Abstract
Crotonaldehyde is a representative α,β-unsaturated aldehyde endowed of mutagenic and carcinogenic properties related to its propensity to react with DNA. Cyclic crotonaldehyde-derived deoxyguanosine (CrA-PdG) adducts can undergo ring opening in duplex DNA to yield a highly reactive aldehydic moiety. Here, we demonstrate that site-specifically modified DNA oligonucleotides containing a single CrA-PdG adduct can form crosslinks with topoisomerase I (Top1), both directly and indirectly. Direct covalent complex formation between the CrA-PdG adduct and Top1 is detectable after reduction with sodium cyanoborohydride, which is consistent with the formation of a Schiff base between Top1 and the ring open aldehyde form of the adduct. In addition, we show that the CrA-PdG adduct alters the cleavage and religation activities of Top1. It suppresses Top1 cleavage complexes at the adduct site and induces both reversible and irreversible cleavage complexes adjacent to the CrA-PdG adduct. The formation of stable DNA–Top1 crosslinks and the induction of Top1 cleavage complexes by CrA-PdG are mutually exclusive. Lastly, we found that crotonaldehyde induces the formation of DNA–Top1 complexes in mammalian cells, which suggests a potential relationship between formation of DNA–Top1 crosslinks and the mutagenic and carcinogenic properties of crotonaldehyde.
INTRODUCTION
Crotonaldehyde is a rather simple α,β-unsaturated aldehyde (enal) that is ubiquitous in the human environment. The general population is exogenously exposed to crotonaldehyde through mobile source emissions, tobacco smoke and other thermal degradation mixtures (1). It is also produced endogenously from lipid peroxidation (2) and from the metabolism of N-nitrosopyrrolidine (3,4). Crotonaldehyde has been shown to be mutagenic in both Salmonella typhimuruim (5,6) and human lymphoblasts (7). Site-specific mutagenesis revealed that the crotonaldehyde-dG adducts are modestly mutagenic in COS7 cells as well (8). In addition, crotonaldehyde has been found to induce liver tumors in rats (9) and is regarded as a possible human carcinogen.
Both the mutagenic and carcinogenic effects of crotonaldehyde exposure have been proposed to be associated with the ability of crotonaldehyde to react covalently with DNA. Crotonaldehyde as well as other structurally related α,β-unsaturated aldehydes produce cyclic l,N2-deoxyguanosine (dG) adducts in vitro. The reaction mechanism involves nucleophilic Michael addition by the exocyclic N2 atom of dG to the carbon–carbon double bond of the enal, and subsequent ring closure by reaction of the carbonyl carbon atom with the N1 position of dG (10–12). The resulting products from a reaction between crotonaldehyde and dG are a pair of diastereomeric adducts, R- and S-α-methyl-γ-hydroxy-1,N2-propano-2′-deoxyguanosine (R- and S-CrA-PdG) (11). Several methods have been developed and used successfully to detect these types of DNA adducts in vivo (13–17). In addition, the recent syntheses of DNA oligonucleotides containing site- and stereo-specific CrA-PdG adducts has enabled their chemistry and biology to be further studied in vitro (18,19). For instance, positioning the cyclic CrA-PdG adduct opposite dC in duplex DNA has been shown to promote ring opening to the corresponding aldehydic form of the CrA-PdG adduct (N2-[3-oxo-1-methyl-propyl]-dG) (20). The unveiling of the aldehyde moiety within a DNA duplex allows for secondary reactions, including the formation of interstrand DNA–DNA crosslinks to the N2-position of an opposing guanine base in a 5′-CpG sequence (20–22) as well as DNA–peptide (23) and DNA–protein crosslinks (24). Interestingly, the formation of interstrand DNA–DNA crosslinks are dependent on the adduct stereochemistry (25,26), whereas the crosslinking efficiency to the tetrapeptide KWKK is similar between the diastereomeric R- and S-CrA-PdG adducts (23).
