Abstract
The eukaryotic RecA homologs Rad51 and Dmc1 are essential for strand exchange between homologous chromosomes during meiosis. All members of the RecA family of recombinases polymerize on DNA to form helical nucleoprotein filaments, which is the active form of the protein. Here we compare the filament structures of the Rad51 and Dmc1 proteins from both human and budding yeast. Previous studies of Dmc1 filaments suggested that they might be structurally distinct from filaments of other members of the RecA family, including Rad51. The data presented here indicate that Rad51 and Dmc1 filaments are essentially identical with respect to several structural parameters, including persistence length, helical pitch, filament diameter, DNA base pairs per helical turn and helical handedness. These data, together with previous studies demonstrating similar in vitro recombinase activity for Dmc1 and Rad51, support the view that differences in the meiotic function of Rad51 and Dmc1 are more likely to result from the influence of distinct sets of accessory proteins than from intrinsic differences in filament structure.
INTRODUCTION
During meiosis, recombination ensures the proper segregation of chromosomes by establishing a physical connection between homologs, thus providing the tension required by the meiotic spindle to accurately separate paired chromosomes. The process of meiotic recombination is initiated by the programmed formation of double-strand breaks (DSBs) in DNA. These DSBs are sites of assembly for the two RecA homologs present in most eukaryotes, the Rad51 recombinase, which functions during mitosis and meiosis and the meiosis-specific recombinase, Dmc1. The RecA homologs form helical filaments on overhanging 3' single-strand DNA ends produced by 5′ nucleolytic resection at DSBs. These nucleoprotein filaments then catalyse strand invasion and exchange between homologous chromosomes during meiosis (1).
During recombination in diploid cells, a nucleoprotein recombinase filament can recombine with either a sister chromatid or one of the two homologous chromatids. During mitosis, when Rad51 is the only recombinase expressed, recombination occurs predominantly between sisters. In contrast, during meiosis when both Rad51 and Dmc1 are expressed, the majority of recombination occurs between homologs (2,3).
The in vivo activities of Rad51 and Dmc1 are regulated by distinct sets of accessory factors. The ability of Dmc1 to contribute to meiotic recombination is influenced by its interactions with accessory factor proteins, including Mei5-Sae3, Hop2-Mnd1 and Tid1/Rdh54. While some of these proteins may interact with Rad51, the primary Rad51 accessory factors include Rad52, Rad55-Rad57 and Rad54. In addition to the unique regulation of Dmc1 by accessory factors during meiosis, a meiotic block to intersister recombination is established by abundant chromosome-associated structural proteins such as Red1 and Hop1 and by the meiosis-specific kinase Mek1 (4–9).
A comparison of joint molecule formation by 2D gel analysis of dmc1 and rad51 mutants provided evidence that Dmc1 specifically promotes interhomolog recombination (6). This study used a red1 background in which the block to intersister recombination is defective and in which Dmc1-independent joints form. In the red1 background, deletion of RAD51 had no effect on the yield of interhomolog interactions but deletion of DMC1 eliminated such interactions, leaving only intersister connections. These findings indicate that Dmc1 protein possesses an interhomolog-specific activity that Rad51 lacks. It is unclear to what extent, if any, the observed difference in interhomolog activity results from an intrinsic difference between Rad51 and Dmc1 proteins.
Rad51 and Dmc1 share 54% amino acid identity in humans and 45% amino acid identity in yeast (10). Both Rad51 and Dmc1 have been found to be capable of catalyzing the key steps of recombination, as detected by the strand exchange and strand assimilation (D-loop) assays (11–14). The optimal conditions for Dmc1 activity appear to be slightly different than those for Rad51 although overall the activities are strikingly similar. On the other hand, previous studies led to the suggestion that Dmc1 and Rad51 forms polymers that are structurally distinct.
Dmc1 differs from Rad51 in that it has a strong tendency to form octomeric rings in vitro and can bind DNA as stacked rings (15–17). This led to the proposal that Dmc1 might promote recombination by a unique ring-based mechanism dramatically different from that of Rad51 and RecA (17). However, it was subsequently shown that conditions which stimulate Dmc1 recombinase activity also stimulate its ability to form filaments that are similar to those formed by Rad51 and RecA (18–21). These findings strongly support the view that the functional form of Dmc1, at least for in vitro recombination reactions, is the helical filament. Whether or not the toroid form of Dmc1 plays a role in vivo remains to be determined.
Two recent papers suggested that helical nucleoprotein filaments formed by the budding yeast Dmc1 (ScDmc1) differed from those formed by other members of the RecA family including Rad51 (20,22). These two studies used atomic force microscopy (AFM) to generate images of ScDmc1 filaments. Analysis of these images indicated that filaments had a helical pitch of 13.4 ± 2.5 nm and a filament width of 13.5 ± 0.8 nm (20). A second study by the same group reported a helical pitch of 16.3 ± 0.8 nm with a width of 14.9 ± 1 nm for filaments formed by the same protein as that used in the first study (22). These values are in contrast to those reported for Rad51 and RecA filaments, which have respective average pitches of 9.9 nm and 9.2 nm and widths of ∼10–11 nm (23,24). In the first of the two AFM studies, a determination of the number of striations for apparently full-length filaments on an 872 bp linear DNA substrate resulted in an estimate of 24 ± 2 nucleotides per helical turn of the ScDmc1 filament (20). Assuming that each protomer in the filament binds three nucleotides, this finding suggested that there could be eight protomers per helical turn in the filament. Alternatively, if Dmc1 binds with six protomers per turn like RecA and Rad51, the value of 24 ± 2 nucleotides per turn would indicate a DNA binding ratio of 1:4 monomers per nucleotide. The second of the two AFM papers on ScDmc1 also reported that the protein can form left-handed helical filaments on DNA (22), in contrast to previous reports on RecA and Rad51 (23,25). In addition to the reported differences between Dmc1 and Rad51, published AFM images of Dmc1 (20,22,26) raised the possibility that the meiosis-specific protein forms straighter, more rigid, filaments than those seen by AFM of Rad51, RadA or RecA (27–31). The possibility that Dmc1 might form more rigid filaments than Rad51 was of interest to us because intrinsic or imposed structural rigidity of recombinase filaments could cause those filaments to extend away from the axial elements that organize sister chromatids into loops (32). Such extended structures could act to prevent intersister recombination and promote interhomolog recombination.
