Abstract
SN1-type alkylating agents, like N-methyl-N-nitrosourea (MNU) and N-ethyl-N-nitrosourea (ENU), are potent mutagens. Exposure to alkylating agents gives rise to O6-alkylguanine, a modified base that is recognized by DNA mismatch repair (MMR) proteins but is not repairable, resulting in replication fork stalling and cell death. We used a somatic mutation detection assay to study the in vivo effects of alkylation damage on lethality and mutation frequency in developing zebrafish embryos. Consistent with the damage-sensing role of the MMR system, mutant embryos lacking the MMR enzyme MSH6 displayed lower lethality than wild-type embryos after exposure to ENU and MNU. In line with this, alkylation-induced somatic mutation frequencies were found to be higher in wild-type embryos than in the msh6 loss-of-function mutants. These mutations were found to be chromosomal aberrations that may be caused by chromosomal breaks that arise from stalled replication forks. As these chromosomal breaks arise at replication, they are not expected to be repaired by non-homologous end joining. Indeed, Ku70 loss-of-function mutants were found to be equally sensitive to ENU as wild-type embryos. Taken together, our results suggest that in vivo alkylation damage results in chromosomal instability and cell death due to aberrantly processed MMR-induced stalled replication forks.
INTRODUCTION
The SN1-type alkylating agents N-methyl-N-nitrosourea (MNU) and N-ethyl-N-nitrosourea (ENU) are strong chemical mutagens, which cause DNA damage by transferring a methyl or ethyl group to the oxygen and nitrogen atoms of nucleotide bases (1,2). The resulting base adducts tend to mispair during semi-conservative replication. If not corrected the following round of replication converts the mismatches to point mutations (1). The recently described structures of normal and O6-methylated guanine (O6-meG) paired with either cytosines or thymines in the replication complex have provided an explanation for why alkylated bases frequently mispair (3). Normally, the relative replication efficiency of unmethylated G paired with C exceeds that of G paired with T by 100 000-fold. In contrast, the replication efficiency for O6-meG paired with T is 10-fold higher than for O6-meG paired with C (3). In addition, the proofreading activity of DNA polymerase cannot distinguish O6-meG paired with C or T, and the mismatch goes unnoticed.
As DNA alkylation does occur naturally, cells have developed repair mechanisms to prevent mutagenesis. The molecular principles of DNA methylation have been studied most extensively and will be discussed here. The cell has several repair pathways to deal with the different types of methylation adducts (2). The O6-meG adduct is specifically removed by the enzyme O6-meG methyltransferase (Mgmt) (4–6). In addition, in the event that O6-meG pairs with thymine during replication, this mismatch is recognized by mismatch repair enzymes (2,4,7–9). DNA mismatch repair (MMR) is the machinery that corrects small replication errors, such as base–base mismatches and insertion/deletion loops (IDLs). In mammals, there are five MMR genes that produce the components of three different heterodimers. The MutS heterodimer is the first to recognize the replication error and exists in two varieties: MutSα and MutSβ. MutSα, consisting of the MMR proteins MSH2 and MSH6, recognizes single base pair mismatches and small IDLs. MutSβ, a heterodimer of MSH2 and MSH3, is primarily involved in the recognition of larger IDLs. Subsequent to MutS binding, a MutL heterodimer is recruited. MutLα, the predominant form functioning in MMR, consists of MLH1 and PMS2 (10). In addition to replication errors, MMR proteins recognize other forms of DNA damage, such as modified basepairs. Upon recognition of O6-meG pairs, the MMR system will induce strand excision of the bases mispaired to the methylated site in order to repair the damage. However, since the methylated base is in the template strand of the replicated DNA, it cannot be replaced. Instead, it will continuously be paired with C or T by the polymerase, followed by MMR recognition and excision. This ‘futile cycle’ of repair will finally result in the stalling of the replication fork. By subsequent activation of checkpoints it may lead to cell cycle arrest and apoptosis. MMR-mediated DNA damage checkpoint signalling has been shown to occur via ATR and phosphorylation of Chk1 (11–13), while ATM plays a non-essential role (13,14). The associated apoptotic response has been shown to be independent of p53 (7).
In the absence of a direct DNA damage response, the O6-meG-induced stalled replication forks may collapse, resulting in double-strand breaks (DSBs) (13,15). A potential mechanism for this was revealed by the observation that MMR-dependent futile cycling results in persistent small stretches of single-stranded DNA in the first round of replication, which become DSBs in the second cycle of replication (16). The DSBs would then be repaired by homologous recombination (HR), resulting in sister chromatid exchanges (2,5,14,17,18). This hypothesis is supported by the fact that HR-deficient yeast strains have increased sensitivity to alkylation damage (19).
For the reasons mentioned above, O6-meG is the most toxic methylation-induced lesion, to which MMR mutants are resistant (4,9,15,20,21). The basic molecular response may be similar for ethylation, although affinities of DNA polymerase, Mgmt, and MMR for O6-ethyl-guanine are probably different. Indeed, Claij et al. (21) found a higher survival rate of msh2 mutant cells compared to wild-type cells after exposure to ENU, albeit to a lesser degree than after exposure to MNNG. Also, the same homozygous msh2 mutant cells had a selective growth advantage over heterozygous cells when treated with ENU (20). However, in two other studies on human and hamster cells, no differences were observed between MMR-deficient cells and their wild-type counterparts in response to ENU treatment (22,23).
When alkylated mispairs in MMR-deficient cells do not induce arrest but are allowed to persist, single base pair mutations can accumulate, resulting in increased genomic instability in MMR mutants. A higher mutation frequency in MMR-deficient cell lines has indeed been reported for methylating agents (4,18) and ethylating agents (21).
