Abstract
Plant viruses are inducers and targets of RNA silencing. Viruses counteract with RNA silencing by expressing silencing-suppressor proteins. Many of the identified proteins bind siRNAs, which prevents assembly of silencing effector complexes, and also interfere with their 3′ methylation, which protects them against degradation. Here, we investigated the 3′ modification of silencing-related small RNAs in Nicotiana benthamiana plants infected with viruses expressing RNA silencing suppressors, the p19 protein of Carnation Italian ringspot virus (CIRV) and HC-Pro of Tobacco etch virus (TEV). We found that CIRV had only a slight effect on viral siRNA 3′ modification, but TEV significantly inhibited the 3′ modification of si/miRNAs. We also found that p19 and HC-Pro were able to bind both 3′ modified and non-modified small RNAs in vivo. The findings suggest that the 3′ modification of viral siRNAs occurs in the cytoplasm, though miRNA 3′ modification likely takes place in the nucleus as well. Both silencing suppressors inhibited the 3′ modification of si/miRNAs when they and small RNAs were transiently co-expressed, suggesting that the inhibition of si/miRNA 3′ modification requires spatial and temporal co-expression. Finally, our data revealed that a HEN1-like methyltransferase might account for the small RNA modification at the their 3′-terminal nucleotide in N. benthamiana.
INTRODUCTION
RNA silencing is a sequence-specific regulatory mechanism conserved in almost all eukaryotes and involved in a wide variety of functions, such as genome stability through the initiation of heterochromatin formation (1), as well as anti-viral defence in plants, insects and possibly Caenorhabditis elegans (2). RNA silencing has also been found to be involved in the regulation of gene expression in plants and animals through microRNAs (miRNA) (3,4). Nonetheless, RNA silencing has diverse functions, and different RNA silencing pathways co-exist in a single species.
The mechanisms of various RNA silencing pathways are quite similar. Generally, RNA silencing is initiated by the cleavage of the double-stranded or extensively folded RNAs into short interfering RNAs (siRNA) or miRNAs by the enzyme DICER, a member of the RNase III family. Small RNAs are then loaded into the RNA-induced silencing complex (RISC). Prior to RISC activation, the passenger strand of the siRNA is removed and only the guide strand incorporated into the Argonaute (AGO) protein containing RISC, which is then capable of sequence-specific inhibition of gene expression (5,6). In plants and C. elegans, RNA-dependent RNA polymerases (RdRp) have also been found to play a role in RNA silencing. Plant RdRp has been implicated in the maintenance step by amplifying siRNAs (7), and the RdRp of C. elegans has been shown to be involved in the production of unprimed secondary siRNAs (8). The 3′ methylation of plant small RNAs was considered to be the sole plant-specific step of RNA silencing. In the Arabidopsis plant, short RNAs are likely methylated on the 2′-hydroxyl group of their 3′-terminal nucleotide by HEN1 methyltransferase (9). HEN1 contains a putative nuclear localization signal (NLS) and is thought to exist and function in the nucleus (10). The methylation appears to protect small RNAs from oligouridilation and subsequent degradation (11), and it is present in all species of known small RNAs (siRNA, miRNA, tasiRNA and sense- and hairpin transgene-derived and transposon- and repeat-derived siRNAs). In fact, in hen1-mutant Arabidopsis plants, the abundance of miRNA is much lower and they are of a different size (10). Recently, it was reported that the piRNAs of zebrafish; rasi-, si-, and piRNAs of Drosophila; and the piRNAs of the mouse and rat also carry similar modifications (12–15).
Plant viruses are both inducers and targets of RNA silencing; therefore, viruses evolved RNA silencing suppressors. Both positive and negative strand RNA viruses encode silencing suppressors, but no sign of a common origin for these proteins has been found (16). In theory, RNA silencing could be inhibited at different steps and, so far, four modes of targeting its various steps have been identified. The p19 protein of Carnation Italian ringspot tombusvirus (CIRV), binds siRNAs, which are the central molecules of RNA silencing (16). Thus, effective inhibition requires sequestration of the viral siRNAs by p19 in vivo (17). The same strategy is used by HC-Pro of Tobacco etch virus (TEV), p21 of Beet yellows virus (BYV) and p122 of Tobacco mosaic virus (cr-TMV). The sequestration of siRNA results in the inhibition of RISC assembly against the virus, and this mechanism is thought to be the most common model for RNA silencing suppressors (18–20). However, alternative models for the suppression of RNA silencing have also been reported. Based on in vitro studies, the 2b protein of Cucumber mosaic virus (CMV) was suggested to inhibit RISC activity, as well as single-stranded siRNA containing active RISC, via physical interaction with the PAZ domain of the plant AGO1 protein (21). Recent studies demonstrated that the P0 RNA silencing-suppressor protein of Beet western yellows polerovirus (BWYV) acts as an F-box protein targeting AGO1 and marking it for ubiquitination and degradation (22–24). Finally, the coat protein (CP) of Turnip crinkle virus (TCV), via its dsRNA-binding activity (2,19), was found to inhibit DCL4-mediated siRNA processing (25).