In order to better characterize the structure of Dmc1 filaments and compare them to Rad51 filaments we used transmission electron microscopy (TEM) following negative staining with uranyl acetate to compare the structural features of filaments side-by-side. We analyzed Rad51 and Dmc1 filaments from both the budding yeast Saccharomyces cerevisiae and human. We also generated a reconstruction of the human Dmc1 (HsDmc1) filament as well as collecting measurements of persistence length, helical pitch, diameter and base pairs per turn for Dmc1 in parallel with Rad51. The helical handedness of Dmc1 was also determined. The data presented indicate the filaments formed by Dmc1 and Rad51 from either yeast or human are strikingly similar to one another and to those formed by their bacterial relative, RecA. Possible explanations for the unusual characteristics of the previously reported AFM images of ScDmc1 filaments are discussed.
MATERIALS AND METHODS
DNA substrates
Single-stranded DNA substrates were created by purification of denatured PCR products. A 1 kb PCR product was prepared using two primers, 5′-biotinylated GAGTTTTATCGCTTCCATGAC-3′ and 5′-AATTTATCCTCAAGTAAGGGG-3′, and PhiX174 New England Biolabs (NEB) as a template. The PCR reaction product was purified using a PCR purification column (Qiagen, Valencia, CA, USA) and then allowed to bind 250 μl of streptavidin magnetic beads (Dynabeads, Carlsbad, CA, USA) at room temperature in buffer containing 10 mM Tris pH 7.5, 1 mM EDTA and 1 M NaCl. The beads were then washed in the same buffer and resuspended in 0.4 M NaOH to denature the double-stranded DNA (dsDNA). The supernatant containing the 1000 nt single-stranded DNA (ssDNA) oligo was collected and neutralized by the addition of 5.75 μl of 3.25 M NaOAc, pH 4.9 followed by ethanol precipitation.
Supercoiled plasmid was converted to the nicked circular form by digestion with deoxyribonuclease I (Amersham, Buckinghamshire, UK) in buffer containing 50 mM Tris HCl, pH 7.5, 10 mM MgCl2, 50 μg/ml BSA. For 10 μg of the pNRB252 plasmid, 3 × 10−5 U/μl DnaseI was added and incubated at 37°C for 15 min. The reaction was stopped with the addition of 500 mM EDTA, purified with phenol/chloroform and subsequently ethanol precipitated and resuspended in dH2o.
The 1312 bp plasmid pNRB252 was constructed by inserting the 429 bp EcoRV-BsrBI fragment from pRS306 (33) into the HincII site of piAN7 (34).
Protein purification
Yeast Dmc1 (ScDmc1) was purified using a modified version of the previously published method (14). Briefly, the plasmid containing HIS6-DMC1 under a T7 promoter, was expressed in beta-lactam region (BLR) (DE3)/pLysS Escherichia coli cells (Novagen, Darmstadt, Germany). The cells were lysed by French press (Amicon, Billerica, MA, USA) in the presence of protease inhibitors (5 μg/ml antipain, 2 μg/ml aprotinin, 100 ng/ml leupeptin, 100 μg/ml pefabloc SC) and cleared by ultracentrifugation. The lysate was passed over a Talon column (Clontech, Mountain View, CA, USA) and the eluted protein was then applied to a heparin column (Amersham). The heparin-eluted protein was then applied to a final Q sepharose (Amersham) column. The eluted His6-Dmc1 was dialyzed into storage buffer and kept at −80°C.
The yeast Rad51 (ScRad51) FL/MBP expression strain, plasmid and purification protocol were generously supplied by the Rice lab (University of Chicago). Rad51 was expressed in the Rosetta (DE3) pLysS cell line (Novagen) using a PET3 vector, sonicated in the presence of protease inhibitors (Complete Mini tablets from Roche, Basel, Switzerland) and cleared by ultracentrifugation. Ammonium sulfate was added to the cleared lysate for a final concentration of 0.24 g/ml and centrifuged again. The pellet was resuspended in buffer containing 20 mM Tris HCl pH 7.5, 0.5 mM EDTA, 1 M NaCl, 1 M urea, 5% glycerol, 1 mM dithiothreitol (DTT) and then dialyzed into buffer P (50 mM NaPhos pH 7.5, 1 M NaCl, 5% glycerol, 1 mM DTT). The dialyzed protein solution was applied to a nickel column (Amersham), washed with buffer P and eluted with an imidazole gradient. The appropriate eluate fractions were pooled and incubated with 10 mM ATP and 10 mM MgCl2 for 15 min at 37°C and then at room temperature for 1 h. The protein was applied again to the nickel column, washed with buffer P containing 10 mM ATP and 10 mM MgCl2 and eluted with an imidazole gradient as before. The appropriate fractions were then applied to an amylose (NEB) column, washed with buffer P and eluted with buffer P + 10 mM maltose (Sigma), pH 7.5. The fractions containing Rad51 were then pooled and digested with tobacco etch virus (TEV) protease at a Rad51 to TEV ratio of 30:1. The protein was applied again to the nickel column and eluted with steps of 10 mM, 15 mM, 20 mM and 25 mM imidazole. The fractions containing Rad51 were pooled, concentrated and dialyzed into Rad51 storage buffer (20 mM Tis HCl pH 7.5, 0.5 mM EDTA, 50 mM NaCl, 30% glycerol, 1 mM DTT). The HsDmc1 protein was purified from High-Five insect cells as previously described (18) and the human Rad51 (HsRad51) protein was generously provided by Phillip P. Connell (University of Chicago).