In zebrafish, ENU mutagenesis has a long history of efficient use in both forward and reverse genetic screens (24–27) where it is applied to adult fish to introduce base pair changes in germ cells. We have applied ENU and MNU treatment to early embryos in order to study the effect of alkylation-induced damage in fast-dividing somatic cells, in vivo. We have found that after treatment with alkylating agents, wild-type zebrafish embryos accumulate high numbers of mutations due to chromosomal instability, and have low survival rates. These effects are dependent on MMR activity and are strongly reduced in embryos deficient in the MMR-component MSH6.
MATERIAL AND METHODS
Zebrafish lines
Msh6 mutant fish (hu1811) were obtained by target-selected mutagenesis, and the initial characterization was described elsewhere (28). Genotyping was done by PCR amplification and resequencing, using exon 10 specific forward (5′-GCTGGTGGCAACTTAAATC-3′) and reverse (5′-GCTCAACAGATACTTGCTT TG-3′) primers or using KASPAR genotyping technology (KBioscience, Hoddesdon, UK) and forward primers for wild type (5′-GAAGGTGACCAAGTTCATGCTCGCTTCTCAGACGTACTTGTGC-3′) and mutant (5′-GAAGGTCGGAGTCAACGGATTCCGCTTCTCAGACGTACTTGTGT-3′) and the common reverse primer (5′-CAGTCTGTTCTACGCGAGCTACTTT-3′). The wild-type embryos that are used as controls in all experiments have a similar genetic background as the msh6−/− embryos, as they are derived from wild-type siblings of the msh6−/− fish that produced the mutant embryos.
To generate msh6, albino double mutant fish, the original msh6 heterozygous founder was crossed with an albino line that carries a point mutation in exon 6 of the zebrafish SLC45A2 gene. This mutation causes a glycine to arginine change at position 461, and although this line has not been documented elsewhere, the phenotype is indistinguishable from other albino lines. The allele is identified as albhu1844 (hu1844), but for clarity it will be called ‘alb’ in the report. Double heterozygous fish (hu1874) were subsequently incrossed to generate both msh6−/−, alb/alb and msh6+/+, alb/alb fish.
The Ku70 line (hu2485) also originates from target-selected mutagenesis and contains a thymine to adenine mutation in exon 10, resulting in a premature stop codon. This line has not been reported yet. Genotyping of the Ku70 allele was done by resequencing, similar to what was done for the msh6 line, with forward (5′-ATGACATACGCACTGTGGAC-3′) and reverse (5′-AATCAGGAGGATAGACC AAATC-3′) primers.
The tp53 line (zdf1), with mutation M214K, has been described elsewhere (29) and will be identified as tp53 throughout the study. Genotyping was done using KASPAR forward primers for wild type (5′-GAAGGTGACCAAGTTCATGCTGAGGATGGGCCTGCGGTTCA-3′) and mutant (5′-GAAGGTCGGAGTCAACGGATTGAGGA TGGGCCTGCGGTTCT-3′) and the reverse primer (5'-CAACTGTGCTACTAAACTACATGTGCAAT-3′).
Somatic mutation frequency
Pair crosses of msh6, albino double mutants were performed to assay the somatic mutation frequency. At three days post-fertilization (dpf), both eyes of all embryos were inspected for pigmentation. Patches of unpigmented cells were considered the result of a somatic mutation in the albino locus. For detailed analysis of these patches, embryos were fixed in 4% PFA, embedded in plastic and sectioned.
Chemical and ionizing radiation treatment of embryos
Embryos were treated at 5–6 h post-fertilization (hpf) for 1 h with ENU or MNU in 10 mM NaPO4 buffer pH 6.6. After treatment they were rinsed once with NaPO4 buffer and once with embryo medium before they were put back to embryo medium at 28.5°C. Embryos were irradiated at 5–6 hpf in a small volume of embryo medium in a Gammacell 1000 (Gammaster, Ede, the Netherlands) and then moved to 28.5°C in fresh embryo medium.
The survival of the embryos was monitored during the first days post-treatment, with final scoring at 3 dpf. Embryos that had died, were unhatched, and/or had severe phenotypes, e.g. curled bodies, oedemas, and reduced body size, were scored as not surviving. Albino cells in the eye were also scored at 3 dpf, as described above.
All experiments were done with multiple independent crosses, and were repeated at least two times. Data were analysed using ANOVA and significance levels P < 0.05, and are represented as mean ± standard error of the mean (SEM).
Expression arrays
RNA samples from 10 embryos coming from five different crosses treated with 0.4 mM ENU and from 10 untreated embryos from five different crosses were used to balance the potential effects of genetic variation. Total RNA was isolated with the mirVana miRNA isolation kit (Ambion, Austin, TX, USA). cDNA synthesis and labeling were done using the low RNA input linear amplification kit (Agilent, Santa Clara, CA, USA) and Cyanine 3-CTP and 5-CTP (Agilent). Subsequent purification was done using the RNeasy Mini kit (Qiagen, Hilden, Germany). Duplicate samples were run in dye-swap. Labelled samples were hybridized overnight to 44K zebrafish expression arrays (Agilent), washed, scanned with scanner (Agilent), and analysed using Feature Extraction and Array-Assist software (Agilent), all according to standard procedures and instructions of the suppliers.
Metaphase spreads and chromosome paints
Twenty-four hours post-fertilization embryos were dechorionated and incubated for 90 min in colchicine to arrest cells in metaphase. After hypotonic treatment in 1.1% sodiumcitrate and fixation in 3:1 methanol:acetic acid, cells were suspended in 50% HAc and spread onto glass slides. Slides were mounted and stained with DAPI vectashield (Vector Labs, Burlingame, CA, USA).
