source parameters were optimized to allow for the observation of intact RNA-RNA and protein-RNA noncovalent complexes, as previously described.53, 70 Analyte solutions were mixed with iso-propanol immediately before analysis to a final concentration of 10% in volume to assist desolvation. Typically, 5 μL samples were loaded into the nano-electrospray needle and a spray voltage of less than 1 kV was applied to the solution through a stainless steel wire inserted from the back end. No solvent pumps were necessary, as the solution flow-rate was dictated by the applied voltage and the size of the nano-ESI needle tip (typically ~1–2 μm). Spectra were acquired in negative ionization mode and processed using XMASS 7.0.2 (Bruker Daltonics, Billerica, MA). Scans were completed in broadband mode that allowed for a typical 150,000 resolving power at m/z 2000. The spectra were externally calibrated using a 1 mg/ml solution of CsI, which produced a series of peaks throughout the mass range of 1000–6000 m/z and enabled to achieve a typical mass accuracy of 20 ppm or better across the range. Each analysis was performed a minimum of three times and only representative spectra were shown.
In tandem mass spectrometry experiments, the precursor ion of interest was isolated in the FTICR cell using correlated rf sweeps (CHEF)89 and then activated through sustained off-resonance irradiation-collision induced dissociation (SORI-CID).75 Frequency offsets below and above the resonant frequency of the precursor ion were sampled to avoid possible “blind spots” in the product spectra.90 Best results were achieved by using irradiation frequencies that were 600 to 2000 Hz below that of the precursor ion. Mild activation regimes were reached by applying off-resonant pulses for 250 msec, using 26 to 31 dB attenuation of the maximum power output allowed by the hardware. Harsher dissociation conditions were achieved by activating the precursor ion of interest for the same time interval, but employing only 17 to 20 dB attenuation. Argon was used as the collision gas in pulsed bursts of 100–250 ms, which resulted in momentary increases from 1×10−11 to 1×10−7 mbar of the background pressure measured by the instrument gauge located underneath the ion optics. No attempt was made to determine the actual pressure within the FTICR cell. Twenty-five to fifty scans were typically averaged for each spectrum.
Data analysis
The different protein-RNA and RNA-RNA species observed in solution by ESI-FTICR or obtained from their gas-phase activation by SORI-CID were unambiguously identified from their observed molecular masses. The typical accuracy afforded by these determinations can be appreciated by comparing the experimental masses with the theoretical ones calculated from sequence, which are reported in Scheme 2 for each construct in the study. The typical high-resolution afforded by these experiments (Figure 1b, inset) allowed for an immediate solution of any ambiguity that might have resulted from nearly overlapping signals.
The partitioning among the different forms assumed by SL1 in solution was estimated in a semi-quantitative fashion from the signal intensity of each species divided by its respective charge state. The use of normalized intensities is legitimated by the fact that NC binding does not induce complete neutralization of the nucleic acid component, but rather produces a mere shift in charge distribution (see also 53, 70 and ref.s therein). With proper precautions and controls, this approach is sufficiently robust to allow for the determination of actual equilibrium binding constants.53, 55, 56 The data provided by the isomerization of wild-type SL1A in the presence of NC (Figure 6) can be used to better exemplify the treatment implemented here. In the SORI-CID spectrum in Figure 6b, the 1:1 product exhibited a −7 charge state and an absolute intensity of 2.13 × 107 arbitrary units, corresponding to a normalized intensity of 3.04 × 106 units. Likewise, the 2:2 product with a −9 charge state and an absolute intensity of 4.22 × 107 arbitrary units provided a normalized intensity of 4.69 × 106 units. However, the fact that two RNA equivalents were present in this species required that its intensity be counted twice, for a final normalized value of 9.38 × 106 units. Each normalized value divided by the sum of the values of all the species in the spectrum provided the percentage of the corresponding species. The partitioning was then completed by attributing monomeric products to the KL conformer and dimeric species to the ED counterpart, as reported in the box of Figure 6b. The same process was carried out for the SORI-CID spectra of all the species in Figure 6a to obtain the KL/ED partitioning included in parenthesis above each signal. Finally, summing the percentage of KL in each ion population, weighed by the respective normalized intensity in this spectrum, provided the overall percentage of KL species in the sample, as reported in the box of Figure 6a.
