Abstract
Ribonuclease P (RNase P), a ribonucleoprotein (RNP) complex required for tRNA maturation, comprises one essential RNA (RPR) and protein subunits (RPPs) numbering one in bacteria, and at least four in archaea and nine in eukarya. While the bacterial RPR is catalytically active in vitro, only select euryarchaeal and eukaryal RPRs are weakly active despite secondary structure similarity and conservation of nucleotide identity in their putative catalytic core. Such a decreased archaeal/eukaryal RPR function might imply that their cognate RPPs provide the functional groups that make up the active site. However, substrate-binding defects might mask the ability of some of these RPRs, such as that from the archaeon Methanocaldococcus jannaschii (Mja), to catalyze precursor tRNA (ptRNA) processing. To test this hypothesis, we constructed a ptRNA-Mja RPR conjugate and found that indeed it self-cleaves efficiently (kobs, 0.15 min−1 at pH 5.5 and 55°C). Moreover, one pair of Mja RPPs (POP5-RPP30) enhanced kobs for the RPR-catalyzed self-processing by ∼100-fold while the other pair (RPP21-RPP29) had no effect; both binary RPP complexes significantly reduced the monovalent and divalent ionic requirement. Our results suggest a common RNA-mediated catalytic mechanism in all RNase P and help uncover parallels in RNase P catalysis hidden by plurality in its subunit make-up.
INTRODUCTION
Ribonuclease P (RNase P) is an endoribonuclease responsible for the 5′ maturation of tRNAs. It is a ribonucleoprotein (RNP) complex containing one essential RNA (RNase P RNA; RPR) in all three domains of life and a varying number of protein subunits (RNase P Protein; RPP): one in bacteria, and at least four and nine in archaea and eukarya, respectively (1–6). The bacterial RPR is catalytically active in the absence of its RPP under in vitro conditions that include a high concentration of Mg2+ (or another suitable divalent metal ion) which is essential for catalysis (1). However, only a few representative archaeal and eukaryal RPRs are weakly active under various conditions tested in vitro (6–9). This result is surprising given the remarkable secondary structure similarity and conservation of nucleotide identity in the putative catalytic core in all RPRs and raises the possibility that not all RPRs are functionally equivalent (Figure 1A–C; 10). We investigate here a euryarchaeal RPR that was reported to be inactive when tested alone (7,8) and examine the basis for its inactivity and its cooperation with cognate RPPs during catalysis.
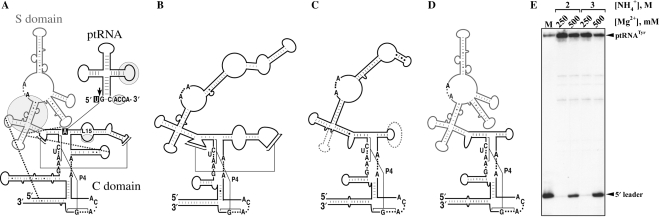
Figure 1.
An RPR hybrid comprising an S and C domain from a bacterial and an archaeal type M RPR, respectively, is catalytically active and can accurately process ptRNATyr. (A) The shaded circles, ovals, and black boxes depict known interactions between the Eco RPR and a ptRNA substrate (11). Arrow indicates the site of cleavage in the ptRNA. In the RPR, the C and S domains are indicated in black and gray, respectively. Thick dotted lines indicate tertiary interactions unique to the bacterial RPR. (B and C) Secondary structures of representative archaeal type A and M RPRs, respectively. The structural elements absent in type M RPR (C) are highlighted with dotted lines. (D) Secondary structure of the bacterial/archaeal hybrid RPR (EcoS-MjaC RPR) used in this study. The S domain of bacterial RPR is indicated in gray. Universally conserved nucleotides are indicated in A–D. (E) EcoS-MjaC RPR (2.5 μM) was tested for RNase P activity by incubating at 50°C for 15 min with 5′ labeled ptRNATyr (∼1 pM) in 50 mM Tris-acetate (pH 8) and different NH4+ and Mg2+ concentrations as indicated. M represents a size marker generated by processing of ptRNATyr by Eco RNase P.
Based on the conserved structural elements of the RPR, euryarchaeal RNase P is classified into types A and M (8). At the secondary structure level, type A RPRs resemble the ancestral bacterial RPRs while type M RPRs lack some of the structural elements implicated in substrate binding in bacterial RPRs (Figure 1A–C; 8,10,11). Consistent with this observation, only a few type A RPRs and none of the type M RPRs are catalytically active in vitro without their RPPs (7). In this study, we employ Methanocaldococcus jannaschii (Mja) RPR as a prototype for type M RPRs. Using a unimolecular enzyme-substrate conjugate expected to alleviate substrate-binding defects in Mja RPR, we establish that Mja RPR (in the absence of RPPs) supports cleavage in cis, thus reaffirming the RPR's pivotal catalytic role.
If the RPR is the catalytic moiety in RNase P from all three domains of life, why do archaeal/eukaryal RNase P holoenzymes require multiple RPPs for function in vivo while bacterial RNase P employs a single RPP, which serves to normalize the binding affinity and rate of cleavage of different precursor tRNAs (ptRNAs) by the RPR (12–15)? Biochemical characterization of a partially purified archaeal RNase P holoenzyme demonstrated that the RPR is associated with at least four RPPs (POP5, RPP30, RPP21 and RPP29), which have eukaryal homologs (2,3). We recently reconstituted Pyrococcus furiosus (Pfu; type A) RNase P in vitro in an effort to develop archaeal RNase P as an experimental surrogate for the more complex eukaryal counterpart (16). In this study, by examining the role of RPPs in aiding the self-processing of the type M RPR-ptRNA cis conjugate, we are beginning to uncover functional parallels between the archaeal RPPs and the sole bacterial RPP.