The effect of silencing-suppressor proteins on the methylation of several miRNAs has been shown in transgenic Arabidopsis plants expressing different RNA silencing suppressors, including HC-Pro of TEV, p21 of BYV and p19 of Tomato bushy stunt virus (TBSV). The inhibition of methylation was due to the physical interaction between the suppressor and the miRNA/miRNA (*) duplex (26). The inhibitory effect of virus infection on miRNA methylation has also been reported (20,27,28).
Similar to 3′-methylation, an unidentified 3′ protection of small RNAs is also known to be affected by RNA silencing suppressors. A study using tobacco plants expressing HC-Pro and infected with Y-Sat and its helper CMV observed a differential affect on small RNA 3′ protection. Though 3′ protection of the CMV- and Y-Sat-derived siRNAs was partially inhibited, no difference in 3′ modification of endogenous small RNAs miR168 and miR166 was found (29).
To better understand the mechanism of small RNA 3′ protection and the effect of silencing-suppressors on small RNA biogenesis in natural viral infection, we examined the 3′ status of small RNAs during the course of viral infections. We found that the 3′ modification of viral siRNAs was variable depending on the virus species. In non-infected plants, miRNAs were fully resistant to a β-elimination reaction, suggesting that they were all methylated. In contrast, viral siRNAs were fully sensitive to β-elimination in TEV-infected plants, and the miRNAs were partially sensitive. However, viral siRNAs derived from CIRV-infected plants were only slightly affected and the 3′ protection of miRNAs was only partially disturbed. Immunoprecipitation (IP) studies also showed that HC-Pro and p19 are responsible for the inhibition of the 3′ protection of small RNAs by binding the small RNA duplexes in virus-infected plants. Furthermore, our data also suggest that there is only partial protection of the 3′-terminus of miRNA in the nucleus, and that it is completed in the cytoplasm. Our model predicts that inhibition of miRNA 3′ protection requires the spatial and temporal co-expression of miRNA and silencing-suppressor proteins. Although we did not identify the precise nature of the 3′ modification of N. benthamiana small RNAs, it is likely that they are methylated by a HEN1-like methyltransferase, as was suggested for small RNAs derived from the closely related N. tabacum plant (29). This phenomenon as a component of RNA-silencing machinery is quite conserved in eukaryotes, including plants. Therefore, we assumed that small RNAs resistant to β-elimination were all methylated at the 2′-hydroxyl group of the 3′-terminal nucleotide.
MATERIALS AND METHODS
RNA extraction, β-elimination and northern-blot analysis
RNA was isolated from the systemic leaves of N. benthamiana infected with TEV (12–14 dpi) and CIRV (7dpi) with TRI Reagent (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer's instructions. A total of 10 μg of RNA was used for oxidation and β-elimination, as previously described (27). The RNA samples were resolved on 12% denaturing PAGE. The denaturing gel and hybridization analysis were performed as previously described (18). Probes used for northern blots were in vitro transcripts of full-length virus clones for the viral siRNAs, DNA oligos for the U6 loading control RNA (gttttatcaagtcccagaccgtatcaaatat) and tRNA (ctacagtcctccgctctaccaactgagctaaggtcgg), and LNA oligos from a manufacturer (Exiqon, Vedbæk, Denmark) for miRNAs. Membranes were exposed to a storage phosphor screen and bands quantified by the Genius Image Analyzer (Syngene, Cambridge, United Kingdom).
Agrobacterium tumefaciens infiltration
For agro-infiltration of N. benthamiana leaves, mixtures of Agrobacterium strains bearing the miR171c precursor (OD600 = 0.1) and different suppressors (OD600 = 0.2) were used as previously described (20). Samples were taken after 2.5 days.
IP
For IP, 0.6 g of systemic leaves from mock and TEV-(containing a His-tagged HC-Pro, a gift from J.J. Lopez-Moya) or CIRV-infected N. benthamiana were collected at 12 dpi (TEV) or 7 dpi (CIRV) to prepare extracts in 1 × IP buffer containing 40 mM Tris (pH 8.0), 100 mM NaCl, 2 mM MgCl2 and 1 mM DTT. A tenth of each of the IP extracts was collected as input samples. The IP was carried out at 4°C for 1 h with anti-His antibody for His-HC-Pro and polyclonal anti-CIRV p19 antibody for CIRV p19 (30). Suppressor–antibody complexes were pulled out with protein A sepharose beads, which was subsequently eluted in 2 × PK buffer containing 100 mM Tris (pH 7.5), 300 mM NaCl, 10mM EDTA (pH 8) and 2% SDS. Mock IP was carried out without antibody. Input, IP eluates and mock IP eluates were used for western blotting and RNA extraction.