Helical reconstructions
The HsDmc1 filaments were reconstructed using the IHRSR (iterative helical real space reconstruction) method (35). Filament images were cut into 26 745 small overlapping segments and the relative orientations of these segments were determined by projection mapping. Each segment was 80 pixels long, with a sampling of 3.9 Å per pixel. Multiple reference models, having different pitches, were used to sort the segments by pitch. The largest bin, corresponding to a pitch of ∼10 nm, contained 9781 segments. Back projection was used to produce a 3D structure upon which the helical symmetry was imposed.
Persistence length calculations
Dmc1 and Rad51 filaments were formed on ssDNA in the presence of adenylyl-imidodiphosphate (AMP-PNP) and 1 mM Mg2+ as described below. RecA filaments were formed using RecA protein (NEB) and identical conditions and substrates as used for Rad51 and Dmc1, except that ATPγS replaced AMP-PNP.
Filaments were measured for end-to-end distance (R) and for contour length (L). The critical parameter describing filament stiffness in this model is persistence length (λ). Lower λ indicates greater flexibility. The persistence length may be determined experimentally by the equation: R2 = 2λ 2 (L/λ − 1 + e− L/λ). Once L and R are determined experimentally, this equation may be used to derive λ by an iterative method (36). This analysis was done using data from Rad51 filaments (n = 34), Dmc1 filaments (n = 36) and RecA filaments (n = 38). Only filaments greater than 250 nm in length were scored. The calculations were done using R: A Language and Environment for Statistical Computing (version 2.4.1).
Filament preparation
Single strand nucleoprotein filaments were prepared by incubating ssDNA oligos at 10 μM with respect to nucleotide with the specified recombinase protein at 3.33 μM in the appropriate reaction buffer. The ScDmc1 buffer contained 20 mM HEPES (pH 7.5), 1 mM DTT and 1 mM MgOAc with 0.5 mM AMP-PNP or 1 mM CaCl2 with 0.5 mM ATP. The ScRad51 buffer is the same with the exception of 20 mM Tris-HCl (pH 7.5) used in place of HEPES. The HsDmc1 buffer contained 25 mM triethanolamine-HCl (pH7.2), 200 mM KCl and 2 mM MgOAc with 2.5 mM AMP-PNP or 1 mM CaCl2, with 0.5 mM ATP. The HsRad51 buffer contained 20 mM Tris-HCl (pH 8), 1 mM DTT and 1 mM MgOAc with 0.5 mM AMP-PNP or 1 mM CaCl2 with 0.5 mM ATP. Filaments were formed on nicked plasmid DNA by incubating 20 μM DNA with 6.67 μM recombinase protein in the appropriate buffer. Incubations were done at 37°C for 5 min for the yeast proteins and 20 min for the human proteins. Samples prepared for persistence length, stoichiometry and pitch length calculations were spread on carbon coated grids and negatively stained with 1% uranyl acetate.
For the determination of the helical handedness of ScDmc1 filaments, 2 μl samples were first adsorbed to small glass coverslips at 37°C for 5 min, in preparation for freeze-drying and platinum replication. The glass was then rinsed vigorously with 10 ml of buffer solution to remove unattached proteins, passed briefly through dH2O to remove buffer and salts and quick-frozen by impact against a copper block cooled to 4°K with liquid helium.
The frozen samples were stored in liquid nitrogen until mounting in a Balzers 301 vacuum evaporator, where they were freeze dried for 20 min at −80°C and then rotary-replicated with 2 nm of platinum deposited from an electron-beam gun mounted at 15° above the horizontal. The replica was then stabilized with a carbon film deposited from a 70° angle. Replicas were floated off the glass onto concentrated hydrofluoric acid, then transferred through several rinses of dH2O and picked up on formvar-coated copper grids.
EM imaging
Images taken for the determination of persistence length, stoichiometry and pitch length were recorded at a magnification of 25 000× –49 000× with a GATAN digital camera on a Tecnai F30 transmission electron microscope operated at 300 kV. The collection method employed for imaging involved an initial scan at 19 500 × to locate a region of the grid with visible filaments or coated plasmids, followed by higher magnification imaging (25 000× –49 000×). Pitch length measurements were taken using the program Digitalmicrograph 3.10.1 (GATAN). Images taken of replicas for determining the helical handedness of ScDmc1 filaments were examined in a JEOL 100CX electron microscope and photographed at 150 000 × with an AMT digital camera.
RESULTS
Persistence length
To determine the relative rigidities of ScDmc1 and ScRad51 filaments we employed a method previously used to determine the persistence length of RecA filaments (36). The structures of ScDmc1- and ScRad51-ssDNA complexes were examined by TEM after negative staining with 1% uranyl acetate. These recombinase-coated DNA molecules were measured for end-to-end distance (R) and for contour length (L). The parameter used to describe filament stiffness is persistence length (λ); a lower value for λ indicates a more flexible filament. The persistence length of filaments observed by TEM was calculated as described in Materials and Methods section. To avoid misunderstanding, we note that different definitions of persistence lengths have been used in different studies. The equation used here and in some previous studies gives a value of the persistence length twice that given by the method used by Rivetti et al. (37) for polymers in equilibrium on a 2D surface).