As mentioned above, earlier studies have established the DNA–protein crosslinking potential of CrA-PdG adducts with a limited selection of short peptides. Therefore, to expand on these previous studies, we initially sought to obtain evidence for the existence of DNA–protein crosslinked species between the crotonaldehyde-derived DNA adduct and cellular proteins, specifically DNA topoisomerase I (Top1). In a second set of experiments, we examine the consequences of the CrA-PdG adducts on the DNA cleavage/religation activity of Top1. Lastly, we assess the ability of crotonaldehyde to stimulate the formation of DNA–Top1 covalent complexes in cultured mammalian cells.
MATERIALS AND METHODS
Synthesis of modified oligonucleotides
Oligonucleotide containing the crotonaldehyde-dG adduct was prepare via a postoligomerization strategy as previously described and characterized by MALDI-MS using a 3-hydroxypicolinic acid matrix containing ammonium hydrogen citrate (7 mg/ml) to suppress multiple sodium and potassium adducts (19). Briefly, a 22-mer oligonucleotide was prepared containing an O6-(2-trimethylsilylethyl)-2-flourohypoxanthine base (5′-AAAAATTTTTCCAAG*TCTTTTT-3′, where G* denotes the modified nucleotide) from the corresponding modified phosphoramidite reagent using standard solid-phase DNA synthesis protocols. This modified oligonucleotide was deprotected as previously described and purified by HPLC using gradient 1 (MALDI-MS: found 6774.8; calc'd 6773.3). Displacement of 2-fluoro group with racemic 2-amino-4, 5-dihydroxypentane provided the corresponding oligonucleotide, where G* is N2-(1-methyl-4,5-dihydroxybutyl)-dG which was purified by HPLC using gradient 2 (MALDI-MS: found 6773.0; calc'd 6772.2). The vicinal diol was oxidized with sodium periodate to afford the 22-mer oligonucleotide containing a mixture of the R- and S-crotonaldehyde-dG adducts. The diastereomeric oligonucleotides were separated by HPLC using gradient 3 (MALDI-MS for the R-isomer: 6742.8.0; calc'd 6740.2; S-isomer: found 6743.3; calc'd 6740.2). The specific isomers were identified by enzymatic digestion and identification of the modified nucleoside by coinjection with authentic standards of known stereochemistry (19).
HPLC
Purification of the modified oligonucleotides were performed on a Beckman HPLC system (32 Karat software version 3.1, pump module 125) with a diode array UV detector (module 168) monitoring at 260 nm using Waters YMC ODS-AQ column 250 mm × 10 mm i.d., 5 ml/min with 0.1 M aqueous ammonium formate and CH3CN for oligonucleotides. HPLC gradient: (i) a linear gradient from 1% to 10% acetonitrile over 15 min, then a linear gradient from 10% to 20% acetonitrile over 10 min, then a linear gradient from 20% to 100% acetonitrile over 3 min, then isocratic at 100% acetonitrile for 2 min, followed by a linear gradient back to 1% acetonitrile over 3 min; (ii) a linear gradient from 1% to 10% acetonitrile over 15 min, then a linear gradient from 10% to 13% acetonitrile over 10 min, then a linear gradient from 13% to 80% acetonitrile over 3 min, then isocratic at 80% acetonitrile for 2 min, followed by a linear gradient back to 1% acetonitrile over 3 min; (iii) a linear gradient from 1% to 9.2% acetonitrile over 5 min, then a linear gradient from 9.2% to 13.2% acetonitrile over 30 min, then a linear gradient from 13.2% to 99% acetonitrile over 2 min, then isocratic at 99% acetonitrile for 1 min, followed by a linear gradient back to 1% acetonitrile over 2 min.