Electrophoretic mobility shift assays (EMSAs)
The labelling and annealing of RNA duplexes was carried out as previously described (17). Purified p19 and W39G proteins and N. benthamiana extracts of HC-Pro and AS3 (18) were incubated with labelled RNAs for 30 min at room temperature in lysis buffer (31) supplemented with 0.02% Tween-20. Complexes were resolved on 6% polyacrylamide 0.5 TBE gels. The gels were dried and exposed to a storage phosphor screen (Molecular Dynamics Typhoon Phosphorimager, Amersham Biosciences, Buckinghamshire, United Kingdom) and bands quantified by the Genius Image Analyzer (Syngene).
Cell fractionation
Two grams of systemic leaves were collected from TEV- and CIRV-infected plants. The Sigma Cell Lytic Plant Nuclei Isolation/Extraction Kit (Sigma, St Louis, Missouri, USA) was used according to the manufacturer's instructions to obtain semi-pure nuclear extracts. All components except the nuclear fraction were considered as part of the cytoplasmic fraction. Ten percent of the fractions were used for western blotting and the remaining 90% for RNA isolation.
RESULTS
Viral infection interferes with the 3′ modification of small RNAs
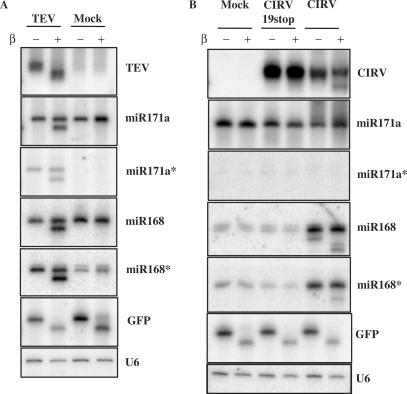
TEV and CIRV both encode well-characterized RNA silencing suppressors that specifically bind small RNAs (18), suggesting that these suppressors may interfere with the 3′ modification of small RNAs. To investigate whether TEV or CIRV interfere with small RNA biogenesis, small RNAs isolated from infected and mock plants were tested by β-elimination followed by northern hybridization. Strikingly, TEV-derived siRNAs in virus-infected plants were completely sensitive to β-elimination, but the endogenous miRNAs were partially resistant. A portion of the miR171 and miR168 mature strands (51.2 and 51.6%, respectively), were not affected by β-elimination (Figure 1A). The result for miR168 was particularly interesting because this molecule regulates the expression of AGO1 in Arabidopsis thaliana (32). Because HC-Pro is a small RNA-binding protein, we expected the accumulation of the star (*) strand of miRNA in TEV infected plants (18,33). Indeed, miR171* accumulated in TEV, but not in mock inoculated plants. In contrast, high levels of both the mature and star strands of miR168 accumulated in both the mock and virus inoculated plants. Both miR171* and miR168* were also partially modified (52.1 and 38.3% were resistant to β-elimination, respectively) in TEV-infected plants, suggesting that HC-Pro may interfere with the 3′ protection of these miRNAs (Figure 1A). The effect of CIRV infection on the biogenesis of small RNAs was different than the effect of TEV. We found that CIRV infection only partially inhibited the virus-derived 3′ protection of siRNAs since most of the viral siRNA (70.1%) was resistant to the β-elimination reaction (Figure 1B). In addition, the viral siRNAs were all protected at the 3′ end if the plants were infected by a mutant virus (CIRV19stop) (30) that does not express the silencing-suppressor p19 protein. In contrast to TEV, CIRV neither have any effect on miR171a nor were we able to detect miR171a*. On the contrary, both miR168 and miR168* were detected in mock and CIRV19stop extracts. We also detected an increase in the amount of miR168/miR168* in CIRV-infected samples compared to mock and CIRV19stop-infected plants, indicating that p19 may play a pivotal role in the accumulation of miR168/miR168* duplexes in the context of viral infection. In the case of miR168, we detected fractions of a shorter (17.3%) and longer transcript (82.7%), likely due to its previously described truncation in the presence of p19 (34). Though the truncated fraction of miR168 (shorter RNA) was unprotected against β-elimination, the major fraction (full-length miR168) was almost fully protected, indicating that CIRV had only a slight effect on the 3′ protection of both strands of miR168 (Figure 1B). Interestingly, the truncation of miR168 occurred only on the mature strand and not the star strand, suggesting an asymmetry of the supposed p19-miR168/miR168* complex (Figure 1B). These results surprisingly indicate that, although p19 efficiently binds miRNA duplexes and efficiently inhibits the 3′ methylation of plant miRNAs when it is expressed transgenically (18,26,33), it interferes only slightly in miRNA biogenesis if expressed by the replicating virus.
Figure 1.