Acknowledgments
This research was funded by the National Institutes of Health (2R01-GM064328) and the National Science Foundation (CHE-0439067). N.A.H. was also supported by an NIH Chemistry Biology Interface Training Fellowship (T32-GM066706).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laughrea M, Jetté L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 2.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci U S A. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clever J, Parslow TG. Mutant Human Immunodeficiency Virus Type I Genomes with Defects in RNA Dimerization or Encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paillart JC, Berthoux L, Ottmann M, Darlix JL, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen N, Jetté L, Liang C, Wainberg MA, Laughrea M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol. 2000;74:5729–5735. doi: 10.1128/jvi.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muriaux D, Girard PM, Bonnet-Mathonière B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 9.Clever J, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci U S A. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muriaux D, Fossé P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 12.Laughrea M, Jetté L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernacchi S, Ennifar E, Toth K, Walter P, Langowski J, Dumas P. Mechanism of hairpin-duplex conversion for the HIV-1 dimerization initiation site. J Biol Chem. 2005;280:40112–40121. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- 15.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci U S A. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muriaux D, De Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 17.Rist MJ, Marino JP. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry. 2002;41:14762–14770. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Baba S, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J Biol Chem. 2001;276:31274–31278. doi: 10.1074/jbc.M104577200. [DOI] [PubMed] [Google Scholar]
- 19.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for retroviral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 20.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 21.Prats AC, Roy C, Wang PA, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greatorex J, Gallego J, Varani G, Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J Mol Biol. 2002;322:543–557. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J Mol Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y, Kerwood DJ, Paoletti AC, Shubsda MF, Borer PN. Stem of SL1 RNA in HIV-1: structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry. 2003;42:5259–5269. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 27.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 28.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveal an extended duplex with two adenine bulges. Struct Fold Des. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 29.Mujeeb A, Parslow TG, Zarrinpar A, Das C, James TL. NMR structure of the mature dimer complex of HIV-1 genomic RNA. FEBS Lett. 1999;458:387–392. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 30.Girard F, Barbault F, Gouyette C, Huynh-Dinh T, Paoletti J, Lancelot G. Dimer initiation sequence of HIV-1Lai genomic RNA: NMR solution structure of the extended duplex. J Biomol Struct Dyn. 1999;16:1145–1157. doi: 10.1080/07391102.1999.10508323. [DOI] [PubMed] [Google Scholar]
- 31.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 32.Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA Structures of the HIV-1 Dimerization Initiation Site in the Kissing-Loop and Extended-Duplex Dimers. J Biochem (Tokyo) 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 33.Ennifar E, Dumas P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. J Mol Biol. 2006;356:771–782. doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 34.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulyanov NB, Mujeeb A, Du Z, Tonelli M, Parslow TG, James TL. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J Biol Chem. 2006;281:16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita M, Fenn JB. Electrospray ion source. Another variation on the free-jet theme. J Phys Chem. 1984;88:4671–4675. [Google Scholar]