MATERIALS AND METHODS
Cloning of RPRs and ptRNA-RPR cis conjugates
Complete details are provided in the Supplementary Data.
RNase P activity assays
The hybrid RPR and cis conjugates were folded as follows: incubation at 50°C for 50 min in water followed by 37°C for 30 min in 50 mM Tris-acetate (pH 8) containing either 100 mM Mg(OAc)2 and 2.5 M NH4OAc (for ptTyr-S5-M RPR) or 10 mM Mg(OAc)2 and 800 mM NH4OAc (for ptTyr-S3-M, ptTyr-S3-ΔS M and EcoS-MjaC RPRs).
In the absence of RPPs, trans cleavage by the hybrid RPR or self-cleavage by the cis conjugates was initiated by addition of 500 mM Mg(OAc)2 (or the amounts indicated if the experiment was a titration analysis).
To ensure that ptTyr-S3-M RPR does not self-cleave during reconstitution with Mja RPPs, we used two different pre-incubation strategies that permit assembly without promoting significant cleavage (Table 1). In the first approach, 50 nM ptTyr-S3-M RPR was mixed with 250 nM of each binary complex in 50 mM MES (pH 5.1 at 55°C), 800 mM NH4OAc, and either 1 mM Mg(OAc)2 (for POP5-RPP30) or 10 mM Mg(OAc)2 (for RPP21-RPP29). In the second approach, which was necessary for the reaction performed in the presence of all four RPPs, 25 mM Ca2+ replaced Mg2+ in the pre-incubation step. In both instances, reactions were initiated by adding an equal volume of 50 mM MES (pH 5.1 at 55°C), 800 mM NH4OAc, and 200 mM Mg(OAc)2 (final [Mg2+] = 100 mM).
Table 1.
Effect of Mja RPPs on the ionic requirements and rate of self-processing of ptTyr-S3-M and ptTyr-S3-ΔS M RPR conjugates at pH 5.1 and 55°C
| RNase P assayed | kobs, min−1 | [NH4+], M | [Mg2+], M |
|---|---|---|---|
| Reconstituted and pre-incubated with low [Mg2+] | |||
| ptTyr-S3-M RPR | 0.05 ± 0.01 | 2.5 | 0.5 |
| +RPP21-RPP29 | 0.06 ± 0.01 | 0.8 | 0.1 |
| +POP5-RPP30 | 4.83 ± 0.02 | 0.8 | 0.1 |
| Reconstituted and pre-incubated with Ca2+ | |||
| ptTyr-S3-M RPR | |||
| + POP5-RPP30 | 5.16 ± 0.08 | 0.8 | 0.1 |
| +4 RPPs | 5.46 ± 0.04 | 0.8 | 0.1 |
| ptTyr-S3-ΔS M RPR* | 0.004* | 2.5 | 0.5 |
| +POP5-RPP30 | 4.86 ± 0.06 | 0.8 | 0.1 |
*Rate at pH 5.1 obtained by extrapolating from values obtained between pH 5.4 and 7.1 (Fig. 3A).
Unless stated otherwise, all assays involved incubation for a specified period at 55°C in a thermal cycler. Reactions were terminated by adding an equal volume of stop solution [10 M urea, 5 mM EDTA, 0.05% (w/v) xylene cyanol, 10% (v/v) phenol]; if the time of incubation was short (e.g. 5 s) to accommodate rapid cleavage, reactions were terminated by first immersing the reaction tubes in liquid nitrogen and subsequently adding stop solution.
Kinetic analysis
The substrate and product RNAs were separated on either 8% or 10% (w/v) polyacrylamide/7 M urea gels and visualized by PhosphorImaging (Molecular Dynamics). The extent of cleavage was quantitated using ImageQuant (Molecular Dynamics) software. To obtain the rate of product formation (kobs), the % product formed at time t (Pt) was fit to Pt = P∞(1 − e−kt) using Kaleidagraph software (Synergy). Background cleavage during the 20-min pre-incubation used for RNP assembly was typically less than 5% and corrected using Pt = Po + (P∞ − Po) (1 − e−kt), where Po refers to product formed at to (i.e. the end of the pre-incubation). In the cis conjugate reactions, the maximum product formed varied from 25 to 95% depending on the catalytic entity tested and assay pH.
Dependence of rate on assay pH
The rate of product formation was determined at 55°C and various pH values: 5.1–6.3 for ptTyr-S3-M RPR, 4.65–5.1 for ptTyr-S3-M RPR + POP5-RPP30, and 5.4–7.1 for ptTyr-S3-ΔS M RPR; 50 mM ammonium acetate (pH<5) or MES (pH>5) or HEPES (>6.5) buffers were employed in these assays. At each pH, at least three replicates were performed to obtain the mean and standard deviation values.