Virus infection interferes with small RNA 3′ modification. RNA for northern blots were isolated systemically from leaves of N. benthamiana plants infected with TEV (A), CIRV, CIRV19stop (B) and mock inoculated plants. A total of 4 μg of RNA was used for β-elimination. In (B), two sets of membranes containing the same samples were used. The membrane, which was used for the miR171a* hybridization resulting in no signal, was used later for miR171a and CIRV hybridizations resulting in strong and specific signals. Hybridizations were performed with the indicated probes. U6 RNA serves as loading control. The 10 pmol of synthetic GFP RNA oligo was used as an internal control for β-elimination.
Analysis of suppressor-bound small RNAs
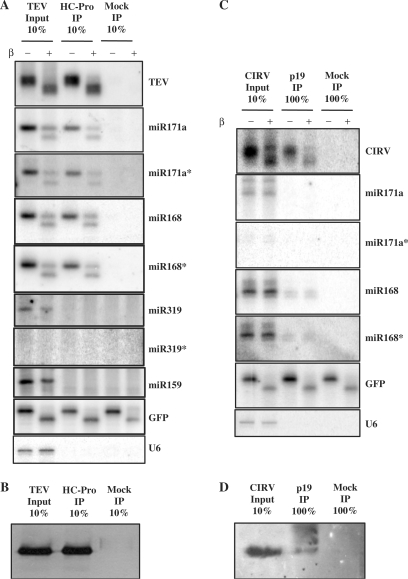
It has been shown that RNA silencing suppressors physically interact with plant virus-derived small RNAs (17–19,33,35). To investigate whether HC-Pro of TEV or p19 of CIRV may interact with 3′ modified or unmodified small RNAs in virus-infected plants, we performed IP against the suppressor proteins from systemically infected leaf extracts. RNA samples isolated from inputs and IP eluates were β-eliminated and probed for viral siRNA and different miRNAs. The results for HC-Pro showed that HC-Pro-bound TEV-derived siRNAs were fully sensitive to β-elimination (Figure 2A), suggesting that siRNA binding by HC-Pro inhibits 3′ modification. Moreover, our results revealed that HC-Pro was complexed with 3′ modified and unmodified miR171a and miR168. The same ratio of 3′ modified and unmodified miRNA was found in the input and IP eluates, indicating that HC-Pro does not have any preference for binding 3′ modified or unmodified miRNA (Table 1). The results for p19 showed that both 3′ modified and unmodified CIRV-derived siRNA were found in the IP eluate (Figure 2C), as well as both strands of miR168 duplexes. In contrast, none of the miR171 duplex strands were found in the IP eluates, suggesting that p19 does not have efficient access to the miR171 duplex.
Figure 2.
Both 3′ protected and unprotected small RNAs are bound by HC-Pro and p19. RNA for northern blots were isolated from IPs performed on extracts made from systemic leaves of TEV (A) and CIRV (C) infected plants. In the case of mock IP, no antibody was used. For TEV, 10% of the input and the eluate of the IP were loaded. In the case of CIRV, 10% of input and 100% of the IP eluate were loaded. Hybridizations were performed with the indicated probes. U6 RNA serves as control for IPs, 10 pmol of synthetic GFP RNA oligo was used as an internal control for β-elimination. Western blots were loaded with protein extracts of inputs and eluates of HC-Pro (B) and p19 (D) IPs.
Table 1.
3′ Protected status of HC-Pro bound siRNAs and miRNAs
| HC-Pro input | HC-Pro IP | |
|---|---|---|
| Small RNA | 3′ Protected (%) | 3′ Protected (%) |
| TEV siRNA | 0 | 0 |
| 171a | 58.5 | 52.4 |
| 171a* | 51 | 54.7 |
| 168 | 50.9 | 51.5 |
| 168* | 50.7 | 47.5 |
To extend our study, we tested other miRNAs in extract from TEV-infected plants and HC-Pro IP eluate (Figure 2A, Table 2). Only the mature strand of the group of tested miRNAs (miR159, miR398, miR319, miR390, miR161, etc.) was detected in the input, though they were not detected in the IP eluate (Figure 2A, Table 2), indicating that the miRNA already underwent strand separation. Therefore, the 3′ modification of these miRNAs was not affected by the double-stranded small RNA binding HC-Pro. The inability to detect other miRNAs (miR162*, miR163 and miR395) in the input may be due to a low level of homology between the N. benthamiana miRNA and the probe LNA oligo designed to detect A. thaliana miRNAs (36) (Table 2). We obtained similar results with p19 IP from CIRV-infected plant extracts (data not shown; Table 2). These findings revealed that, although p19 binds both siRNA and miRNA duplexes, p19 only slightly affects the 3′ modification of siRNA and miRNA. On the other hand, TEV seriously affected the 3′ modification of the small RNAs found to physically interact with HC-Pro.
Table 2.