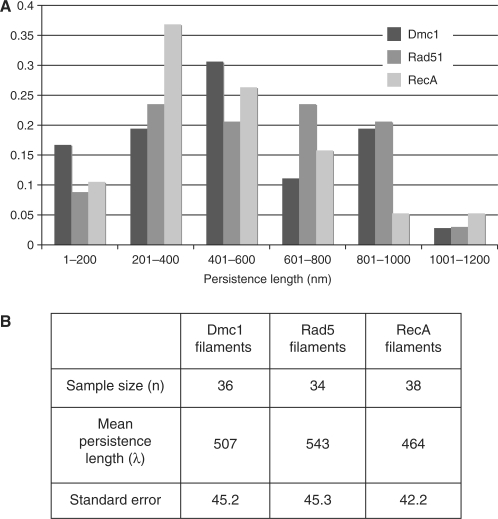
Helical filaments of ScRad51 and ScDmc1 were formed on 1000 nt ssDNA in the presence of AMP-PNP and Mg2+. RecA filaments were formed under the same conditions with the exception of using ATPγS as the non-hydrolyzable ATP analog in place of AMP-PNP. The persistence length of ScDmc1 was determined to be 507 ± 45.2 nm and that of ScRad51 543 ± 45.3 nm (Figure 1). The persistence length of RecA was also quite similar to the two eukaryotic proteins, 464 ± 42.2 nm. The relative values of Rad51 and Dmc1 persistence length indicate that there is no significant difference in filament stiffness between these recombinase filaments under the conditions examined. A recently reported HsRad51 persistence length value of 190 ± 12 nm, while lower than the values we have determined for the yeast proteins, is still consistent with our findings due to the fact that the filaments examined by magnetic tweezer analysis do not fully saturate the DNA substrate on which they were formed (38).
Figure 1.
Persistence length of ScRad51 and ScDmc1 filaments. Persistent lengths were determined for negatively stained filaments as described in Materials and Methods section. (A) Distribution of persistence lengths for each recombinase protein. (B) Mean persistence length values and standard errors for data from (A).
The persistence length values obtained here for the three recombinases are slightly lower than the previously determined value of ∼600 nm for RecA (36). This modest difference is likely to reflect different size distributions in filaments included in the measurements. We emphasize that the most significant conclusion from this study is that analysis of the three recombinases did not reveal a significant difference in filament rigidity.
Pitch and width of ScDmc1 and HsDmc1 filaments
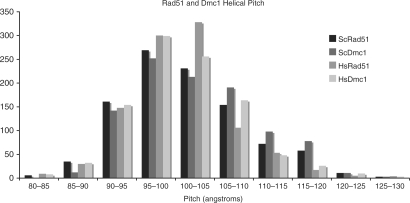
As discussed above, a previous AFM study determined the helical pitch of ScDmc1 filaments to be substantially larger than the range of values obtained by TEM analysis of RecA, Rad51 and HsDmc1 filaments (18,23,24,39). Additionally, the two measurements of filament diameter are larger than the published diameters of RecA and Rad51 filaments (23,24). To further examine these parameters, we conducted a side-by-side comparison of Rad51 and Dmc1 using both yeast and human protein, with respect to filament pitch and diameter. As shown in Figure 2, the measurements of Dmc1- and Rad51-ssDNA filaments formed in the presence of AMP-PNP and Mg+2 indicate that the distributions of pitch values are the same for all four proteins. The average pitch values for the yeast proteins were 10.2 ± 0.8 nm for Rad51 and 10.4 ± 0.8 nm for Dmc1. The average pitch values for the human proteins were 10.1 ± 0.7 nm for Rad51 and 10.1 ± 0.7 nm for Dmc1. All four nucleoprotein filaments have pitch values of ∼10 nm in agreement with previous analyses of ScRad51 and both human recombinases (18,24,40). These measurements demonstrate striking similarity between Dmc1 and Rad51 filaments.
Figure 2.
Helical pitch values. Pitch length comparison of 1000 individual striations from ScDmc1, ScRad51, HsDmc1 and HsRad51 ssDNA filaments. Filaments were formed in the presence of 1–2 mM Mg2+ and 0.5–2.5 mM AMP-PNP, as indicated in Materials and Methods section.
The average filament diameters for the yeast proteins are 11.9 ± 0.8 nm for Rad51 and 11.2 ± 0.7 nm for Dmc1. The average filament diameters for the human proteins are 11.8 ± 0.8 nm for Rad51 and 11.9 ± 0.9 nm for Dmc1. The values for Dmc1 and Rad51 diameter are similar to, though slightly larger than the published values of 10–11 nm for RecA and Rad51 filaments (23,24). These data also indicate structural conservation between the helical filaments formed by RecA-like proteins.
Base pairs per helical turn of ScDmc1 and HsDmc1 filaments
The previous analysis of Dmc1 DNA-binding stoichiometry had been conducted using linear DNA molecules (20). This analysis was based on the assumption that the observed filaments contained intact DNA molecules that were fully saturated. Under this assumption, the results indicated that Dmc1 formed filaments with a stoichiometry of ∼24 nucleotides per helical turn in contrast to previous measurements of Rad51 and RecA filaments which yielded values of ∼19 bp per helical turn (23,24).
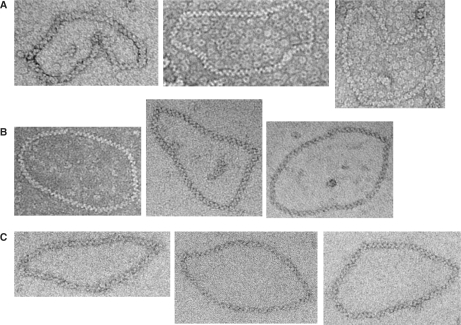
To clarify the stoichiometry of DNA binding by the helical filament we analyzed HsDmc1 filaments formed on relaxed circular dsDNA molecules and compared them to HsRad51 filaments formed on the same substrates (Figure 3). Our results show that for a fully coated a 1312 bp plasmid pNRB253 (see MATERIALS AND METHODS section) there is an average of 67.7 ± 0.8 striations (n = 6) for HsRad51 filaments and an average of 68.8 ± 0.8 striations (n = 6) for HsDmc1 filaments. These values correspond to a DNA binding stoichiometry of 18.9–19.3 bp/turn for HsDmc1. A comparison of ScRad51 filaments on the 1312 bp circular relaxed plasmid (n = 4) to the human protein filaments was also performed demonstrating that the helical stoichiometry of ScRad51 (65.8 ± 1.5 striations/1312 bp or 19.5–20.4 bp/turn) is similar to the human protein (24).
Figure 3.