For chromosome paints, slides with metaphase spreads were refixed in 3:1 methanol:acetic acid, dehydrated in an ethanol series, and dried. Then the slides were washed with 2× SSC and incubated with 8 * 10−5% pepsin in 0.01 M HCl for 5 min, both at 37°C. After three washes with 2x SSC, slides were fixed in 1% formaldehyde in PBS for 5 min, rinsed three times with PBS, dehydrated and dried. The slides were denatured in 70% formamide and probes were hybridized overnight at 37°C. Paints for chromosome 4 labelled with biotin, chromosome 5 labelled with FITC and chromosome 7 labeled with Cy3 were kindly provided by Dr Fengtang Yang (30). The slides were washed two times with 50% formamide, once with 2× SSC, and once with 4× SSC, 0.5% Tween-20, all at 42°C. To detect the biotin label, the slides were exposed to avidin Cy5-conjugated antibodies (Amersham, Buckinghamshire, UK). To enhance FITC staining, the slides were exposed to rabbit Alexa Fluor® 488-conjugated anti-fluorescein/Oregon Green® antibodies (Molecular Probes, Invitrogen, Eugene, OR, USA). Antibody stainings were done in one reaction in 4× SSC, 0.5% Tween-20 for 15 min at 37°C, and subsequently the slides were washed three times with 4× SSC, 0.5% Tween-20 at 42°C. Slides were mounted and stained with DAPI SlowFade (Invitrogen, Eugene, OR, YSA).
RESULTS
Msh6−/− embryos have an increased spontaneous somatic mutation frequency
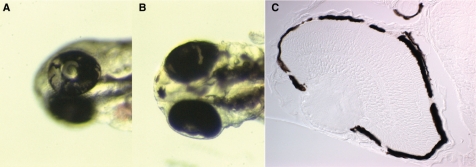
An assay was set up for the in vivo detection of somatic mutations in zebrafish embryos using loss of heterozygosity of the albino gene in the pigmented cells of the eye. Pigmentation of zebrafish embryos is visible from day 2 of development as individual cells on the trunk and in a uniformly pigmented layer of cells in the eye. Pigmentation is totally absent in albino mutants. Because pigmentation requires only one functional allele of the albino gene to be present, we were able to score every inactivating mutation. To this end, msh6−/− zebrafish were crossed with msh6−/−, alb/alb animals to obtain msh6 mutant embryos that are albino heterozygotes, and msh6+/+ with msh6+/+, alb/alb animals to obtain embryos that are wild type for msh6 in an albino heterozygous background. In these embryos, a mutation that causes loss of heterozygosity at the albino locus will result in a cell that lacks pigmentation. When this cell divides a patch of unpigmented cells will arise, which can easily be scored in the pigmented layer of the embryonic eye (Figure 1A and B). In order to validate the assay, the spontaneous somatic mutation frequency of msh6−/− and msh6+/+ embryos was determined. The frequency of embryos with albino patches was 17-fold higher in the msh6 mutant background (Table 1) compared to wild type. In the progeny of msh6−/− females crossed with msh6+/+ males, the mutation frequency was similar to that of msh6 mutant embryos. In contrast, the progeny from msh6+/+ females crossed with msh6−/− males had a mutation frequency similar to wild type embryos, irrespective of which of the two parents was albino (Table 1). The fact that the maternal genotype determines the mutator phenotype of the progeny indicates that the mutations arise early in development, before the midblastula transition when zygotic transcription is switched on (31). During this period, the embryonic phenotype reflects the transcripts and proteins that are provided in the yolk by the mother; therefore, if the mother is mutant for msh6, up to the midblastula transition her progeny will also be devoid of the enzyme. To more closely investigate the number of mutant cells in each embryo, some eyes with albino patches were sectioned. In most cases, multiple small patches rather than a single unpigmented patch were seen (Figure 1C). This is likely due to the extensive cell migration that occurs during eye development (32), as the overall mutation frequency indicates that all patches in one eye were caused by a single mutation. Altogether, the albino loss-of-heterozygosity assay in zebrafish proves to be a very powerful system for in vivo somatic mutation detection.
Figure 1.
Loss of heterozygosity at the albino locus in zebrafish embryos. (A, B) Examples of unpigmented patches of cells in the pigment layer of the eye caused by mutations in the wild-type allele of the albino gene. (C) Example of a cross-section of the eye showing the typical multiple small patches.
Table 1.
Spontaneous non-pigmented patches in the eyes of heterozygous albino zebrafish in wild-type and MMR-deficient backgrounds
| Cross | No. embryos scored | No. with patches | Frequency |
|---|---|---|---|
| msh6+/+F X msh6+/+M | 2068 | 2 | 0.00097 |
| msh6−/−F X msh6−/−M | 1434 | 24 | 0.017 |
| msh6+/+F X msh6−/−M | 1043 | 1 | 0.00096 |
| msh6−/−F X msh6+/+M | 1612 | 32 | 0.020 |
F, female; M, male.