Preparation and end-labeling of oligonucleotide substrates
Oligonucleotides were either 5′-end labeled using T4 polynucleotide kinase and [γ-32P] ATP (crosslinking assay) or 3′-end labeled using terminal deoxynucleotidyl transferase (TdT) and [α-32P] ddATP (Top1 cleavage assay), respectively. Unincorporated radioactive nucleotides were removed using a mini Quick Spin Oligo column (Roche, Indianapolis, IN, USA) after inactivation of the kinase or TdT by heating for 5 min at 95°C. For construction of double-stranded oligonucleotides, labeled single-stranded oligonuceotides were annealed with their complementary strand by heating for 5 min at 95°C and slowly cooling to room temperature.
Borohydride trapping of DNA–Top1 crosslinks
Both crotonaldehyde-derived deoxyguanosine adducted DNA substrates and nonadducted DNA substrates were incubated with recombinant Top1 in the presence or absence of 50 mM NaCNBH3 for 15 min at 25°C in a reaction buffer containing 10 mM Tris–HCl (pH ∼7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 15 µg/ml BSA and 0.2 mM DTT. NaCNBH3 was prepared on the day of use and added to each reaction immediately preceding the addition of Top1. Reactions were terminated by the addition of one volume of Tris/Glycine/SDS loading buffer. Samples were resolved on a 4–20% sodium dodecyl sulfate (SDS)–polyacrylamide gel in the presence of 1 × Tris/Glycine/SDS buffer for ∼1 h at 100 V. Results were visualized by PhosphorImager analysis.
Top1-mediated DNA cleavage reactions
DNA substrates were incubated with recombinant Top1 in the presence or absence of camptothecin and/or 50 mM NaCNBH3 for 20 min at 25°C in a reaction buffer containing 10 mM Tris–HCl (pH ∼7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 15 µg/ml BSA and 0.2 mM DTT. Reactions were terminated by the addition of SDS (final concentration of 0.5%). An equal volume of loading buffer (80% formamide, 1 mM EDTA, 10 mM NaOH, 0.1% xylene cyanol, 0.1% bromphenol blue) was added. The samples were subsequently heated to 95°C for 5 min and analyzed on a 20% sequencing polyacrylamide gel. For reversal experiments, the addition of SDS was preceded by the addition of NaCl (final concentration of 0.35 M at 25°C for the indicated times).
Detection of DNA–Top1 covalent complexes in MCF7 cells
DNA–Top1 covalent complexes were isolated using the immunocomplex of enzyme (ICE) bioassay as previously described (27,28). Briefly, 1 × 106 MCF7 cells treated with drug or untreated were immediately lyzed with 1% sarkosyl. After homogenization with a Dounce homogenizer, cell lysates were gently layered on CsCl step gradients and centrifuged at 165 000g for 20 h at 20°C. Half-milliliter fractions were collected, diluted with an equal volume of 25 mM sodium phosphate buffer (pH 6.5) and applied to Immobilon-P membranes by using a slot-blot vacuum manifold. DNA–Top1 complexes were detected using the C21 Top1 monoclonal antibody and standard western blotting procedures. Experiments were done independently at least twice.
RESULTS
Covalent trapping of topoisomerase I at the CrA-PdG adduct in duplex DNA
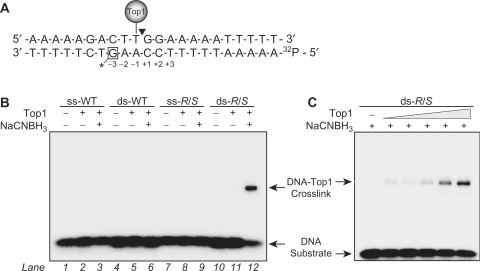
To evaluate the propensity of the crotonaldehyde-derived dG adducts to crosslink to Top1, we incorporated a single CrA-PdG adduct into a previously identified 22-bp DNA oligonucleotide sequence that contains a single high-affinity Top1 cleavage site (29–31). As shown in Figure 1A, the CrA-PdG adduct was placed on the nonscissile strand at the −3 position relative to the Top1 cleavage site. In order to assay for crosslink formation between Top1 and oligonucleotides carrying such an adduct, we adapted the borohydride trapping approach previously employed by Krutz and Lloyd (23). In this assay, 32P-labeled DNA substrates are incubated with the protein of interest under reducing conditions, i.e. in the presence of sodium cyanoborohydride (NaCNBH3). The samples are then separated using denaturing SDS–polyacrylamide gel electrophoresis. DNA–protein crosslinks appear as radiolabeled species that migrate slower than the substrate DNA.