Base pairs per helical turn of HsDmc1, HsRad51 and ScRad51 filaments. Sample of micrographs of (A) HsDmc1, (B) HsRad51 and (C) ScRad51 filaments on relaxed 1312 bp plasmids.
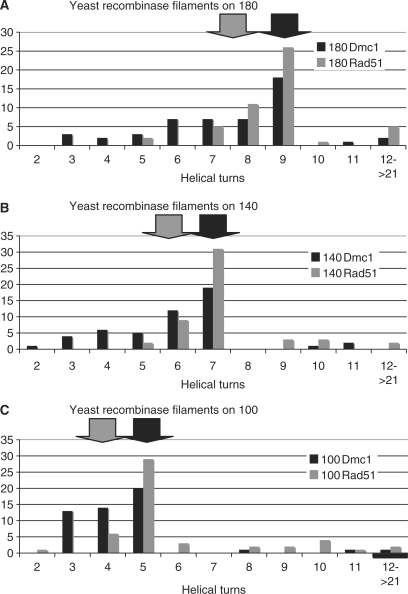
Due to difficulties in obtaining ScDmc1 filaments of sufficient length to completely coat a 1312 bp plasmid, we used short linear dsDNA substrates of 100, 140 and 180 bp to compare the DNA binding of yeast Rad51 and Dmc1. The canonical helical stoichiometry of six protomers per turn predicts ∼9 turns/180 bp, ∼7 turns/140 bp and ∼5 turns/100 bp (indicated by the black arrows in Figure 4) while a stoichiometry of 24 bp/turn predicts ∼7.5 turns/180 bp, ∼5.8 turns/140 bp and ∼4.2 turns/100 bp (indicated by grey arrows in Figure 4). The samples of ScRad51 and ScDmc1 filaments formed on these three linear substrates were used to compare the distribution of striation number and filament lengths of each protein, allowing for a relative comparison of helical stoichiometry. The striation number averages for ScDmc1 filaments (7.6 ± 2.4 turns/180 bp, 6.0 ± 1.9 turns/140 bp and 4.5 ± 1.7 turns/180 bp) were lower than the ScRad51 striation numbers (9.2 ± 2.7 turns/180 bp, 7.2 ± 1.5 turns/140 bp, 6.1 ± 2.7 turns/bp). This appears to be due to a tendency of our preparations of ScDmc1 to form shorter filaments that do not saturate the substrate. Importantly, the distributions displayed in Figure 4 have identical peaks for both ScRad51 and ScDmc1 on all three substrates (9 turns/180 bp, 7 turns/140 bp, 5 turns/100 bp). We interpret the filaments having the peak value to be those in which the DNA substrate is saturated. This conclusion is supported by concordance of the ∼19 bp/striation value obtained from ScRad51 filaments formed on all three small linear DNAs and the circular DNA substrate. Interestingly, the detection of a small number of very long filaments raises the possibility that the recombinase filaments may be capable of extending beyond the end of the DNA substrate. Regardless, all filaments formed were of the predicted length for the given filament's striation number, providing further support of conserved pitch length as evidenced by the linear distribution of each sample (Supplementary Figure 1). The helical stoichiometry of ScDmc1 binding at saturation to each of the three linear molecules gives the predicted values for filaments with ∼19 bp/turn. These numbers indicate that ScDmc1 filaments unwind the DNA substrate to which they are bound to the same extent as ScRad51, HsDmc1 and all other RecA-like protein filaments.
Figure 4.
Distribution of helical turns for ScDmc1 and ScRad51 filaments. Number of striations per filament. ScDmc1 and ScRad51 filaments formed on linear fragments of (A) 180 bp, (B) 140 bp and (C) 100 bp. Filaments were formed in the presence of 1 mM Mg2+ and 0.5 mM AMP-PNP. Black arrows indicate predicted turns based on 19 bp/turn, grey arrows indicate predicted turns based on 24 bp/turn.
ScDmc1 filament helicity
Chen et al. (22) presented TEM images that they interpreted as showing recombinase filaments with left-handed helicity on DNA. Although negatively stained TEM images can appear to reveal helical handedness, the depth of focus of TEM is larger than the height of the filaments precluding reliable determination of handedness by negative staining or cryo-EM (41,42). In addition, Chen et al. (22) included an AFM image that appears to show a left-handed helix of ScDmc1 on DNA formed in presence of Ca+2 and ATP (Supplementary Figure 1A of ref. 22). This is of particular interest since RecA and Rad51 have been observed to form only right-handed filaments on DNA (23,25). Further interest stems from the finding that RadA can be crystallized in a left-handed form, albeit in the absence of DNA (22).
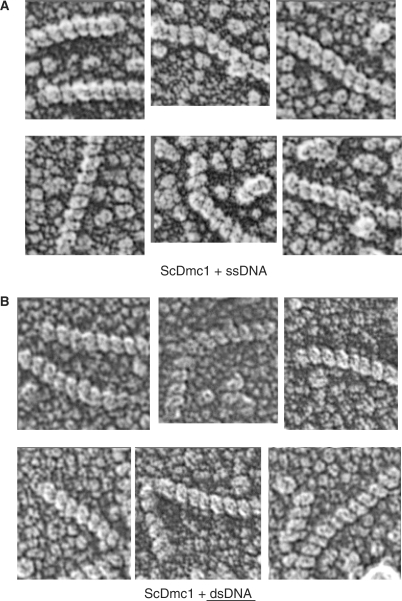
To characterize the handedness of Dmc1-DNA, we used quick-freeze/deep-etch EM with shadowing to examine filaments formed on either ssDNA or dsDNA. The filament samples were prepared using 1000 nt ssDNA oligos or linear dsDNA in the presence of buffer containing Ca+2 and ATP (Figure 5). Absorbed samples were overlaid with F-actin filaments, as an internal standard (data not shown). For the Dmc1-dsDNA filaments we collected a sample size of n = 86 filaments. All of the observed filaments displayed a clear right-handed helicity. We repeated this same experiment but replaced the dsDNA substrate with a 1000 nt ssDNA linear fragment. For this condition we collected a sample size of n = 61 filaments. Again, all the filaments that we observed displayed a clear right-handed helicity. These data suggest that under the conditions examined, ScDmc1 predominantly or exclusively forms right-handed filaments, as has been observed for both Rad51 and RecA (23,25).