Msh6−/− embryos are more resistant to ENU and MNU
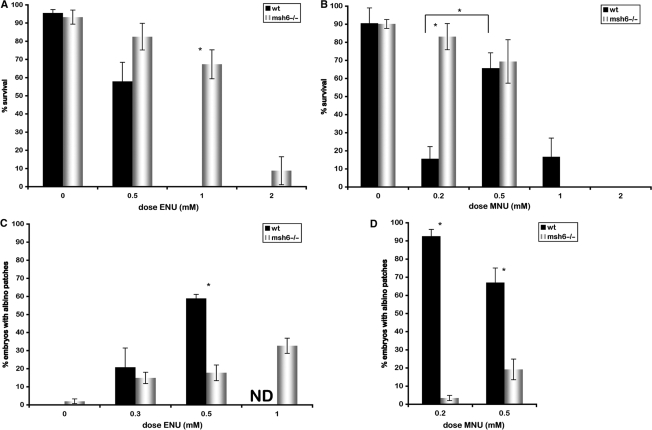
In the developing zebrafish embryo, the first 10 cell divisions occur synchronously, rapidly, and without checkpoints. After the midblastula transition, cell cycles become longer with more extensive G1 and G2 phases, which supplies time for checkpoint signalling, cell cycle arrest and DNA repair (31). To test the sensitivity of zebrafish embryos to alkyating agents, we chose to treat embryos of midblastula stage, 5 hpf, with the ENU and MNU. As ENU is the most efficient mutagen in zebrafish, we first exposed groups of embryos to increasing concentrations of ENU (24,25). Consistent with what has been observed in other systems, msh6−/− embryos had increased survival rates at high concentrations of ENU relative to wild-type embryos (Figure 2A). To compare the effects of ethylation and methylation, a second group of embryos was exposed to increasing concentrations of MNU. Once again, we observed a higher rate of survival for msh6−/− embryos relative to wild-type embryos. However, to our surprise, this difference was reproducibly only seen at low concentrations (0.2 mM) of MNU, due to the extreme killing of MNU in wild-type embryos (Figure 2B). At higher concentrations, msh6−/− and wild-type embryos had similar survival rates. The survival rate of wild-type embryos was significantly lower in 0.2 mM than in 0.5 mM MNU (Figure 2B). In addition, we observed that embryonic phenotypes were much more severe at 0.2 mM compared to 0.5 and 1.0 mM MNU (data not shown).
Figure 2.
Msh6−/− embryos are more resistant to alkylation damage and have reduced mutation frequencies. (A, B) Survival of wild-type (black) and msh6−/− (white) embryos at increasing doses of ENU (A) and MNU (B). (C, D) Mutation frequency measured by the albino loss-of-heterozygosity assay in wild-type (black) and msh6−/− (white) embryos induced by increasing doses of ENU (C) and MNU (D). Asterisks indicate significant differences. ND, not done.
The ENU- and MNU-induced mutation frequency is reduced in msh6−/− embryos
We used the albino loss-of-heterozygosity assay to study the frequency of chemical-induced mutation after exposure to low doses of ENU. The frequency of mutation induced by ENU was found to be one order of magnitude higher than the frequency of spontaneous mutation in MMR mutants. Unexpectedly, the mutation frequency after ENU exposure was greatly increased in wild-type embryos compared to msh6−/− embryos (Figure 2C). The same experiment was done using MNU as the alkylating agent, resulting in an even higher increase in mutation frequency in the wild-type relative to the msh6−/− embryos (Figure 2D). These results indicate that the absence of MMR recognition of alkylation damage does not result in an increased accumulation of point mutations. However, comparing the frequencies of mutation and survival after treatment with ENU or MNU, the results do suggest a correlation between mutation load and lethality. Therefore, the mutations that accumulate after exposure to alkylating agents might be caused by lesions other than point mutations, and could result from MMR-dependent replication fork stalling following O6-alkylguanine (O6-alkG) recognition.
Alkylation-induced lethality is caused by MMR-dependent chromosomal aberrations
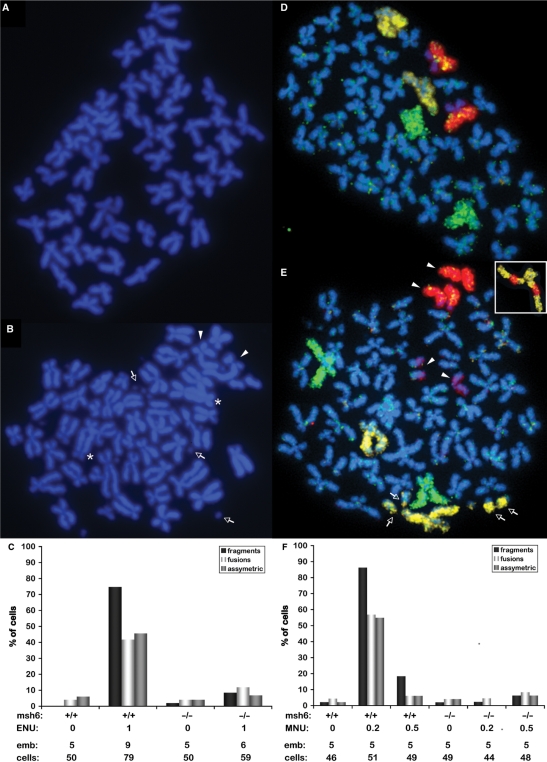
MMR-mediated stalled replication forks have the potential to turn into DSBs, which can cause chromosomal aberrations when they are not properly repaired. Metaphase spreads were performed to investigate whether aberrant chromosomes were the source of the high frequency of mutations in embryos treated with ENU. Since it takes two rounds of replication for the alkylation damage to corrupt replication forks, treated embryos were allowed to develop until 24 hpf before chromosomal analysis [cells in the developing embryo undergo on average two to three cell divisions between 6 and 24 hpf; (33)]. Several types of aberrant chromosomes that were extremely rare in untreated cells, such as fragments, fusions and asymmetric chromosomes (Figure 3A) were frequently observed in ENU-treated wild-type cells (Figure 3B). When quantified, the levels of chromosomal instability were much higher in cells of ENU-treated wild-type embryos than in those of untreated embryos. On the other hand, msh6−/− cells had only a mild increase in the frequency of chromosomal aberrations with ENU-treatment (Figure 3C). To confirm that the observed aberrations were indeed random chromosomal rearrangements, we painted three individual chromosomes with previously described zebrafish paints (30). The three pairs of chromosomes were easily distinguished in cells from untreated wild-type embryos (Figure 3D). However, in wild-type cells that had been exposed to 1 mM ENU, we observed painted chromosome fragments (Figure 3E), as well as chromosomes that were detected by two different paints, indicating chromosome rearrangements (Figure 3E, inset).