- 37.Aleksandrov ML, Gall LN, Krasnov VN, Nikolaev VI, Pavlenko VA, Shkurov VA. Extraction of ions from solutions under atmospheric pressure: a method of mass spectrometric analysis of bioorganic compounds. Doklady Akademii Nauk. 1984;277:379–383. [Google Scholar]
- 38.Comisarow MB, Marshall AG. Fourier transform ion cyclotron resonance. Chem Phys Lett. 1974;25:282–283. [Google Scholar]
- 39.Hendrickson CL, Emmett MR, Marshall AG. Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Annu Rev Phys Chem. 1999;50:517–536. doi: 10.1146/annurev.physchem.50.1.517. [DOI] [PubMed] [Google Scholar]
- 40.Ganem B, Li Y-T, Henion JD. Detection of oligonucleotide duplex forms by ion-spray mass spectrometry. Tetrahedron Lett. 1993;34:1445–1448. [Google Scholar]
- 41.Light-Wahl KJ, Springer DL, Winger BE, Edmonds CG, Camp DG, II, Thrall BD, Smith RD. Observation of a small oligonucleotide duplex by electrospray ionization mass spectrometry. J Am Chem Soc. 1993;115:803–804. [Google Scholar]
- 42.Hofstadler SA, Griffey RH. Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 43.Hofstadler SA, Sannes-Lowery KA, Hannis JC. Analysis of nucleic acids by FTICR MS. Mass Spectrom Rev. 2005;24:265–285. doi: 10.1002/mas.20016. [DOI] [PubMed] [Google Scholar]
- 44.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 45.Kapur A, Beck JL, Sheil MM. Observation of daunomycin and nogalamycin complexes with duplex DNA using electrospray ionzation mass spectrometry. Rapid Commun Mass Spectrom. 1999;13:2489–2497. doi: 10.1002/(SICI)1097-0231(19991230)13:24<2489::AID-RCM816>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 46.Gabelica V, De Pauw E, Rosu F. Interaction between antitumor drugs and a double-stranded oligonucleotide studied by electrospray ionization mass spectrometry. J Mass Spectrom. 1999;34:1328–1337. doi: 10.1002/(SICI)1096-9888(199912)34:12<1328::AID-JMS889>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 47.Sannes-Lowery KA, Mei H-Y, Loo JA. Studying aminoglycoside antibiotic binding to HIV-1 TAR RNA by electrospray ionization mass spectrometry. Int J Mass Spectrom Ion Proc. 1999;193:115–122. [Google Scholar]
- 48.Hofstadler SA, Sannes-Lowery KA, Crooke ST, Ecker DJ, Sasmor H, Manalili S, Griffey RH. Multiplexed screening of neutral mass-tagged RNA targets against ligand libraries with electrospray ionization FTICR MS: a paradigm for high-throughput affinity screening. Anal Chem. 1999;71:3436–3440. doi: 10.1021/ac990262n. [DOI] [PubMed] [Google Scholar]
- 49.Wan KX, Shibue T, Gross ML. Non-Covalent Complexes between DNA-Binding Drugs and Double-Stranded Oligodeoxynucleotides: A Study by ESI Ion-Trap Mass Spectrometry. J Am Chem Soc. 2000;122:300–307. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 50.Griffey RH, Hofstadler SA, Sannes-Lowery KA, Ecker DJ, Crooke ST. Determinants of aminoglycoside-binding specificity for rRNA by using mass spectrometry. Proc Nat Acad Sci USA. 1999;96:10129–10133. doi: 10.1073/pnas.96.18.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sannes-Lowery KA, Griffey RH, Hofstadler SA. Measuring dissociation constants of RNA and aminoglycoside antibiotics by electrospray ionization mass spectrometry. Anal Biochem. 2000;280:264–271. doi: 10.1006/abio.2000.4550. [DOI] [PubMed] [Google Scholar]
- 52.Rosu F, Gabelica V, Houssier C, De Pauw E. Determination of affinity, stoichiometry and sequence selectivity of minor groove binder complexes with double-stranded oligodeoxynucleotides by electrospray ionization mass spectrometry. Nucleic Acids Res. 2002;30:e82. doi: 10.1093/nar/gnf081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagan N, Fabris D. A direct mass spectrometric determination of the stoichiometry and binding affinity of the complexes between HIV-1 nucleocapsid protein and RNA stem-loops hairpins of the HIV-1 Ψ-recognition element. Biochemistry. 2003;42:10736–10745. doi: 10.1021/bi0348922. [DOI] [PubMed] [Google Scholar]