Characteristic of miRNAs in virus-infected plants
| miRNA | TEV input | HC-Pro IP | 3′ Protection affected | CIRV input | p19 IP | 3′ Protection affected | Homology (%) | |
|---|---|---|---|---|---|---|---|---|
| 1. | 171 | + | + | + | + | − | − | 100 |
| 2. | 171* | + | + | + | − | − | − | 80 |
| 3. | 168 | + | + | + | + | + | + | 100 |
| 4. | 168* | + | + | + | + | + | + | 100 |
| 5 | 159 | + | − | − | + | − | − | 100 |
| 7. | 398 | + | − | − | − | − | − | 50 |
| 8. | 319 | + | − | − | + | − | − | 95 |
| 11. | 390 | + | − | − | − | − | − | 90 |
| 12. | 161 | + | − | − | − | − | − | 67 |
| 13. | 160 | + | − | − | − | − | − | 100 |
| 14. | 162 | + | − | − | + | − | − | 100 |
| 15. | 162* | ND | − | ND | − | 66 | ||
| 9. | 163 | ND | − | ND | − | 42 | ||
| 10. | 395 | ND | − | ND | − | 42 |
3′ Modification of small RNAs occurs in the cytoplasm
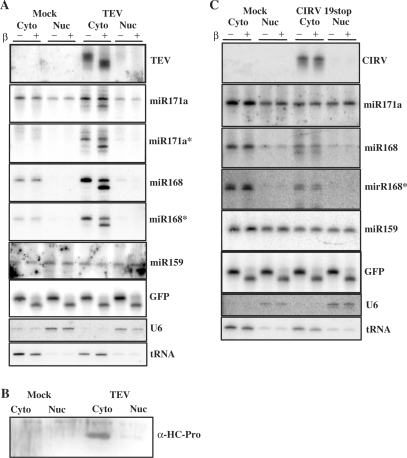
In Arabidopsis plants, the processing of miRNA primary transcripts occurs in the nucleus and requires Dicer-like 1 (DCL1) enzyme activity (37). Most of the miRNA duplexes investigated are likely exported to the cytoplasm by HASTY, the Arabidopsis ortholog of Exportin 5 (38). It has been reported that the small RNA methyltransferase, HEN1, is localized exclusively in the nucleus (39). However, we have recently shown that siRNAs derived from cytoplasmically replicating cr-TMV were partially methylated (20) in wild-type, but not in hen1 mutant, Arabidopsis plants. This suggests that HEN1 is also active in the cytoplasm. In fact, viral siRNAs from the CIRV19stop virus, which replicates exclusively in the cytoplasm, were also completely resistant to β-elimination and TEV-derived siRNAs were completely sensitive, indicating that their 3′-termini were not protected. These results raise a question about the compartmentalization of 3′ protection/methylation of small RNA. To this end, plant extracts from systemically infected leaves of TEV and CIRV19stop inoculated plants (9 dpi and 7 dpi, respectively) were used for cell fractionation followed by the isolation of RNA from both the nuclear and cytoplasmic fractions. The 3′ protection status of small RNAs was tested by β-elimination and visualized by northern blotting. As expected, viral siRNAs were found in the cytoplasmic fraction of the infected cells, because both TEV and CIRV19stop replicates in the cytoplasm (Figure 3A and C). Similar to previous observations, the patterns of 3′ protection were completely different for siRNAs derived from TEV or CIRV19stop. Though TEV-derived siRNAs were fully unprotected, the siRNAs derived from CIRV19stop virus were fully resistant to β-elimination. Consistently, the majority of HC-Pro co-localized with viral siRNA in the cytoplasmic fraction (Figure 3A and B), suggesting that the 3′ modification of viral siRNAs predominantly occurs in the cytoplasm.
Figure 3.
Subcellular localization of small RNAs and HC-Pro. Total RNA for northern blots were isolated from nuclear and cytoplasmic fractions of TEV (A) and CIRV19stop (C) infected N. benthamiana. Hybridizations were performed with the indicated probes. Oligo probes for U6 and tRNA were used to check the purity of the nuclear and cytoplasmic fractions. The 10 pmol of synthetic GFP RNA oligo was used as an internal control for β-elimination. Western blot shows the distribution of HC-Pro of TEV (B).
Next, we assessed the cellular localization and 3′ modification of miRNAs in infected and mock inoculated plants. In the nuclear fractions from TEV, CIRV19stop and mock inoculated plants, we detected the mature, but not the star strands, of miR171, which were fully protected against β-elimination (Figure 3). On the other hand, we found that both strands of miR171 and miR168 from the cytoplasmic fraction of TEV-infected plants were partially (about 50%) resistant to β-elimination (Figure 3A), suggesting that HC-Pro can interact with miRNA duplexes in the cytoplasm and inhibit their 3′ modification. However, both strands of miR168 from the cytoplasm of mock and CIRV19stop infected cells were fully resistant to β-elimination, suggesting that the 3′ modification of miRNAs likely occurs in the cytoplasm (Figure 3C). Finally, we checked the compartmentalization of miR159, whose 3′ protection was not affected by either TEV or CIRV19stop infection. Our results show that fully protected miR159 RNAs were present in both the nucleus and cytoplasm. Because only the mature strands of miR159 were found in the cytoplasm of TEV-infected plants, we concluded that the transcription of pre-miRNA, processing, 3′ modification, export, strand separation and RISC loading of miR159 in the cytoplasm likely occurred before these cells became infected by the invading virus.