RESULTS
Archaeal type M RPR supports ptRNA processing independently of its cognate RPPs
Under prolonged incubation at pH 6 in the presence of 0.2 M Mg2+ and 0.8 M NH4+, human RPR was recently demonstrated to process a ptRNA-based model substrate (kobs ∼10−5 min−1, the pseudo-first order rate constant under single-turnover conditions; 9). Subsequently, the Cyanophora paradoxa cyanelle RPR, which is part of an RNase P with high protein content, was also reported to exhibit ptRNA cleavage at pH 6, 0.1 M Mg2+ and 0.1M NH4+ (kobs ∼ 10−3 min−1; 17). However, when we examined if the euryarchaeal type M RPRs would be active under analogous assay conditions with Escherichia coli (Eco) ptRNATyr as the substrate, we observed no activity with the Archaeoglobus fulgidus (Afu) and Mja RPRs (data not shown). No ptRNA processing activity was evident even with a variety of other assay conditions tested (37–55°C; 0.1–0.4 M Mg2+, 0.8–3 M NH4+, pH 6–7.5; and RPR concentrations up to 20 μM; data not shown). The failure of type M RPRs to catalyze ptRNA cleavage in trans contrasts with the human RPR and might reflect differences in their structures.
If the inability of Afu and Mja RPRs to catalyze ptRNA processing is due to the absence of specific RNA structural elements (e.g. P8, L15; Figure 1C) implicated in ptRNA binding in the bacterial RPR (Figure 1A; 8,11,18–21), alleviation of substrate-binding defects might render the type M RPR active. Towards this objective, we constructed a bacterial-archaeal (type M) hybrid RPR based on three observations. First, biochemical studies delineated the presence of two independently folding modules in bacterial RPRs: a substrate-specificity (S) domain that has conserved nucleotides for recognition of the T stem-loop of the ptRNA; and a catalytic (C) domain which can (i) recognize the leader, acceptor stem, and the 3′ terminal CCA sequence, and (ii) cleave the ptRNA (18–24). Second, we recently demonstrated that in Pfu RPR (euryarchaeal type A), the C domain was capable of cleaving ptRNA in the presence of two of its cognate RPPs (16). Lastly, mitochondrion-encoded RPRs in jakobid flagellates (e.g. Reclinomonas americana) became active upon fusion of their C domains with the S domain of Eco RPR (25). Therefore, guided in part by the recent high-resolution structures of bacterial RPRs (26,27) and prior biochemical studies in which similar hybrids were generated, we constructed a hybrid RPR with the S domain of Eco RPR fused to the putative C domain of Mja RPR (EcoS-MjaC RPR; Figure 1D and Supplementary Data). Indeed, EcoS-MjaC RPR accurately processes ptRNATyr (Figure 1E). Therefore, Mja RPR supports trans cleavage of ptRNATyr if its S domain, probably defective in substrate binding, is replaced with a bacterial S domain that does not suffer from this limitation.
Since the above experiment revealed that the Mja RPR's inactivity is likely due to defects in ptRNA recognition/binding, we inquired if an enzyme-substrate (ES) covalent conjugate in which a ptRNA is covalently tethered to the Mja RPR would facilitate its processing. However, we did not observe any cis cleavage when we tested conjugates in which ptRNATyr was attached to the 3′ end of either Afu or Mja RPR [with spacers of either 50 nts (Afu) or 25 nts (Mja) to the cleavage site; data not shown].
As there was precedence for bacterial and archaeal type A RPRs supporting cis cleavage of a ptRNA attached to the L15 loop (7,28), we made similar constructs with Mja RPR especially since L15 is likely to be part of the active site in bacterial RPR (Figure 2A; see Supplementary Data; 20,21,26,27). When we tested such conjugates with spacers of 0, 5 and 10 nts, only the latter two were cleaved. However, since the 10-nt-spacer conjugate also exhibited some mis-cleavage, only the 5-nt-spacer conjugate was characterized further. The accuracy of cleavage in ptRNATyr-S5-Mja RPR (abbreviated as ptTyr-S5-M RPR; Figure 2A) was confirmed by comparing the migration of the 5′ leader with that of a size marker generated by partial digestion of 5′ labeled ptTyr-S5-M RPR with RNase T1 (Figure 2B).
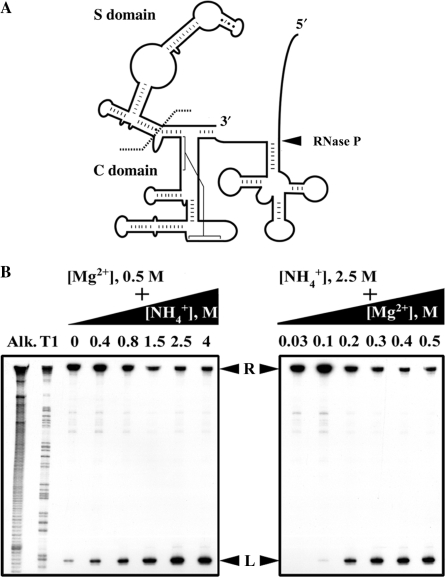
Figure 2.
Design and optimization of self-cleavage conditions for an active-site tethered ES conjugate. (A) Illustration of ptTyr-S3 (and S5)-M RPR in which ptRNATyr is attached to L15 of Mja RPR with either a 3- or 5-nt spacer. The C and S domains are demarcated to indicate what was deleted in ptTyr-S3-ΔS M RPR. (B) Titration of monovalent and divalent cations to identify the optimal conditions for maximal self-cleavage of 5′ labeled ptTyr-S5-M RPR (see text for details). R and L indicate RPR cis conjugate and the 5′ leader, respectively.
To identify conditions that permit efficient and accurate self-cleavage of ptTyr-S5-M RPR, we varied different parameters. Either the Mg2+ concentration was held constant at 0.5 M while the NH4+ concentration was increased from 0 to 4 M, or the NH4+ concentration was maintained at 2.5 M while the Mg2+ concentration was raised from 0 to 0.5 M (Figure 2B). Such experiments together with others where the assay pH or temperature was altered, established the optimal condition as 55°C in 50 mM Tris-acetate (pH 8), 500 mM Mg(OAc)2 and 2.5 M NH4OAc.