Figure 3.
Alkylation-induced lethality is caused by MMR-dependent chromosomal aberrations. (A) Metaphase spread of an untreated embryo showing the 50 regular chromosomes of a single cell. (B) Metaphase spread of a typical cell from a wild-type embryo treated with 1 mM ENU, showing chromosome fragments (arrows), fused chromosomes (asterisks), and asymmetric chromosomes (arrowheads). (C) Quantitation of the three types of aberrant chromosomes mentioned in (B). Below the graph are indicated the msh6 genotype, ENU treatment (mM), number of embryos scored, and total number of cells scored. (D) Metaphase spread from an untreated embryo with chromosome paints for chromosomes 4 (yellow), 5 (green) and 7 (red), showing two of each chromosome per cell. (E) Metaphase spread of a typical cell from a wild-type embryo treated with 1 mM ENU, stained with chromosome paints for chromosome 4 (yellow), 5 (green) and 7 (red); indicated aberrations include fragments of chromosome 4 (arrows), and both chromosomes 7 having broken into halves (arrowheads) [note that in both (D) and (E) the short arms of chromosome 7 show weaker staining than the long arms]. Inset in (E): example of a rearranged chromosome 4 with fragments of chromosome 7. (F) Quantitation of the three types of aberrant chromosomes mentioned in B after MNU exposure. Below the graph are indicated the msh6 genotype, MNU treatment (mM), number of embryos scored, and total number of cells scored.
When the same experiment was performed with MNU-treated embryos, the mutation frequency was also low in treated msh6−/− embryos. The frequency of aberrant chromosomes was higher in wild-type embryos exposed to 0.2 mM MNU than in those exposed to 0.5 mM MNU. Therefore, the frequency of chromosomal rearrangements observed after MNU exposure correlates well with the degree of lethality and frequency of mutation (Figure 3F). Altogether, these results strongly indicate that chromosomal instability is the source of MMR-dependent mutations and lethality induced by alkylation.
To systematically determine which signalling and repair pathways are triggered by these chromosomal aberrations in vivo, we used microarrays to look for changes in transcriptome expression in low-dose ENU-treated embryos. TP53 expression was increased 3-fold in treated wild-type embryos, but not in treated msh6−/− embryos. No other significant changes in the expression of DNA metabolizing or signal transduction genes were found, including ATR and ATM pathway genes and factors downstream of TP53 (data not shown).
ENU-induced chromosomal instability arises at replication
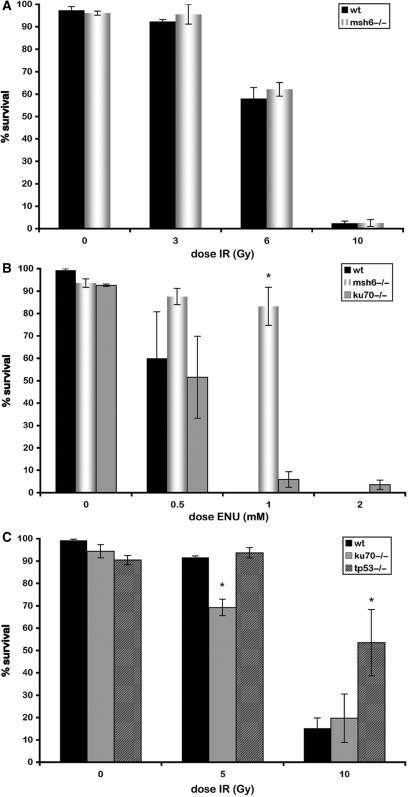
To investigate whether wild-type and MMR mutant cells respond differently to DSBs that arise through means other than alkylation, we tested the ability of 5 hpf wild-type and msh6−/− embryos to survive increasing doses of ionizing radiation (IR). Ionizing radiation induces random DSBs, independent of cell cycle phase. We observed no differences in the survival rates of msh6−/− mutant and wild-type embryos for any of the IR doses tested (Figure 4A).
Figure 4.
MMR-dependent ENU-induced chromosomal instability arises during replication. (A) Induction of random DSBs during all phases of the cell cycle by IR does not result in surivival differences in between msh6−/− and wild-type embryos. (B) ENU sensitivity of wild-type and ku70 mutant. (C) IR sensitivity of wild type, tp53 mutant (control) and homozygous ku70 mutant. Asterisks indicate significant differences.
DSBs can be repaired via two different mechanisms that act in different phases of the cell cycle. Homologous recombination (HR) is active during the S and G2 phases when sister chromatids are available. Non-homologous end joining (NHEJ) is the predominant pathway for the repair of DSBs during G1 phase. To determine which repair pathway is important for the repair of alkylation damage in vivo, we treated a NHEJ-defective mutant (ku70−/−) with ENU and assayed survival. This experiment was performed only in the wild-type background, as ENU-induced damage only results in the stalling of replication forks and DSBs in the presence of functional MMR. No difference in sensitivity to ENU was observed for ku70 knockouts as compared to wild-type embryos (Figure 4B), indicating that HR, not NHEJ, is most likely involved in the repair of ENU-induced DSBs. To confirm that ku70−/− is a loss-of-function mutant, a range of IR doses was applied to ku70−/− mutant embryos. A small but significant decrease in survival was observed with increasing irradiation (Figure 4C), indicating that the ku70 mutant is defective for DSB repair by NHEJ. In addition, these results confirm that NHEJ is active in developing embryos. The observed IR sensitivity of our ku70 mutant embryos is analogous to morpholino knockdown experiments of ku70 and ku80, which have previously been shown to result in increased apoptosis in response to irradiation (34,35). As a control, tp53 mutant embryos, which have been shown to have reduced levels of apoptosis upon irradiation, were exposed to similar doses of IR (29). The tp53 mutant embryos indeed had increased survival frequencies at the highest IR dose (Figure 4B).