- 54.Gooding KB, Higgs R, Hodge B, Stauffer E, Heinz B, McKnight K, Phipps K, Shapiro M, Winkler M, Ng WL, Julian RK. High throughput screening of library compounds against an oligonucleotide substructure of an RNA target. J Am Soc Mass Spectrom. 2004;15:884–892. doi: 10.1016/j.jasms.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 55.Turner KB, Hagan NA, Fabris D. Inhibitory effects of archetypical nucleic acid ligands on the interactions of HIV-1 nucleocapsid protein with elements of Ψ-RNA. Nucl Acids Res. 2006;34:1305–1316. doi: 10.1093/nar/gkl004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int J Mass Spectrom Ion Proc. 2002;216:1–27. [Google Scholar]
- 57.Griffey RH, Greig MJ, Haoyun A, Sasmor H, Manalili S. Targeted Site-Specific Gas-Phase Cleavage of Oligoribonucleotides. Application in Mass Spectrometry-Based Identification of Ligand Binding Sites. J Am Chem Soc. 1999;121:474–475. [Google Scholar]
- 58.Turner KB, Hagan NA, Kohlway A, Fabris D. Mapping noncovalent ligand binding to stemloop domains of the HIV-1 packaging signal by tandem mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1401–1411. doi: 10.1016/j.jasms.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Liu C, Pasa-Tolic L, Hofstadler SA, Harms AC, Smith RD, Kang C, Sinha N. Probing RegA/RNA interactions using electrospray ionization-fourier transform ion cyclotron resonance-mass spectrometry. Anal Biochem. 1998;262:67–76. doi: 10.1006/abio.1998.2753. [DOI] [PubMed] [Google Scholar]
- 60.Rostom AA, Fucini P, Benjamin DR, Juenemann R, Nierhaus KH, Hartl FU, Dobson CM, Robinson CV. Detection and selective dissociation of intact ribosomes in a mass spectrometer. Proc Nat Acad Sci USA. 2000;97:5185–5190. doi: 10.1073/pnas.97.10.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosu F, Gabelica V, Houssier C, Colson P, De Pauw E. Triplex and quadruplex DNA structures studied by electrospray mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:1729–1736. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- 62.Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. Evaluation of binding of perylene diimide and benzannulated perylene diimide ligands to DNA by electrospray mass spectrometry. J Am Soc Mass Spectrom. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 63.Gale DC, Smith RD. Characterization of noncovalent complexes formed between minor groove binding molecules and duplex DNA by electrospray ionization-mass spectrometry. J Am Soc Mass Spectrom. 1995;6:1154–1164. doi: 10.1016/1044-0305(95)00530-7. [DOI] [PubMed] [Google Scholar]
- 64.Schnier PD, Klassen JS, Strittmatter EF, Williams ER. Activation Energies for Dissociation of Double Strand Oligonucleotide Anions: Evidence for Watson-Crick Base Pairing in Vacuo. J Am Chem Soc. 1998;120:9605–9613. doi: 10.1021/ja973534h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gabelica V, Rosu F, Houssier C, De Pauw E. Gas phase thermal denaturation of an oligonucleotide duplex and its complexes with minor groove binders. Rapid Commun Mass Spectrom. 2000;14:464–467. doi: 10.1002/(SICI)1097-0231(20000331)14:6<464::AID-RCM895>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 66.Wan KX, Gross ML, Shibue T. Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J Am Soc Mass Spectrom. 2000;11:450–457. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 67.Gabelica V, De Pauw E. Comparison between solution-phase stability and gas-phase kinetic stability of oligodeoxynucleotide duplexes. J Mass Spectrom. 2001;36:397–402. doi: 10.1002/jms.141. [DOI] [PubMed] [Google Scholar]
- 68.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Evaluation of complexation of metal-mediated DNA-binding drugs to oligonucleotides via electrospray ionization mass spectrometry. Nucleic Acids Res. 2001;29:E103–103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.David WM, Brodbelt JS, Kerwin SM, Thomas PW. Investigation of quadruplex oligonucleotide-drug interaction by electrospray ionization mass spectrometry. Anal Chem. 2002;74:2029–2033. doi: 10.1021/ac011283w. [DOI] [PubMed] [Google Scholar]