Figure 5.
Right-hand helicity of ScDmc1 filaments on single and double strand DNA. Filaments were formed on ssDNA (n = 61) or dsDNA (n = 86) in the presence of calcium and ATP and then examined by deep-etch shadowing EM as described in the Materials and Methods section.
Three dimensional reconstruction of a Dmc1-DNA helical filament
To further examine the structure of the HsDmc1 filament, the IHRSR method (35) of EM image analysis was used to reconstruct a HsDmc1-dsDNA filament formed in the presence of AMP-PNP and Mg+2. Based on the determination of helicity described above, the model assumes that the helix was right handed. Different initial helical symmetries (six through nine subunits/turn) were used for the IHRSR approach with a convergence to a symmetry of ∼6.4 subunits per turn providing the best fit to the crystal structure of the Dmc1 ATP-binding core (43).
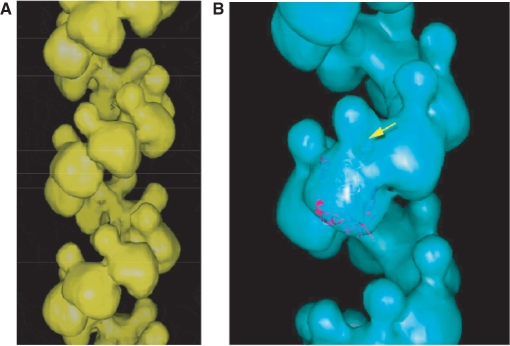
The reconstructed HsDmc1 filament structure is remarkably similar to the previously published structure of the HsRad51 filament (40). The HsDmc1 filament image (Figure 6A) indicates that, as with the HsRad51 reconstruction, the protomers of HsDmc1 are composed of a large lobe located along the helical screw and a smaller lobe that extends into the helical groove of the filament. The reconstruction of the HsRad51 filament provided evidence, based on the fit of the crystal structure (44) that this small lobe represents the N-terminus of the protein (40). In the present reconstruction, the larger lobe (Figure 6B) is the size predicted by the volume of the previously published crystal structure of the ATPase domain of HsDmc1 (43) implying that the small lobe is very likely to represent that N-terminal region of the protein. While the data presented here indicates that the consensus helical symmetry is about six protomers, we note that HsDmc1 helical turns display considerable variation in pitch and width, properties likely to reflect filament elasticity.
Figure 6.
Helical reconstructions of HsDmc1 filaments. The filament was reconstructed using the iterative helical real space reconstruction method. (A) Filament formed on dsDNA in the presence of KCl and AMP-PNP. (B) The large lobe of the HsDmc1 protomer can accommodate the atomic structure of the C-terminal region of HsDmc1.
DISCUSSION
Structural conservation of the eukaryotic recombinases
The data presented in this report and summarized in Table 1 demonstrate the high degree of structural conservation between the eukaryotic recombinase filaments. This conservation of structure indicates that the Dmc1 filament is comparable to that of other members of the RecA family including Rad51. Reports on the structures of Thermus aquaticus RecA (45), archeal RadA (27) and T4 bacteriophage UvsX (46) indicate that all of these proteins form helical filaments with characteristics that are very similar to those of filaments formed by E. coli RecA as well as ScRad51 and HsRad51. An interesting exception to the overall conservation of these structures appears to be the C. elegans Rad51 filaments, which reconstruction experiments indicate contain eight protomers per turn (47).
Table 1.
Helical parameters for budding yeast and human recombinases
| RecA | ScDmc1 | ScRad51 | HsDmc1 | HsRad51 | |
|---|---|---|---|---|---|
| Persistence length (nm) | 464 ± 42.2 (630a) | 507 ± 45.2 | 543 ± 45.3 | – | (190 ± 12f) |
| Helical pitch (Å) | 96b | 101 ± 7.0 | 101 ± 6.5 (99c) | 103 ± 7.5 (100e) | 102 ± 7.7 (99g) |
| Filament diameter (Å) | 110b | 119 ± 9.1 | 118 ± 7.8 | 120 ± 7.3 | 119 ± 8.2 |
| Helical stoichiometry (bp/turn) | 18.6b | ∼19 | 19.9 ± 0.5 (18.6c) | 19.1 ± 0.2 | 19.2 ± 0.4 (18.6h) |
| Helical handedness | Rightb | Right | Rightd | – | – |
aRef. (36).
bRef. (23).
cRef. (24).
dRef. (25).
eRef. (18).
fRef. (38).
gRef. (40).
hRef. (39).
Values determined in this study are indicated in bold. Previously reported values are given in non-bold font with sources indicated by footnote.
The helical reconstruction of the HsDmc1 filament is striking in its similarity to the previously published HsRad51 reconstruction and supports numerous observations of structural conservation (40). The molecular structure of the ATPase domain of HsDmc1 is accommodated by the volume of the large lobe of protomers in the filament reconstruction. In light of similarity to the Rad51 filament, the HsDmc1 reconstruction suggests that the smaller lobes are N-terminal domains. Both the HsRad51 and HsDmc1 reconstructions suggest they have an average of ∼6 protomers per helical turn. In the case of HsDmc1 this finding is supported by the fact that alternate reconstructions, including one that assumed eight protomers per turn, cannot accommodate the volume of the ATPase domain crystal structure. Although the consensus structure appears to be ∼6 protomers per turn, the pitch and twist of Dmc1 filaments are quite variable. This finding makes it likely that regions of the coil can contain either more or fewer than six protomers per turn.