Taken together, the observations that the msh6 mutant is equally sensitive to IR as wild type and the ku70 mutant is equally sensitive to ENU as wild type indicate that MMR-dependent ENU-induced DNA damage arises during replication and not during interphase.
DISCUSSION
Although the in vitro effects of alkylating agents have been studied extensively, in vivo studies in vertebrate species are still lacking. Here, for the first time, we have studied the in vivo effects of alkylation-induced damage in wild-type and MMR-deficient zebrafish embryos. Msh6 mutant zebrafish have previously been shown to display microsatellite instability and have a predisposition to cancer (28), illustrating that they are MMR deficient. To study somatic mutation accumulation in vivo, an assay was developed for the detection of loss-of-heterozygosity at the albino locus in the pigmented cells of the embryonic eye. Using this assay, the background somatic mutation frequency in the msh6 mutant was found to be 17-fold above that of wild-type embryos. This is consistent with studies in other organisms, where 10–100-fold increases in the mutation frequency were observed in MMR-deficient backgrounds (21,36–38). Since the zebrafish albino gene does not contain obvious repeat sequences, the underlying mutations are expected to be point mutations, although their nature has not been determined experimentally.
Mutant and wild-type embryos were treated with SN1 alkylating agents. Previous studies using cell culture experiments did not give conclusive evidence that ethylation lethality is dependent on MMR. Some of these studies reported an increased resistance to alkylation in MMR mutants (20,21), but others reported no differences compared to wild type (22,23). Here, we show that msh6 mutant embryos survive ENU treatment significantly better than do wild-type embryos. Surprisingly, MNU reproducibly induced severe MMR-dependent lethality only at the lowest concentration tested (0.2 mM). It is difficult to explain why a given compound is more lethal at lower than at higher concentrations. We speculate that this may be related to the fact that DNA methylation is itself a natural regulatory mechanism, and therefore a threshold exists above which the DNA damage response and repair pathways are triggered. In this scenario, low-dose methylation damage could still deregulate cellular processes and result in cell death. Alkylating agents and DNA demethylating agents are currently used in chemotherapy. In that respect, this type of in vivo study may be clinically significant, as epigenetic changes are frequent in cancers (39).
According to this study, the alkylation damage response in zebrafish is similar to what has been reported for the cells of other organisms. However, more detailed studies will be needed to firmly establish zebrafish as a general model for studying human DNA repair processes. For example, it is not known whether zebrafish has a functional mgmt homologue. The Ensembl database (www.ensembl.org) suggests an orthologue (ENSDARG00000043275), but data on how and when it functions and whether it is maternally provided are lacking.
As discussed, O6-alkG-induced stalled replication forks can be converted to DSBs. These DSBs arise in a MMR-dependent manner, which has been confirmed by the observation that msh6 mutant embryos are not resistant to the DSBs induced by IR. Additionally, DSBs that arise during replication are normally repaired by HR and not by NHEJ (5,14,17,18). Ku70 mutant zebrafish, which were shown to be NHEJ-deficient, were as sensitive to ENU as wild-type zebrafish, indicating that NHEJ does not repair ethylation-induced DSBs. Previously, it was shown that Ku80 mutant hamster cells are able to repair MNNG-damage but not IR-damage (40). These data confirm that DSBs resulting from MMR-recognized O6-alkG arise at replication.
The observed decrease in ENU- and MNU-induced mutation rate of msh6 mutants can now be explained by the fact that the albino loss-of-heterozygosity mutations are large chromosomal lesions. Chromosomal aberrations have also been reported after MNU treatment of human, mouse, and hamster cells (14,22,23). They occur when DSBs caused by O6-alkG are aberrantly repaired or unrepaired, which subsequently results in cell death. Although DNA checkpoint signalling induced by O6-alkG-mediated stalled replication forks was shown to depend on ATR/Chk1 signalling (11–13), secondary lesions may induce a different DNA damage response. This could explain why we found TP53 to be upregulated in embryos with high levels of chromosomal instability, while MMR-dependent alkylation DNA damage signalling was reported to be independent of TP53 (7). Similarly, it explains the involvement of ATM-signalling after alkylation damage (14). In our in vivo zebrafish experiments, the alkylation response appears to be mostly directed towards chromosome instability. It should be mentioned that while DNA damage checkpoints are absent during the first cell divisions of zebrafish development, the ability to undergo checkpoint-induced apoptosis was found to start even later (41), which may explain why the O6-alkG damage is able to cause high levels of chromosomal aberrations in zebrafish embryos.