The extent of processing of 5′ labeled ptTyr-S5-M RPR was determined by quantitating formation of the 5′ leader. A single exponential function adequately describes the rate of product formation (data not shown). We next inquired if the phosphodiester bond-breaking step is rate limiting. Deprotonation of a hydrated Mg2+ ion in the RPR's active site is believed to generate the hydroxide nucleophile for attacking the scissile phosphodiester linkage in the ptRNA substrate (29,30). Therefore, evidence for the chemical step being rate limiting is generally derived from correlating the rate of product formation with the hydroxide ion concentration (i.e. specific base catalysis; 30). A slope of ∼1 in plots of log(kobs) versus pH reflects cleavage being the sole rate-limiting determinant; absence or decrease in such a dependence typically implies that other factors (e.g. conformational rearrangements, substrate docking) also contribute to the rate-limiting step.
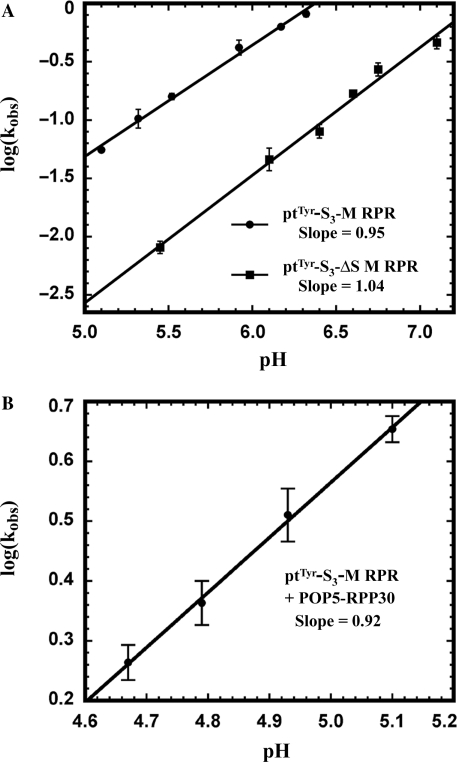
When we assayed ptTyr-S5-M RPR at pH 5.5 and 7.5, we observed only a 2-fold increase in kobs as opposed to the 100-fold enhancement expected if the bond-breaking step is solely rate limiting. Based on the premise that there might be slower steps preceding the chemical step during self-cleavage of ptTyr-S5-M RPR, we inquired if alterations in the length of the spacer (3, 4, 8 or 9 nts) might yield a cis conjugate whose rate is determined only by phosphodiester hydrolysis. Indeed, log(kobs) versus pH for the self-cleavage of a 3-nt-spacer conjugate, ptTyr-S3-M RPR, exhibited a slope of 0.95 in the pH range 5.1–6.3 (Figure 3A). As expected for a first-order reaction, we found that the kobs values at various ptTyr-S3-M RPR concentrations are nearly identical.
Figure 3.
Dependence of kobs on assay pH for self-cleavage of ptTyr-S3-M RPR and ptTyr-S3-ΔS M RPR (A) and ptTyr-S3-M RPR + POP5-RPP30 (B).
The kobs value of 0.15 min−1 at pH 5.5 and 55°C for ptTyr-S3-M RPR compares favorably to 0.75 min−1 at pH 5.5 and 50°C reported for ptAsp-S5-Eco RPR (31). This parallel in kobs values is striking given that the type M RPR lacks the functional S domain present in Eco RPR. Tethering of the substrate to the type M RPR appears to have rendered the S domain less essential. Importantly, the similar kobs values, despite the different ptRNA substrates, sites of conjugation, and assay temperatures used in the two studies, might reflect comparable ptRNA cleavage potential for the C domains of bacterial and archaeal RPRs.
Only a subset of Mja RPPs influences the kobs of ptTyr-S3-M RPR self-cleavage
To examine the contribution of Mja RPPs to ptTyr-S3-M RPR catalysis, we cloned the genes encoding Mja POP5, RPP21, RPP29 and RPP30 into vectors that allow T7 RNA polymerase-based overexpression in E. coli BL21(DE3) cells. Our previous studies on in vitro reconstituted Pfu RNase P revealed that the four RPPs function as two binary complexes (POP5-RPP30 and RPP21-RPP29; 16); therefore, we co-overexpressed the Mja RPPs as binary complexes in E. coli. By exploiting the high pI values of Mja RPPs (see Supplementary Data), we were able to purify the complexes to near homogeneity by cation-exchange chromatography. Details of the cloning, overexpression and purification of Mja RPPs will be described elsewhere. Purified Mja RPPs when combined with the Mja RPR generated a functional Mja RNase P holoenzyme that exhibited multiple turnover (kcat ∼ 6–24 min−1) at 55°C with Eco ptRNATyr or Synechocystis ptRNAGln (32) as the substrate. A kcat value of 34 min−1 was reported for cleavage of Bacillus subtilis ptRNAAsp at 50°C by partially purified native Mja RNase P (33).