Figure 1.
Covalent trapping of the DNA–protein crosslink formed between Top1 and a double-stranded DNA oligonucleotide containing a CrA-PdG adduct. (A) Shown is the sequence of the 22-bp oligonucleotide used in the study. The position of the CrA-PdG adduct is indicated by a box and asterisk on the lower strand. The positions of the high affinity Top1 cleavage site and the 32P-radiolabel are also shown. (B) Both single-stranded (ss) and double-stranded (ds) control (WT) or R/S-CrA-PdG (R/S) adducted oligonucleotides were incubated at 25°C for 15 min in the presence or absence of Top1 (250 nM). NaCNBH3 (50 mM) was added to reactions as indicated. (C) Reactions were carried out exactly as described in (B) with the double-stranded R/S-CrA-PdG (R/S) adducted oligonucleotide in the presence of increasing amounts of Top1 (25, 50, 100, 250 and 500 nM) and the presence of NaCNBH3 (50 mM). All reaction products were resolved through SDS–PAGE (4–20%) analysis. Arrows indicate the positions of the DNA–Top1 crosslink and the free DNA substrate.
The data in Figure 1B show that Top1 did not yield detectable crosslinks with the DNA substrates lacking the CrA-PdG adduct (lanes 1–6). The single-stranded CrA-PdG-adducted DNA substrate also did not generate DNA–Top1 crosslinks (Figure 1B, lanes 7–9), which is to be expected given that recent studies have shown the CrA-PdG adduct exists in the nonreactive ring-closed structure in single-stranded DNA (26). In contrast, the formation of the reactive ring-open conformation of the CrA-PdG adduct occurs in the duplex DNA substrate and is thus able to form DNA–Top1 crosslinks, as evidenced by the appearance of a new band having retarded mobility in Figure 1B (lane 12). The band ascribed to the DNA–Top1 crosslink can only be observed after reduction of the complex by NaCNBH3 treatment, suggesting that Top1 forms a transient Schiff base intermediate with the CrA-PdG adduct. In addition, the yield of the band corresponding to the DNA–Top1 crosslink was also dependent on the amount of Top1 in the reaction (Figure 1C). These results support as well as extend previous studies, which observed that the ring opening of the CrA-PdG adduct to form a reactive aldehyde was required for crosslink formation (20,23).
The DNA–Top1 crosslinking capacity of CrA-PdG adducts in duplex DNA is independent of stereochemistry
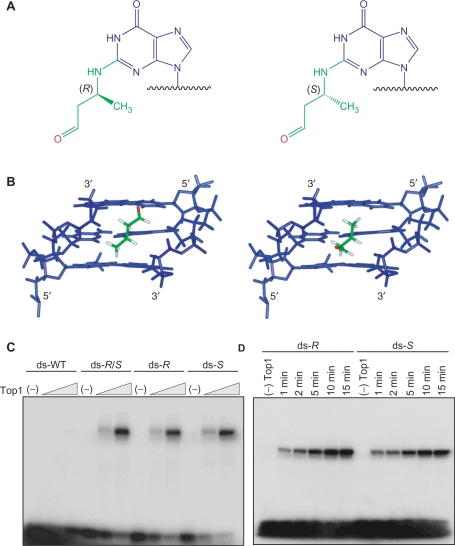
Recent NMR studies of the individual R- and S-CrA-PdG adducts have shown that the ring open conformation of the CrA-PdG adduct in duplex DNA is located in the minor groove with the aldehyde functionality oriented in the 5′- and 3′-direction, respectively (Figure 2A and 2B) (26). The initial borohydride trapping studies were performed using DNA substrates containing a mixture of the R- and S-CrA-PdG diastereomers. In order to investigate the possible stereospecific formation of DNA–Top1 crosslinks by CrA-PdG adducts, the R- and S-CrA-PdG adducted oligonucleotides were separated and independently subjected to the borohydride trapping assay. As shown in Figure 2C, no significant differences were detected in the formation of DNA–Top1 crosslinks between the mixture and the DNA substrates containing the individual diastereomers. In addition, similar results were obtained when comparing the rates of crosslink formation for the R- and S-CrA-PdG adducted DNA substrates (Figure 2D). These results also corroborate previous studies, which demonstrated the formation of DNA–peptide crosslinks were independent of the CrA-PdG adduct stereochemistry (20).