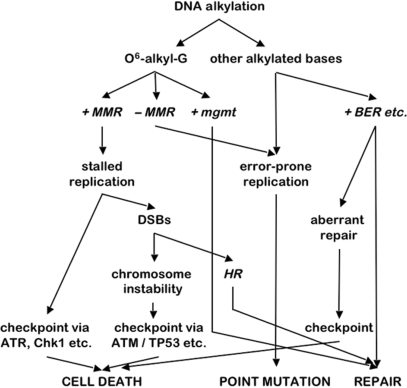
At high concentrations, alkylating damage kills MMR-deficient embryos just as it does wild-type embryos. This is probably due to the accumulation of additional types of alkylation-induced DNA damage, such as N-alkylation products, which can also induce apoptosis through incomplete base excision repair (2). The potential effects of alkylation treatment are schematically summarized in Figure 5. The three different outcomes of alkylation damage, (i) cell death, (ii) point mutation and (iii) repair, can be reached via different repair and signalling routes. This indicates that absence of one function, such as MMR, will shift the equilibrium of the three potential outcomes rather than result in one specific outcome.
Figure 5.
Schematic overview of the responses to alkylation damage and the three possible outcomes, ‘cell death’, ‘point mutation’ and ‘repair’ (capitals) via routes involving different types of DNA repair (italics). Loss of functions, such as MMR, will shift the balance of these three outcomes rather than result in one specific outcome. MMR, mismatch repair; mgmt, O6-methylguanine methyltransferase; BER, base excision repair; DSBs, double-strand breaks; HR, homologous recombination.
According to the observed mutation frequency, the induction of DSBs by alkylation far exceeds the number of point mutations in developing wild-type embryos. Considering that ENU and MNU have primarily been used to induce germ-line point mutations, this suggests that the effect of these drugs may be different for embryos and germ cells. Nevertheless, DSB-related mutations have been observed in several mutagenesis studies aiming for point mutations, indicating that this process also occurs in the germ line. In the mutagenesis treatment of post-meiotic zebrafish germ cells, two out of six resulting mutations were found to be dominant lethal in embryos, suggesting that they represent larger lesions than point mutations (42). Of five mutants from a similar mutagenesis experiment, the mutation was found to be a translocation in one line and a large deletion in the four others (43). ENU and MNU germ-line mutagenesis in mice has also caused large lesions in many cases (44).
Taken together, the results of this study show that alkylation damage induces high levels of chromosomal aberrations in early zebrafish embryos in an MMR-dependent fashion, which results in reduced embryonic survival. Overall, the data indicate that the basic responses to alkylation damage in zebrafish are similar to other organisms. However, the observed high frequency of chromosome rearrangements compared to point mutations in these embryos is clearly different from what has been reported from germ line mutagenesis studies and in vitro experiments. These experiments show that in somatic cells chromosomal instability is a more physiologically relevant outcome of alkylation damage than point mutations. These findings are relevant in considering the role of alkylating events in carcinogenesis, as well as for alkylation chemotherapy.
ACKNOWLEDGEMENTS
The authors thank Fengtang Yang for providing zebrafish chromosome paints and for experimental advice, Marcel Tijsterman for critically reading the manuscript and the Wellcome Trust Sanger Center Danio rerio Sequencing Project for genomic sequence information. This work was supported by Cancer Genomics Center (Nationaal Regie Orgaan Genomics); and European Union-funded FP6 Integrated Project ZF-models. Funding to pay the Open Access publication charges for this article was provided by the Hubrecht Institute, The Netherlands.
Conflict of interest statement. None declared.
REFERENCES
- 1.Noveroske JK, Weber JS, Justice MJ. The mutagenic action of N-ethyl-N-nitrosourea in the mouse. Mamm. Genome. 2000;11:478–483. doi: 10.1007/s003350010093. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O(6)-methyl-guanine lesions. Proc. Natl Acad. Sci. USA. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bignami M, O'Driscoll M, Aquilina G, Karran P. Unmasking a killer: DNA O(6)-methylguanine and the cytotoxicity of methylating agents. Mutat. Res. 2000;462:71–82. doi: 10.1016/s1383-5742(00)00016-8. [DOI] [PubMed] [Google Scholar]
- 5.Kaina B, Fritz G, Coquerelle T. Contribution of O6-alkylguanine and N-alkylpurines to the formation of sister chromatid exchanges, chromosomal aberrations, and gene mutations: new insights gained from studies of genetically engineered mammalian cell lines. Environ. Mol. Mutagen. 1993;22:283–292. doi: 10.1002/em.2850220418. [DOI] [PubMed] [Google Scholar]
- 6.Zak P, Kleibl K, Laval F. Repair of O6-methylguanine and O4-methylthymine by the human and rat O6-methylguanine-DNA methyltransferases. J. Biol. Chem. 1994;269:730–733. [PubMed] [Google Scholar]
- 7.Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc. Natl Acad. Sci. USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 9.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell. Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl Acad. Sci. USA. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debiak M, Nikolova T, Kaina B. Loss of ATM sensitizes against O6-methylguanine triggered apoptosis, SCEs and chromosomal aberrations. DNA Repair. 2004;3:359–368. doi: 10.1016/j.dnarep.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Nowosielska A, Marinus MG. DNA mismatch repair-induced double-strand breaks. DNA Repair. 2008;7:48–56. doi: 10.1016/j.dnarep.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Tsujimura T, Bhattacharyya NP, Maher VM, McCormick JJ. O6-methylguanine induces intrachromosomal homologous recombination in human cells. Carcinogenesis. 1996;17:2229–2235. doi: 10.1093/carcin/17.10.2229. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Marra G, Jiricny J, Maher VM, McCormick JJ. Mismatch repair is required for O(6)-methylguanine-induced homologous recombination in human fibroblasts. Carcinogenesis. 2000;21:1639–1646. doi: 10.1093/carcin/21.9.1639. [DOI] [PubMed] [Google Scholar]