Both Mja POP5-RPP30 and RPP21-RPP29 decrease the concentration of monovalent and divalent ions required for cis cleavage of ptTyr-S3-M RPR; the Mg2+ requirement decreases from 500 to 100 mM with either binary complex and decreases further to 20 mM with both (Figure 4A; Table 1). In addition, Mja POP5-RPP30, but not RPP21-RPP29, enhances by 100-fold the maximal kobs for self-processing of ptTyr-S3-M RPR at pH 5.1 and 55°C (0.05 to 4.8 min−1; Table 1). A plot of log (kobs) versus pH for the ptTyr-S3-M RPR + POP5-RPP30 exhibited a slope of 0.92 (Figure 3B). Slopes close to unity in plots of log(kobs) versus pH for the self-cleavage of ptTyr-S3-M RPR with and without POP5-RPP30 indicate that the chemical step is rate limiting in both instances.
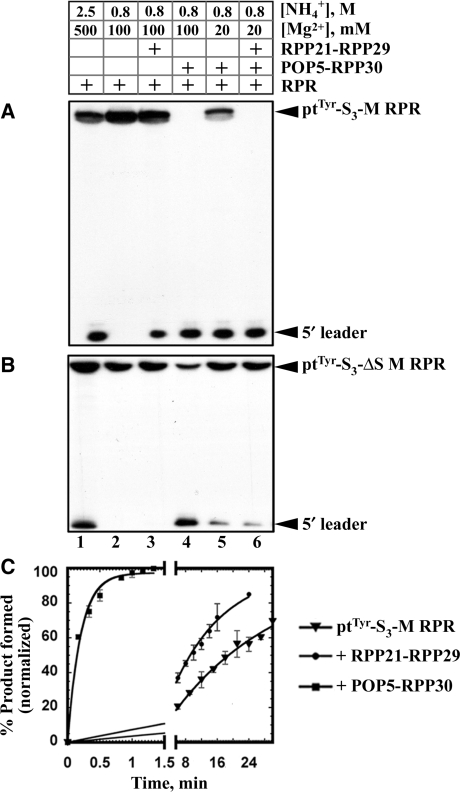
Figure 4.
Effect of Mja RPPs on the self-processing rate of ptTyr-S3-M RPR and ptTyr-S3-ΔS M RPR. All assays were performed in 50 mM MES (pH 5.1) at 55°C. 5′ labeled ptTyr-S3-M RPR (A) or ptTyr-S3-ΔS M RPR (B) was reconstituted with binary complexes of Mja RPPs at various concentrations of monovalent and divalent cations as indicated. (C) Rate of self-cleavage of the ptTyr-S3-M RPR without RPPs (triangles), with RPP21-RPP29 (circles) or with POP5-RPP30 (squares); assay conditions are listed in Table 1.
We next determined whether all four RPPs together will enhance kobs to a level greater than that observed with just POP5-RPP30. However, technical issues prevented us from doing this experiment under conditions employed for the binary complexes. For reactions involving the RPR reconstituted with RPPs, we typically employ a pre-incubation step (10 min at 37°C followed by 10 min at 55°C) in a Mg2+-containing buffer to permit RNP assembly prior to substrate addition (16). Since such an approach with the cis conjugate and four RPPs triggers significant self-cleavage during the pre-incubation step, we devised an alternative strategy.
As kinetic and thermodynamic studies using bacterial RPRs and ptRNAs revealed that the RPR adopts the correct tertiary fold for optimal ES complex formation with Ca2+, a divalent cation that promotes efficient ptRNA binding but not processing (30), we pre-incubated ptTyr-S3-M RPR either with all four RPPs or just POP5-RPP30 in a buffer containing 25 mM Ca2+ instead of Mg2+; cleavage was then initiated by adding an excess of Mg2+ to displace the Ca2+. To estimate the concentration of Mg2+ required for displacing the Ca2+ and promoting maximal cleavage, the rate of self-processing was determined with a Mg2+-titration analysis from 10 to 150 mM Mg2+, which revealed that 100 mM Mg2+ is optimal.
Despite the viability of the above approach, it is conceivable that the calculated rate is influenced adversely by the pre-incubation with Ca2+ and its subsequent displacement by Mg2+. Therefore, we compared the kobs for self-processing of ptTyr-S3-M RPR + POP5-RPP30 under two conditions: assembled in a pre-incubation buffer containing either 1 mM Mg2+ or 25 mM Ca2+ and subsequently assayed in the presence of 100 mM Mg2+. In both cases, the result was similar (4.8 versus 5.2 min−1), thus validating the Ca2+-based approach. Using this method, we determined that the kobs for ptTyr-S3-M RPR self-cleavage with just POP5-RPP30 (5.2 min−1) was not appreciably enhanced by addition of RPP21-RPP29 (5.5 min−1; Table 1).
In the studies described above, due to technical issues thwarting the use of a rapid quench flow apparatus to perform transient kinetic assays at near-neutral pH and 55°C, we used an assay pH of 5.1 to enable manual determination for the rate of self-cleavage of ptTyr-S3-M RPR. Such an approach might be subject to the limitation that protein–protein interactions at pH 5.1 might differ from those occurring at physiological pH.