- 70.Hagan NA, Fabris D. Dissecting the protein-RNA and RNA-RNA interactions in the nucleocapsid-mediated dimerization and isomerization of HIV-1 stemloop 1. J Mol Biol. 2007;365:396–410. doi: 10.1016/j.jmb.2006.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casa-Finet JR, Medaglia N, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vuilleumier C, Bombarda E, Morellet N, Gérard D, Roques BP, Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 73.Urbaneja MA, Wu M, Casas-Finet JR, Karpel RL. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J Mol Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 74.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DJ, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucl Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gauthier JW, Trautman TR, Jacobson DB. Sustained off-resonance irradiation for collision-activated dissociation involving Fourier transform mass spectrometry. Collision-activated dissociation technique that emulates infrared multiphoton dissociation. Anal Chim Acta. 1991;246:211–225. [Google Scholar]
- 76.McLafferty FW. Tandem mass spectrometry. Science. 1981;214:280–287. doi: 10.1126/science.7280693. [DOI] [PubMed] [Google Scholar]
- 77.Cooks RG. Collision-induced Dissociation: Readings and Commentary. J Mass Spectrom. 1995;30:1215–1221. [Google Scholar]
- 78.Ding J, Anderegg RJ. Specific and nonspecific dimer formation in the electrospray ionization mass spectrometry of oligonucleotides. J Am Soc Mass Spectrom. 1994;6:159–164. doi: 10.1016/1044-0305(94)00102-6. [DOI] [PubMed] [Google Scholar]
- 79.Gabelica V, De Pauw E. Comparison of the collision-induced dissociation of duplex DNA at different collision regimes: evidence for a multistep dissociation mechanism. J Am Soc Mass Spectrom. 2002;13:91–98. doi: 10.1016/s1044-0305(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 80.McLuckey SA, Habibi-Goudarzi S. Decompositions of multiply charged oligonucleotide anions. J Am Chem Soc. 1993;115:12085–12095. [Google Scholar]
- 81.Nordhoff E, Kirpekar F, Roepstorff P. Mass spectrometry of nucleic acids. Mass Spectrom Rev. 1996;15:67–138. doi: 10.1002/(SICI)1098-2787(1996)15:2<67::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 82.Heck AJR, van den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 83.Theilleux-Delalande V, Girard F, Huynh-Dinh T, Lancelot G, Paoletti J. The HIV-1(Lai) RNA dimerization. Thermodynamic parameters associated with the transition from the kissing complex to the extended dimer. Eur J Biochem. 2000;267:2711–2719. doi: 10.1046/j.1432-1327.2000.01292.x. [DOI] [PubMed] [Google Scholar]
- 84.Windbichler N, Werner M, Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–6427. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aci S, Mazier S, Genest D. Conformational pathway for the kissing complex-->extended dimer transition of the SL1 stem-loop from genomic HIV-1 RNA as monitored by targeted molecular dynamics techniques. J Mol Biol. 2005;351:520–530. doi: 10.1016/j.jmb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 86.Mihailescu MR, Marino JP. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc Natl Acad Sci U S A. 2004;101:1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B, editors. HIV Sequence Compendium 2005. Theoretical Biology and Biophysics Group; Los Alamos National Laboratory: 2005. [Google Scholar]
- 88.Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Convergence of natural and artificial evolution on an RNA loop-loop interaction: the HIV-1 dimerization initiation site. Rna. 2000;6:1267–1276. doi: 10.1017/s1355838200000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Koning LJ, Nibbering NMM, van Orden SL, Laukien FH. Mass selection of ions in a Fourier transform ion cyclotron resonance trap using correlated harmonic excitation fields (CHEF) Int J Mass Spectrom Ion Proc. 1997;165/166:209–219. [Google Scholar]
- 90.Senko MW, Speir JP, McLafferty FW. Collisional activation of large multiply charged ions using Fourier transform mass spectrometry. Anal Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 91.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]