Taken together, our cell fractionation studies strongly suggest that the 3′ modification of small RNA occurs not only in the nucleus, but also in the cytoplasm, and that this is especially true for viral siRNAs.
Inhibition of miRNA 3′ modification requires spatial and temporal co-expression with suppressor proteins
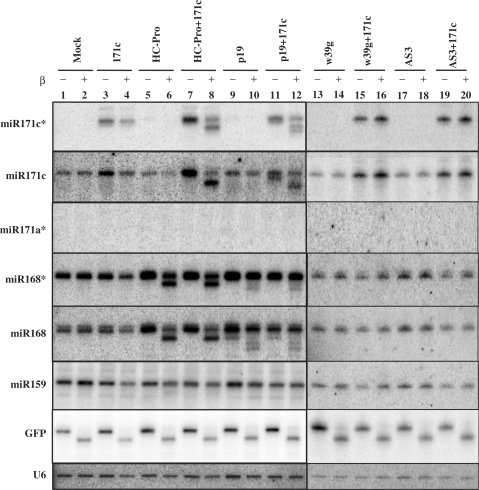
Previous results obtained in stable HC-Pro or TBSV p19 transgenic Arabidopsis plants showed that the suppressors inhibit the methylation of all miRNAs tested (26). However, in the current study, inhibition of 3′ protection was observed only for two miRNAs (miR171 and miR168) out of 11 tested from TEV-infected N. benthamiana (Table 2). These results led to the hypothesis that inhibition of miRNA 3′ protection requires the overlapping spatiotemporal presence of miRNA duplexes and HC-Pro. To test this, we evaluated the 3′ protection status of different miRNAs in samples taken from fully differentiated leaves co-infiltrated with HC-Pro or p19, with or without pre-miR171c. The star strand of miR171c could only be detected when pre-miR171c was expressed at high levels using agro-infiltration, but the mature strand of miR171c was detected in all RNA samples and in increased amounts when it was over-expressed (Figure 4, lanes 3–4, 7–8, 11–12, 15–16 and 19–20). Strikingly, the 3′ protection of both strands of miR171c was only affected when pre-miR171c was co-infiltrated with HC-Pro or p19. In addition, we observed that HC-Pro and p19 inhibited miR171c* 3′ protection with similar efficiency (Figure 4, lanes 8 and 12). However, when mutant suppressors W39G of p19 and AS3 of HC-Pro, those lacking the ability to bind small RNA (Figure 5B), were expressed, the 3′ protection of miRNAs was not altered (Figure 4, lanes 13–20). Though miR171c could be detected only when pre-miR171c was transiently over-expressed, both strands of endogenous miR168 accumulated in all samples (Figure 4), indicating that miR168 is constitutively expressed at high levels. The 3′ modification of endogenous miR168 was also inhibited in the presence of transiently expressed HC-Pro or p19 (Figure 4, lanes 5–12). We noted that although 3′ modification of over-expressed miR171c by HC-Pro and p19 was inhibited to similar extents, the effect of HC-Pro was more pronounced on miR168 and miR168* than the effect of p19. In contrast, we could not detect changes in the 3′ protection pattern of mature miR159 in the agro-infiltrated cells, suggesting that miR159 was not simultaneously present in duplex form (the target structure of HC-Pro and p19) with silencing suppressors in the same compartment. These findings support our hypothesis that the presence of the suppressor and miRNA duplex in the same cell at the same time is the prerequisite for inhibition of miRNA 3′ protection.
Figure 4.
Co-expression of silencing suppressors with miRNA precursor. RNA samples for northern blots were isolated from agroinfiltrated N. benthamiana leaves with the designated constructs. Two sets of membranes containing the same samples were used. The membranewhich was used for the miR171a* hybridization resulting in no signal, was used later for miR171c* and miR171c hybridizations resulting in strong and specific signals. Hybridizations were performed with the indicated probes. U6 RNA serves as loading control. The 10 pmol of synthetic GFP RNA oligo was used as an internal control for β-elimination.
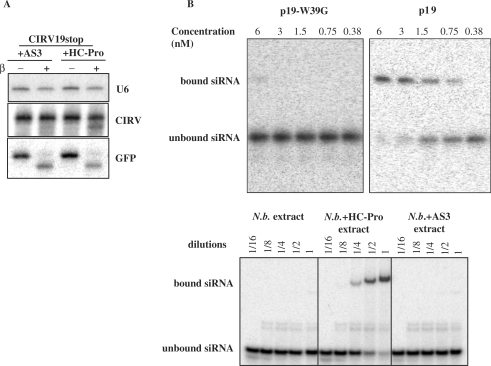
Figure 5.