The helical stoichiometry of both yeast and HsDmc1 filaments has been shown here to be approximately 19 bp/helical turn. This comparison demonstrated the same base pair per helical turn ratio for each protein and therefore suggests the same monomer to DNA binding ratio. By extension, these data indicate that all four proteins, including ScDmc1, bind to DNA with the same helical stoichiometry. Although the current data do not represent a direct determination of the stoichiometry of DNA binding per protomer, the observation of filaments with ∼19bp/turn and goodness of fit of the six-protomer-per-turn reconstruction makes it highly likely that Dmc1 binds to DNA at a ratio of 1 protomer to 3 bp.
Our investigation of the helicity of yeast Dmc1 detected only right-handed filaments RecA and Rad51 as has been previously reported (23,25). Two previous AFM studies of RadA proteins reported different results on filament helicity with one study finding only right-handed filaments (27) and a second reporting both right- and left-handed filaments (48). As mentioned above, another report included an AFM image that appears to be a left-handed filament of Dmc1 on DNA (22). An explanation for the apparent discrepancy is provided below.
Comparison with observations by AFM
In the published images of ScDmc1 from AFM studies (20,22), filaments are particularly straight, in contrast to virtually all EM analyses of other recombinase proteins, including HsDmc1, HsRad51, ScRad51 and RecA (18,19,23,24,39) and in contrast to AFM images of Rad51, RadA and RecA (27–31). However, our persistence length calculations for long ScDmc1 and ScRad51 filaments showed no difference in the rigidity of these filaments. An examination of the previously published Dmc1 AFM images reveals what appears to be irregularities in the mica upon which the filaments were deposited and the parallel alignment of multiple filaments within a single field (20). It is possible that these irregularities are the result of lattice steps in the mica substrate acting to trap filaments along straight lines (C. Wyman, personal communication).
The alignment of filaments may also be due to the movement of the air–liquid interface (meniscus) along the mica surface during preparation of samples for AFM analysis (49–55). Large pitch values obtained by AFM could have resulted from the stretching of the filaments by meniscus movement or they could reflect tip diameter as has been previously reported (27). We therefore suspect that, if the striations seen by AFM reflect helical turns, such striations were from a subset of filaments that were stretched during sample preparation. It is possible, therefore, that the unusual properties of the filaments seen in these images reflect filament elasticity rather than structural differences between ScDmc1 and other members of this protein family.
Although tip diameter and sample deposition effects might explain most discrepancies between previous AFM analyses and the present TEM study, they do not explain the observation of left-handed helical filaments. We examined the helicity of over 60 ssDNA filaments and more than 80 dsDNA filaments formed in the presence of calcium and ATP, the same conditions in which Chen et al. report left-handed helices of Dmc1. Our observations indicate that if Dmc1 can form left-handed helices, it does so only rarely under these conditions. An alternative explanation for the source of the AFM image interpreted as showing left handedness (Supplementary Figure S1A in ref. 22 for Dmc1 and Figure 3 of ref. 48 for SsoRadA) is filament bundling. Filament bundling results in formation of both left- and right-handed structures (56,57). We therefore argue that there is no clear evidence at present for left-handed nucleoprotein filament formation of any RecA-like recombinase. In this context it is important to note that one member of this protein family has been crystallized as a left-handed helix with four protomers per turn (22). However, this form was obtained in the absence of DNA and its biological relevance remains to be determined. The ability of RadA to form right- and left-handed helices as well as toroids demonstrates the structural flexibility of this class of proteins (22,27,58).
Implications of structural and functional conservation
The helical structure of the RecA-like recombinases is very highly conserved. These proteins significantly underwind and extend the DNA substrate within the filament. It is this distortion of the DNA that has been proposed to be the essential structural condition for the pairing and exchange reactions (46). All available data, including those presented here indicate that this requirement is met by the meiosis-specific Dmc1 filament.
As mentioned earlier, there is little difference between the biochemical activities of Rad51 and Dmc1. Both recombinases can promote strand assimilation and exchange. While there are some distinctions in optimal in vitro reaction conditions, it is not clear to what extent these differences are biologically relevant.
In light of the conserved filament structure and the growing body of data indicating conserved intrinsic biochemical activity for Dmc1 and Rad51, it seems likely that the factors that differentiate these proteins in vivo are their distinct protein–protein interactions. In particular, these data provide no evidence for any intrinsic difference in filament rigidity between Rad51 and Dmc1. This result argues against the possibility that this intrinsic rigidity underlies the difference in the ability of Rad51 and Dmc1 to promote interhomolog as opposed to intersister recombination. Nonetheless, it is still possible that Dmc1's interhomolog preference involves projection of filaments away from axial elements. For example, projection from axial element may require Dmc1-specific interactions with one or more accessory factor (32). Further study of the structure of recombination complexes in vivo will be required to determine how interhomolog recombination preference is achieved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Phoebe Rice for helpful advice as well as for the ScRad51 expression plasmid and ScRad51 purification protocol, Phillip P. Connell for providing HsRad51 protein and Don Conrad for assistance with computation. D.K.B. and S.D.S. are particularly grateful to Bob Josephs and the staff of the University of Chicago Electron Microscopy facility for technical advice and assistance. This work was supported by NIGMS grant GM050936. Funding to pay the Open Access publication charges for this article was provided by the National Institute of Health grant GM50936.
Conflict of interest statement. None declared.