- 19.Cejka P, Mojas N, Gillet L, Schar P, Jiricny J. Homologous recombination rescues mismatch-repair-dependent cytotoxicity of S(N)1-type methylating agents in S. cerevisiae. Curr. Biol. 2005;15:1395–1400. doi: 10.1016/j.cub.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 20.Borgdorff V, van Hees-Stuivenberg S, Meijers CM, de Wind N. Spontaneous and mutagen-induced loss of DNA mismatch repair in Msh2-heterozygous mammalian cells. Mutat. Res. 2005;574:50–57. doi: 10.1016/j.mrfmmm.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Claij N, van der Wal A, Dekker M, Jansen L, te Riele H. DNA mismatch repair deficiency stimulates N-ethyl-N-nitrosourea-induced mutagenesis and lymphomagenesis. Cancer Res. 2003;63:2062–2066. [PubMed] [Google Scholar]
- 22.Armstrong MJ, Galloway SM. Mismatch repair provokes chromosome aberrations in hamster cells treated with methylating agents or 6-thioguanine, but not with ethylating agents. Mutat. Res. 1997;373:167–178. doi: 10.1016/s0027-5107(96)00234-5. [DOI] [PubMed] [Google Scholar]
- 23.Galloway SM, Greenwood SK, Hill RB, Bradt CI, Bean CL. A role for mismatch repair in production of chromosome aberrations by methylating agents in human cells. Mutat. Res. 1995;346:231–245. doi: 10.1016/0165-7992(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 24.Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 25.Solnica-Krezel L, Schier AF, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136:1401–1420. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wienholds E, Plasterk RH. Target-selected gene inactivation in zebrafish. Methods Cell. Biol. 2004;77:69–90. doi: 10.1016/s0091-679x(04)77004-1. [DOI] [PubMed] [Google Scholar]
- 27.Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feitsma H, Kuiper RV, Korving J, Nijman IJ, Cupper E. Cancer Res., in press. Zebrafish with mutations in mismatch repair genes develop neurofibromas and other tumors. [DOI] [PubMed] [Google Scholar]
- 29.Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl Acad. Sci. USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman JL, Adeniyi A, Banerjee R, Dallaire S, Maguire SF, Chi J, Ng BL, Zepeda C, Scott CE, Humphray S, et al. Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. BMC Genomics. 2007;8:195. doi: 10.1186/1471-2164-8-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 32.Streisinger G, Coale F, Taggart C, Walker C, Grunwald DJ. Clonal origins of cells in the pigmented retina of the zebrafish eye. Dev. Biol. 1989;131:60–69. doi: 10.1016/s0012-1606(89)80038-7. [DOI] [PubMed] [Google Scholar]
- 33.Kozlowski DJ, Murakami T, Ho RK, Weinberg ES. Regional cell movement and tissue patterning in the zebrafish embryo revealed by fate mapping with caged fluorescein. Biochem. Cell. Biol. 1997;75:551–562. [PubMed] [Google Scholar]
- 34.Bladen CL, Lam WK, Dynan WS, Kozlowski DJ. DNA damage response and Ku80 function in the vertebrate embryo. Nucleic Acids Res. 2005;33:3002–3010. doi: 10.1093/nar/gki613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bladen CL, Navarre S, Dynan WS, Kozlowski DJ. Expression of the Ku70 subunit (XRCC6) and protection from low dose ionizing radiation during zebrafish embryogenesis. Neurosci. Lett. 2007;422:97–102. doi: 10.1016/j.neulet.2007.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrew SE, Xu XS, Baross-Francis A, Narayanan L, Milhausen K, Liskay RM, Jirik FR, Glazer PM. Mutagenesis in PMS2- and MSH2-deficient mice indicates differential protection from transversions and frameshifts. Carcinogenesis. 2000;21:1291–1295. [PubMed] [Google Scholar]
- 37.Denver DR, Feinberg S, Estes S, Thomas WK, Lynch M. Mutation rates, spectra and hotspots in mismatch repair-deficient Caenorhabditis elegans. Genetics. 2005;170:107–113. doi: 10.1534/genetics.104.038521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tijsterman M, Pothof J, Plasterk RH. Frequent germline mutations and somatic repeat instability in DNA mismatch-repair-deficient Caenorhabditis elegans. Genetics. 2002;161:651–660. doi: 10.1093/genetics/161.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider-Stock R, Ocker M. Epigenetic therapy in cancer: molecular background and clinical development of histone deacetylase and DNA methyltransferase inhibitors. IDrugs. 2007;10:557–561. [PubMed] [Google Scholar]
- 40.Jeggo PA, Kemp LM. X-ray-sensitive mutants of Chinese hamster ovary cell line. Isolation and cross-sensitivity to other DNA-damaging agents. Mutat. Res. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 41.Ikegami R, Hunter P, Yager TD. Developmental activation of the capability to undergo checkpoint-induced apoptosis in the early zebrafish embryo. Dev. Biol. 1999;209:409–433. doi: 10.1006/dbio.1999.9243. [DOI] [PubMed] [Google Scholar]
- 42.Riley BB, Grunwald DJ. Efficient induction of point mutations allowing recovery of specific locus mutations in zebrafish. Proc. Natl Acad. Sci. USA. 1995;92:5997–6001. doi: 10.1073/pnas.92.13.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai Y, Feldman B, Schier AF, Talbot WS. Analysis of chromosomal rearrangements induced by postmeiotic mutagenesis with ethylnitrosourea in zebrafish. Genetics. 2000;155:261–272. doi: 10.1093/genetics/155.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell LB, Hunsicker PR, Russell WL. Comparison of the genetic effects of equimolar doses of ENU and MNU: while the chemicals differ dramatically in their mutagenicity in stem-cell spermatogonia, both elicit very high mutation rates in differentiating spermatogonia. Mutat. Res. 2007;616:181–195. doi: 10.1016/j.mrfmmm.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]