Interaction of silencing-suppressor proteins with small RNAs in vitro and in virus-infected leaves. AS3 is not able to interfere with viral siRNA 3′ protection, while the wt HC-Pro partially inhibits the 3′ protection of cytoplasmic viral siRNAs. RNA samples were collected from inoculated leaves, which were infiltrated with the suppressors 6 h post-inoculation. The 20 µg of total RNA were used for each sample and 10 pmol of synthetic GFP RNA oligo was used as an internal control for β-elimination (A). RNA-binding activity of wt and mutant silencing suppressor proteins (B). The 21-nt siRNA duplexes were incubated with a dilution series of purified p19 and p19-W39G (upper panels) or A3 and HC-Pro infiltrated plant extracts and analysed on 5% native 0.5 × TBE gel.
We showed that CIRV19stop viral siRNAs accumulate in the cytoplasm and are fully protected at the 3′-terminus. However, if we transiently express HC-Pro in the Cym19stop-infected leaves, the 3′ protection of viral siRNA is partially inhibited, though the mutant HC-Pro (AS3) expression did not lead to any alteration in siRNA 3′ modification (Figure 5A). This observation further supports the existence of cytoplasmic small RNA modification.
DISCUSSION
In this report, we studied the effect of TEV and CIRV infection on the 3′ modification of small RNAs. The results revealed that both viral infections interfere with this process, though TEV had a more pronounced effect. IP analysis of HC-Pro-bound small RNAs clearly showed that inhibition of the 3′ modification requires physical interaction between the suppressor protein and small RNA duplexes. This conclusion was further supported by experiments using mutant silencing suppressors, which exhibited lost binding activity. Neither p19-W39G nor HC-Pro AS3 was able to interfere with the 3′ modification of viral siRNAs and plant miRNAs. The inhibition of small RNA 3′ modification likely corresponds to the inhibition of the 3′ terminal nucleotide methylation of the 2′-hydroxyl group, catalysed by HEN1 methyltransferase, as was suggested for small RNAs derived from the closely related N. tabacum plant (29). Though the methylation of small RNAs was not confirmed directly, we showed previously that the terminal nucleotides of small RNAs have a free 3′-hydroxyl group by efficiently cloning viral siRNAs (40) and miRNAs (J.B., unpublished data). In addition, viral siRNAs extracted from Cym19stop-infected plants were protected against the extension of polyA tails by PolyA polymerase, suggesting the presence of a methyl group on the 2′-OH of the 3′-terminal nucleotide of the viral siRNAs (29).
Thus, the inhibition of the methylation of small RNAs could be considered as an additional layer of silencing by the viral suppressors since the non-methylated small RNAs are unstable and much less available for the small RNA-mediated silencing pathways. In fact, in hen1 mutant plants, the level of miRNAs is much lower (10). Moreover, our study suggests that the methylation of siRNA and miRNA duplexes is strongly inhibited by HC-Pro in TEV-infected plants, further indicating that the formation of short RNA duplexes represents a common step in the miRNA and siRNA-driven RNA silencing pathways in plants. CIRV had a modest and slightly different effect on the 3′ protection of both si- and miRNAs. In the case of viral siRNAs, this difference can not be due to a lack of physical interaction between p19 and small RNAs because, as our IP study showed, p19 binds both protected and un-protected small RNAs. The miRNA 3′ protection was also disturbed when pre-miR171c and p19 were co-expressed using agroinfiltration. However, during natural virus infection, the p19 is expressed in a more spatially separated environment from miRNA duplexes, which may explain why the p19 does not markedly interfere with the miRNA pathway. In fact, the replication of CIRV occurs in the well-separated vesicles derived from the outer membrane of mitochondria (41). TEV replication is also restricted to the membrane of endoplasmic reticulum (ER) (42), but is not as separated from the cytoplasm as CIRV. Alternatively, the different effect of TEV and CIRV on the small RNA 3′ protection can also be explained by the different modes of small RNA binding utilized by the two RNA silencing suppressors. HC-Pro binds the 2 nt 3′ overhangs of small RNAs, but p19 binds the 5′ phosphates (18,30). Accordingly, HC-Pro may cover the 3′ overhangs of small RNAs; thus, inhibiting the 3′ modification by blocking the HEN1 methyltransferase, which has been shown to bind the 2 nt of the 3′-end overhang (9). Therefore, our observation likely suggests that HC-Pro competes with HEN1 methyltransferase, which is probably also active in N. benthamiana cells. The interaction of HC-Pro with small RNAs might occur in the cytoplasm, since HC-Pro was detected almost exclusively in the cytoplasmic fraction where TEV replicates and it does not contain an identifiable NLS signal.