Removal of the S domain from ptTyr-S3-M RPR does not eliminate self-cleavage activity
Deletion mutagenesis studies on type A bacterial (e.g. Eco) and archaeal (e.g. Pfu) RPRs established that their C domains support catalysis albeit only in the presence of cognate RPPs (16,22,23,34). With the C domain of B. subtilis RPR (bacterial type B), the cleavage rate was 25 000-fold lower compared to the wild-type RPR and 40-fold lower in the presence of the RPP (35). These observations, together with results from footprinting and kinetic studies (18,19), suggest that the S domain contributes to ptRNA recognition and helps position the substrate for optimal cleavage. Reasoning that the decreased efficiency of ptRNA processing with the C domains of bacterial/archaeal RPRs is likely due to substrate-binding/positioning defects caused by removal of the S domain, we hypothesized that the Mja RPR-ptRNA cis conjugate should be active even without the S domain since the substrate is covalently tethered to the enzyme. Indeed, ptTyr-S3-ΔS M RPR, the cis conjugate lacking the S domain, is active (Figure 4B, lane 1). Interestingly, instead of the 25 000-fold defect observed in the trans cleavage scenario with the C domain of B. subtilis RPR (35), cleavage of ptTyr-S3-ΔS M RPR is only 12-fold slower than ptTyr-S3-M RPR at pH 5.1 (0.004 versus 0.05 min−1; Figure 3A; Table 1). Clearly, removal of the S domain has a more adverse impact on the trans compared to the cis cleavage reaction used here. The activity of the ΔS M RPR also attests to the ptRNA processing ability of the C domains of all RPRs, even those bereft of the functionally important L15 and the neighboring P16/P17 helices present in the bacterial RPR (9). The Mja ΔS RPR in ptTyr-S3-ΔS M RPR also represents the smallest functional RPR yet identified (135 nts) and could be used in high-resolution structural studies.
We examined the effect of RPPs on the self-cleavage rate of ptTyr-S3-ΔS M RPR. Addition of POP5-RPP30 resulted in a kobs value of 4.9 min−1, similar to 5.2 min−1 observed with the wild type and indicates that this binary complex can compensate for the missing S domain. In contrast, addition of RPP21-RPP29 to ptTyr-S3-ΔS M RPR impacts neither the rate nor the ionic requirement. This finding suggests that RPP21-RPP29 asserts its effect on the RPR by binding to the S domain, a claim confirmed by our ongoing footprinting analysis which indicates that RPP21-RPP29 binds to the S domain (unpublished observations).
DISCUSSION
RPR is the catalytic subunit of RNase P in all three domains of life
Although the bacterial RPR alone is catalytically active under in vitro conditions of high ionic strength, until recently RPRs from many archaeal and all eukaryal sources were reported as incapable of catalysis without their cognate RPPs. These archaeal and eukaryal RPRs were not rendered active even in the presence of high concentrations of monovalent and divalent cations, which occasionally mitigate structural defects that prevent generation of an active RNA tertiary fold (6–8,36,37). The decreased activity in these RPRs might reflect a natural course towards a more complex RNP in which protein subunits provide functional groups that make up the active site. However, results from a recent study and the data reported here do not support such a premise. First, Kikovska et al. (9) showed that the human RPR can process different ptRNAs and model substrates, albeit 106-fold slower than that of bacterial RPR. Second, we have demonstrated here through engineering strategies intended to overcome substrate-binding defects that a euryarchaeal type M RPR, thus far found to be incapable of ptRNA processing, can catalyze this reaction (Figures 1 and 2).
Since the human and Mja (archaeal type M) RPRs can support ptRNA processing in the absence of RPPs when 150–500 mM Mg2+ is provided, it is evident that they possess the crucial structural elements required for generating the active site. These findings together with the fact that all characterized RPPs, either individually or in combination, cannot promote ptRNA processing without their RPRs confirm that the RPR is the catalytic moiety in all three domains of life. While such a broad inference might seem inconsistent with reports of failure to detect archaeal/eukaryal RPR-alone-catalyzed ptRNA processing, these results might reflect either an inability to form an active RPR fold in vitro or a masking of the cleavage capability due to extremely weak ptRNA binding (37).
Cleavage by Mja RPR is significantly enhanced by POP5-RPP30 at lower Mg2+ concentrations
Addition of the sole bacterial RPP normalizes the binding affinity and rate of cleavage of different ptRNAs by the cognate RPR; moreover, it enhances the affinity for Mg2+ ions in the active site thereby making a vital contribution to rate enhancement at physiological Mg2+ concentrations (14,15). What then are the roles played by the multiple archaeal/eukaryal RPPs?
Our studies on Pfu RNase P, where we examined the trans cleavage of a ptRNA under multiple turnover conditions, revealed that POP5-RPP30 (but not RPP21-RPP29) enhances the kcat of the RPR by nearly 40-fold, in fact to the same extent observed with all four RPPs (16). This finding indicated that POP5-RPP30 plays a vital role in cleavage and/or product release. Although a comprehensive description of the kinetic scheme requires determining the rate constants for individual steps, we rationalized that studying the Mja RPR cis conjugate in the absence and presence of its cognate RPPs might permit us to focus on the chemical step (akin to a single-turnover reaction) without influence from substrate binding or product release. Even for a cis conjugate, however, kobs (the apparent rate of product formation) need not be the rate of the chemical step as steps prior to cleavage could be rate limiting (see scheme below).
The maximal kobs for self-cleavage of ptTyr-S3-M RPR is accelerated ∼100-fold by Mja POP5-RPP30 but not at all by RPP21-RPP29 (Table 1). Similarly, only POP5-RPP30 promotes the trans cleavage of ptRNA by Mja RPR under multiple turnover conditions (unpublished observations). These findings are also consistent with our earlier observation that Pfu RPP21-RPP29 leaves the kcat of the Pfu RPR-catalyzed reaction unaltered while decreasing the Km five-fold and lowering the Mg2+ requirement (16).