REFERENCES
- 1.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson JA, Fink GR. Meiotic recombination between duplicated genetic elements in Saccharomyces cerevisiae. Genetics. 1985;109:303–332. doi: 10.1093/genetics/109.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 7.Pecina A, Smith KN, Mezard C, Murakami H, Ohta K, Nicolas A. Targeted stimulation of meiotic recombination. Cell. 2002;111:173–184. doi: 10.1016/s0092-8674(02)01002-4. [DOI] [PubMed] [Google Scholar]
- 8.Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 9.Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masson JY, West SC. The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem. Sci. 2001;26:131–136. doi: 10.1016/s0968-0004(00)01742-4. [DOI] [PubMed] [Google Scholar]
- 11.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 12.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Golub EI, Gupta R, Radding CM. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl Acad. Sci. USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- 15.Masson JY, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A, West SC. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passy SI, Yu X, Li Z, Radding CM, Masson JY, West SC, Egelman EH. Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl Acad. Sci. USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, Shibata T, Kurumizaka H, Yokoyama S. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol. Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 18.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 19.Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 20.Lee MH, Chang YC, Hong EL, Grubb J, Chang CS, Bishop DK, Wang TF. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 2005;280:40980–40984. doi: 10.1074/jbc.M505896200. [DOI] [PubMed] [Google Scholar]
- 21.Sauvageau S, Stasiak AZ, Banville I, Ploquin M, Stasiak A, Masson JY. Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell. Biol. 2005;25:4377–4387. doi: 10.1128/MCB.25.11.4377-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LT, Ko TP, Chang YC, Lin KA, Chang CS, Wang AH, Wang TF. Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins. Nucleic Acids Res. 2007;35:1787–1801. doi: 10.1093/nar/gkl1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stasiak A, Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 25.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 26.Chang YC, Lo YH, Lee MH, Leng CH, Hu SM, Chang CS, Wang TF. Molecular visualization of the yeast Dmc1 protein ring and Dmc1-ssDNA nucleoprotein complex. Biochemistry. 2005;44:6052–6058. doi: 10.1021/bi048897q. [DOI] [PubMed] [Google Scholar]
- 27.Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umemura K, Komatsu J, Uchihashi T, Choi N, Ikawa S, Nishinaka T, Shibata T, Nakayama Y, Katsura S, Mizuno A, et al. Atomic force microscopy of RecA—DNA complexes using a carbon nanotube tip. Biochem. Biophys. Res. Commun. 2001;281:390–395. doi: 10.1006/bbrc.2001.4333. [DOI] [PubMed] [Google Scholar]
- 29.Shi WX, Larson RG. Atomic force microscopic study of aggregation of RecA-DNA nucleoprotein filaments into left-handed supercoiled bundles. Nano Lett. 2005;5:2476–2481. doi: 10.1021/nl051783v. [DOI] [PubMed] [Google Scholar]
- 30.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang HV, Little PF, Seed B. Improved suppressor tRNA cloning vectors and plasmid-phage recombination. Biotechnology. 1988;10:269–283. doi: 10.1016/b978-0-409-90042-2.50020-9. [DOI] [PubMed] [Google Scholar]
- 35.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 36.Egelman EH, Stasiak A. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J. Mol. Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 37.Rivetti C, Guthold M, Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 38.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl Acad. Sci. USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos. Trans. R. Soc. Lond. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 42.Belnap DM, Olson NH, Baker TS. A method for establishing the handedness of biological macromolecules. J. Struct. Biol. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]
- 43.Kinebuchi T, Kagawa W, Kurumizaka H, Yokoyama S. Role of the N-terminal domain of the human DMC1 protein in octamer formation and DNA binding. J. Biol. Chem. 2005;280:28382–28387. doi: 10.1074/jbc.M503372200. [DOI] [PubMed] [Google Scholar]
- 44.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Angov E, Camerini-Otero RD, Egelman EH. Structural polymorphism of the RecA protein from the thermophilic bacterium Thermus aquaticus. Biophys. J. 1995;69:2728–2738. doi: 10.1016/S0006-3495(95)80144-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Egelman EH. DNA conformation induced by the bacteriophage T4 UvsX protein appears identical to the conformation induced by the Escherichia coli RecA protein. J. Mol. Biol. 1993;232:1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- 47.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc. Natl Acad. Sci. USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MH, Leng CH, Chang YC, Chou CC, Chen YK, Hsu FF, Chang CS, Wang AH, Wang TF. Self-polymerization of archaeal RadA protein into long and fine helical filaments. Biochem. Biophys. Res. Commun. 2004;323:845–851. doi: 10.1016/j.bbrc.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 49.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 50.Cai W, Aburatani H, Stanton VP, Jr, Housman DE, Wang YK, Schwartz DC. Ordered restriction endonuclease maps of yeast artificial chromosomes created by optical mapping on surfaces. Proc. Natl Acad. Sci. USA. 1995;92:5164–5168. doi: 10.1073/pnas.92.11.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 52.Yokota H, Johnson F, Lu H, Robinson RM, Belu AM, Garrison MD, Ratner BD, Trask BJ, Miller DL. A new method for straightening DNA molecules for optical restriction mapping. Nucleic Acids Res. 1997;25:1064–1070. doi: 10.1093/nar/25.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Bai C, Wang C, Zhu C, Lin Z, Li Q, Cao E. A convenient method of aligning large DNA molecules on bare mica surfaces for atomic force microscopy. Nucleic Acids Res. 1998;26:4785–4786. doi: 10.1093/nar/26.20.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Lin J, Schwartz DC. Scanning force microscopy of DNA molecules elongated by convective fluid flow in an evaporating droplet. Biophys. J. 1998;75:513–520. doi: 10.1016/S0006-3495(98)77540-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otobe K, Ohtani T. Behavior of DNA fibers stretched by precise meniscus motion control. Nucleic Acids Res. 2001;29:E109. doi: 10.1093/nar/29.22.e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Egelman EH, Stasiak A. Structure of helical RecA-DNA complexes. II. Local conformational changes visualized in bundles of RecA-ATP gamma S filaments. J. Mol. Biol. 1988;200:329–349. doi: 10.1016/0022-2836(88)90245-8. [DOI] [PubMed] [Google Scholar]
- 57.Shi WX, Larson RG. RecA-ssDNA filaments supercoil in the presence of single-stranded DNA-binding protein. Biochem. Biophys. Res. Commun. 2007;357:755–760. doi: 10.1016/j.bbrc.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 58.McIlwraith MJ, Hall DR, Stasiak AZ, Stasiak A, Wigley DB, West SC. RadA protein from Archaeoglobus fulgidus forms rings, nucleoprotein filaments and catalyses homologous recombination. Nucleic Acids Res. 2001;29:4509–4517. doi: 10.1093/nar/29.22.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.