Our cell fractionation studies also strongly suggested that the 3′ modification of small RNAs occurs both in the nucleus and the cytoplasm. Although it was not obvious for TEV-derived siRNAs because we could not detect any 3′-modified siRNA in extracts from TEV-infected plants. However, fully protected (100%) viral siRNAs were found in the cytoplasm of CIRV19stop-infected plants. Since CIRV replicates exclusively in the cytoplasm, we predict that CIRV-derived siRNAs are likely 3′ modified in the cytoplasm, suggesting the presence of HEN1-like enzyme in the cytoplasm as well. Although earlier reports showed that HEN1 localizes in the nucleus (39), a recent report showed that HEN1 is also present in the cytoplasm (43). In addition, it was recently shown that methylated siRNAs derived from hen1-mutant plants infected with cr-TMV, which replicates in the cytoplasm, were completely absent (20). This further suggests that HEN1 is responsible for both nuclear and cytoplasmic methylation of small RNAs. Alternatively, virus-derived siRNAs should be first transported to the nucleus for methylation by HEN1 and then back to the cytoplasm. However, this is an unlikely scenario since no viral siRNAs were detected in the nuclear fractions from Cym19stop-infected plants, but the cytoplasmic fractions had high levels of detected viral siRNA.
We observed a subset of mature miR171and miR159 in the nucleus, which may indicate that those duplexes underwent methylation and strand separation in the nucleus. Alternatively, we can not exclude that miR171- and miR159-containing effector complexes are first assembled in the cytoplasm and then transported back to the nucleus. In contrast, almost all miR168 molecules were found to be exported to the cytoplasm (Figure 3C). Previously, miRNAs were found in both AGO1- and AGO4-containing complexes in A. thaliana inflorescent tissue. Furthermore, there were some miRNAs exclusively found in association with AGO4 (44). Because AGO4 was found to have a function in RNA-directed DNA methylation in the nucleus, it raises the possibility that certain miRNAs regulate gene expression in the nucleus, as well as the cytoplasm.
Consistent with our observation, Oilseed rape mosaic tobamovirus (ORMV) and crucifer infecting TMV (cr-TMV), which have small RNA-binding RNA silencing suppressors, were shown previously to be able to counteract small RNA methylation (20,27). On the other hand, the methylation pattern of plants infected with Cabbage leaf curl begomovirus (CaLCuv) and African cassava mosaic virus (ACMV) were not changed. The silencing suppressor of ACMV, AC4, was shown to bind mature (single-stranded) miRNAs; therefore, the lack of any effect on methylation may be due to the fact that AC4 acts after the methylation and strand separation steps (45).
Taken together, these observations suggest that the silencing suppressors able to bind small RNA duplexes likely compromise the methylation of these small RNAs. However, this effect is modulated by the nature of the virus, which expresses the silencing-suppressor proteins, such as the spatial and temporal co-expression or separation of suppressor protein and small RNAs.
ACKNOWLEDGEMENTS
This research was supported by grants from the Hungarian Scientific Research Fund (OTKA; T046728, OTKA; T048852 and OTKA; NK60352) and the ‘SIROCCO’ EU project LSHG-CT-2006-037900. L.L. is a recipient of the Bolyai János fellowship. Funding to pay the Open Access publication charges for this article was provided by ‘SIROCCO’ EU project LSHG-CT-2006-037900.
Conflict of interest statement. None declared.
REFERENCES
- 1.Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 2.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 7.Baulcombe DC. Molecular biology. Amplified silencing. Science. 2007;315:199–200. doi: 10.1126/science.1138030. [DOI] [PubMed] [Google Scholar]
- 8.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 9.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silhavy D, Burgyan J. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004;9:76–83. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Lakatos L, Szittya G, Silhavy D, Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopez-Moya JJ, Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merai Z, Kerenyi Z, Kertesz S, Magna M, Lakatos L, Silhavy D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Csorba T, Bovi A, Dalmay T, Burgyan J. The p122 subunit of Tobacco mosaic virus replicase is a potent silencing suppressor and compromises both siRNA and miRNA mediated pathways. J. Virol. 2007;81:11768–11780. doi: 10.1128/JVI.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl Acad. Sci. USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. The Polerovirus F Box Protein P0 Targets ARGONAUTE1 to Suppress RNA Silencing. Curr. Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 24.Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. The Polerovirus Silencing Suppressor P0 Targets ARGONAUTE Proteins for Degradation. Curr. Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 25.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 26.Yu B, Chapman EJ, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F, Jr, Hohn T, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogler H, Akbergenov R, Shivaprasad PV, Dang V, Fasler M, Kwon MO, Zhanybekova S, Hohn T, Heinlein M. Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J Virol. 2007;81:10379–10388. doi: 10.1128/JVI.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vargason J, Szittya G, Burgyan J, Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 31.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 32.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgyan J, Rubino L, Russo M. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 1996;77:1967–1974. doi: 10.1099/0022-1317-77-8-1967. [DOI] [PubMed] [Google Scholar]
- 42.Schaad MC, Jensen PE, Carrington JC. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for MicroRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007;17:1–6. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl Acad. Sci. USA. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]