We interpret our current results within a framework based on the kinetic scheme put forth by Harris and coworkers for trans cleavage of ptRNAs by Eco RNase P (14). When the role of the Eco RPP in RPR-mediated cleavage of ptRNAs with and without consensus recognition sequences was examined in single-turnover reactions, the RPP increased by 100- to 1000-fold the kobs for non-consensus ptRNA processing by the cognate RPR. The scheme below was used to explain the role of the bacterial RPP in catalysis. Subsequent to substrate binding, a conformational change from ES to ES*, which helps position the ptRNA and catalytic metal ions optimally for cleavage, had already been proposed based on results from various kinetic and structural studies (38,39). Because the bacterial RPR's kobs with tight-binding, consensus ptRNAs increases only three fold by the RPP, the possibility of the RPP contributing functional groups to catalysis and thereby increasing kc (the rate of the chemical step) was discounted. What then accounts for the dramatic RPP-facilitated increase in kobs with non-consensus substrates? Since only Kconf and kc contribute to kobs in a single-turnover trans cleavage reaction, the RPP was then inferred to influence the equilibrium (Kconf) that precedes the slower bond-breaking step. By shifting the equilibrium from ES to ES* for the atypical ptRNAs, the bacterial RPP was postulated to stabilize ES* and enhance catalysis (14).
In the case of ptTyr-S3-M RPR, the tethered substrate renders it similar to the ES complex in the trans cleavage scheme. Mja POP5-RPP30 (but not RPP21-RPP29) enhances kobs for self-cleavage of ptTyr-S3-M RPR with a concomitant reduction in the requirement for Mg2+. The kobs for the cis conjugate is dictated only by Kconf and kc. A direct increase in kc upon addition of Mja POP5-RPP30 is unlikely since it would necessitate an alternative mechanism from that employed by the Mja RPR, for which there is no evidence. Moreover, both Mja and Eco RPR cis conjugates exhibit a comparable efficiency suggesting that the functional groups for catalysis are present in the RPR (31). Therefore, the 100-fold increase in kobs upon addition of POP5-RPP30 must arise from increased conversion of ES to ES*. Additional support for this functional parallel between POP5-RPP30 and bacterial RPP stems from the findings that the tertiary structures of archaeal POP5 and bacterial RPP are strikingly similar (40,41), and POP5-RPP30 and Eco RPP footprint at similar locations in the C domains of their respective RPRs (16,42–44).
RPP21-RPP29 does not impact the kobs for self-cleavage but probably stabilizes the RPR's tertiary fold and substitutes for some of the RNA–RNA tertiary interactions that strengthen the structural core in bacterial RPRs. In this fashion, it might mirror protein cofactors that facilitate the self-splicing reactions of certain group I introns by tertiary structure capture (36,45; for a comparison of models of the bacterial and archaeal RNase P holoenzymes, see Supplementary Data).
Although RPP21-RPP29 does not contribute to the kobs for self-processing of ptTyr-S3-M RPR, it renders ptTyr-S3-M RPR active at 100 mM Mg2+ (Figure 4A, lanes 2 and 3). The maximal kobs determined for this reaction is similar to that observed for the RPR-alone reaction in the presence of its optimal Mg2+ concentration of 500 mM (0.06 versus 0.05 min−1; Table 1). Therefore, RPP21-RPP29 enables the RPR to attain its maximal kobs at a lower Mg2+ concentration perhaps by enhancing the affinity of the RNP for Mg2+ without affecting ES*. If these RPPs and Mg2+ bind to the folded state of the RPR, their binding will be thermodynamically coupled.
Since the Mja RPR does not exhibit trans cleavage, we used the rate of self-processing of a cis conjugate as the baseline for its catalytic potential and then determined how RPPs influenced this rate. In the cis construct, the substrate is docked on the enzyme though likely not optimally positioned. If RPP21-RPP29 plays a role in substrate binding, its effect on the catalytic efficiency of a trans cleavage reaction might be more pronounced than in a cis reaction where the substrate is already docked. In fact, RPP21-RPP29 enhances by two fold the kobs for self-cleavage of a cis conjugate with a 5-nt spacer in contrast to a 3-nt-spacer construct where it has no effect (Table 1; data not shown). We are investigating if the pattern of activation by RPPs differs between a cis and a trans cleavage reaction.
The C domain of type M RPR is sufficient for processing a ptRNA provided in cis
Despite the identification of a conserved C domain in all RPRs, previous studies suggested that this domain by itself was either not functional in the absence of cognate RPPs or that its activity was nearly 25 000-fold weaker than the wild type (22,34,35). If this defect is attributable to weak substrate binding, covalent attachment of the substrate should remedy it. Indeed, our study shows that self-cleavage of ptTyr-S3-ΔS M RPR is only 12-fold slower than ptTyr-S3-M RPR at pH 5.1 (Table 1); moreover, the decreased activity suggests that the S domain does play a role in cleavage even in the cis conjugate (perhaps, by influencing Kconf). Such a function of the S domain is clearly redundant with POP5-RPP30 since both ptTyr-S3-M RPR and ptTyr-S3-ΔS M RPR display near identical rates of self-cleavage in the presence of POP5-RPP30 (Table 1). This result parallels the rescue of the bacterial RPR's C domain by its RPP (35) and once again reveals similarities in the functioning of bacterial and archaeal RNase P.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to members of the Gopalan laboratory for useful suggestions and reagents during the course of this investigation, especially to Dr Lien Lai for her advice on cloning strategies and invaluable critique that improved the manuscript. We are indebted to Dr Mark Foster (OSU) for his insightful and important comments on the interpretation of kinetic data. We thank Dr Tim Eubank (OSU) for generously designing the graphics for Figure 2 in the Supplementary Data, and Drs Edward J. Behrman (OSU), Mark Foster and Roland Hartmann (University of Marburg) for comments on the manuscript. This work was supported by National Institutes of Health Grant R01 GM067807 (to Mark Foster and V.G.) and American Heart Association Pre-Doctoral Fellowship 0515218B (to D.K.P.). Funding to pay the Open Access publication charges for this article was provided in part by private funds and the 2008 US Economic Stimulus payment.
Conflict of interest statement. None declared.
REFERENCES
- 1.Guerrier-Takada C, Gardiner K, Marsh T, Pace NR, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 2.Jarrous N, Altman S. Human ribonuclease P. Methods Enzymol. 2001;342:93–100. doi: 10.1016/s0076-6879(01)42538-9. [DOI] [PubMed] [Google Scholar]
- 3.Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8:296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Gopalan V, Altman S. Gesteland RF, Cech TR, Atkins JF The RNA World. Cold Spring Harbor, NY: CSH Laboratory Press; 2006. Chapter 6.1 (only online at http://rna.cshl.edu) [Google Scholar]
- 6.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris JK, Haas ES, Williams D, Frank DN, Brown JW. New insights into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001;7:220–232. doi: 10.1017/s1355838201001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JW. The ribonuclease P database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christian EL, Zahler NH, Kaye NM, Harris ME. Analysis of substrate recognition by the ribonucleoprotein endonuclease RNase P. Methods. 2002;28:307–322. doi: 10.1016/s1046-2023(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh J, Andrews AJ, Fierke CA. Roles of protein subunits in RNA-protein complexes: lessons from ribonuclease P. Biopolymers. 2004;73:79–89. doi: 10.1002/bip.10521. [DOI] [PubMed] [Google Scholar]
- 13.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Harris ME. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007;13:1505–1515. doi: 10.1261/rna.571007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl Acad. Sci. USA. 2006;103:16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Willkomm DK, Schon A, Hartmann RK. RNase P of the cyanophora paradoxa cyanelle: A plastid ribozyme. Biochimie. 2007;89:1528–1538. doi: 10.1016/j.biochi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl Acad. Sci. USA. 1995;92:2510–2514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 20.Oh BK, Pace NR. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22:4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerrier-Takada C, Altman S. Reconstitution of enzymatic activity from fragments of M1 RNA. Proc. Natl Acad. Sci. USA. 1992;89:1266–1270. doi: 10.1073/pnas.89.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loria A, Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 24.Zahler NH, Christian EL, Harris ME. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seif E, Cadieux A, Lang BF. Hybrid Escherichia coli-mitochondrial ribonuclease P RNAs are catalytically active. RNA. 2006;12:1661–1670. doi: 10.1261/rna.52106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 27.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank DN, Harris ME, Pace NR. Rational design of self-cleaving pre-tRNA-ribonuclease P RNA conjugates. Biochemistry. 1994;33:10800–10808. doi: 10.1021/bi00201a030. [DOI] [PubMed] [Google Scholar]
- 29.Guerrier-Takada C, Haydock K, Allen L, Altman S. Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli. Biochemistry. 1986;25:1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- 30.Smith D, Pace NR. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 31.Kaye NM, Christian EL, Harris ME. NAIM and site-specific functional group modification analysis of RNase P RNA: magnesium-dependent structure within the conserved P1-P4 multihelix junction contributes to catalysis. Biochemistry. 2002;41:4533–4545. doi: 10.1021/bi012158h. [DOI] [PubMed] [Google Scholar]
- 32.Pascual A, Vioque A. Substrate binding and catalysis by ribonuclease P from cyanobacteria and Escherichia coli are affected differently by the 3′ terminal CCA in tRNA precursors. Proc. Natl Acad. Sci. USA. 1999;96:6672–6677. doi: 10.1073/pnas.96.12.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews AJ, Hall TA, Brown JW. Characterization of RNase P holoenzymes from Methanococcus jannaschii and Methanothermobacter thermoautotrophicus. Biol. Chem. 2001;382:1171–1177. doi: 10.1515/BC.2001.147. [DOI] [PubMed] [Google Scholar]
- 34.Green CJ, Rivera-Leon R, Vold BS. The catalytic core of RNase P. Nucleic Acids Res. 1996;24:1497–1503. doi: 10.1093/nar/24.8.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loria A, Pan T. The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry. 1999;38:8612–8620. doi: 10.1021/bi990691f. [DOI] [PubMed] [Google Scholar]
- 36.Mohr G, Caprara MG, Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 37.Marquez SM, Chen JL, Evans D, Pace NR. Structure and function of eukaryotic ribonuclease P RNA. Mol. Cell. 2006;24:445–456. doi: 10.1016/j.molcel.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loria A, Pan T. Recognition of the 5′ leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry. 1998;37:10126–10133. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 39.Pomeranz Krummel DA, Altman S. Multiple binding modes of substrate to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1999;5:1021–1033. doi: 10.1017/s1355838299990416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme. J. Mol. Biol. 2006;357:583–591. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 41.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc. Natl Acad. Sci. USA. 2006;103:873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 43.Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNase P RNA by protein. Nat. Struct. Mol. Biol. 2005;12:958–964. doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- 44.Niranjanakumari S, Day-Storms JJ, Ahmed M, Hsieh J, Zahler NH, Venters RA, Fierke CA. Probing the architecture of the Bacillus subtilis RNase P holoenzyme active site by cross-linking and affinity cleavage. RNA. 2007;13:521–535. doi: 10.1261/rna.308707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.