Figure 2.
DNA–Top1 crosslinking capacity of the isolated diastereomeric R- and S-CrA-PdG adducts. (A) Chemical structures of the crotonaldehyde derived R- and S-CrA-PdG adducts. (B) Molecular model and NMR structrure of the R- and S-CrA-PdG adducts, respectively, showing the orientations of the reactive aldehyde moieties and α-carbon methyl groups within the minor groove of the DNA. DNA shown in blue; CrA portion of adduct shown in green; oxygen from the aldehyde moiety shown in red. (C) Double-stranded (ds) control (WT), R/S-, R-, or S-CrA-PdG adducted oligonucleotides were incubated with increasing amounts of Top1 (50 and 250 nM) at 25°C for 15 min in the presence of NaCNBH3 (50 mM). (D) Double-stranded R- and S-CrA-PdG adducted oligonucleotides were incubated with Top1 (250 nM) at 25°C for 1, 2, 5, 10 and 15 min in the presence of NaCNBH3 (50 mM). All reaction products were resolved through SDS–PAGE (4–20%) analysis.
The presence of a CrA-PdG adduct redirects the Top1-mediated DNA cleavage
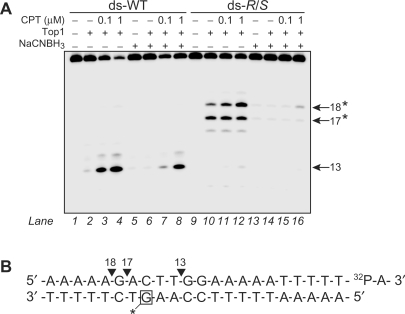
The consequences of the CrA-PdG adduct on Top1-mediated DNA cleavage were examined. Since Top1 becomes covalently linked to the 3′-DNA end, the DNA was labeled at the 3′-terminus with [α-32P]ddATP in order to unambiguously assign the positions of the cleavage products generated by Top1 (shown in Figure 3B). As previously demonstrated (32,33), Top1 did not produce detectable cleavage in the unmodified DNA substrate (Figure 3A, lane 2); however, in the presence of the Top1 inhibitor camptothecin (CPT), the expected 13-mer cleavage product was observed (Figure 3A, lanes 3 and 4) (32). When the CrA-PdG adduct was present at the −3(G) position on the nonscissile strand, an alteration in the Top1-mediated DNA cleavage was detected. Specifically, two new Top1 cleavage products, which corresponded to 17-mer and 18-mer fragments labeled at the 3′-end (see sequence in Figure 3B), were produced that did not require CPT (Figure 3A, lane 10). The CrA-PdG adduct also completely suppressed the expected CPT-induced 13-mer cleavage product (Figure 3A, lanes 11 and 12). Of the two new Top1 cleavage products generated by the CrA-PdG adduct, only the 18-mer fragment was enhanced upon the addition of CPT (Figure 3A, lanes 11 and 12). Similar results were obtained with the DNA substrates containing the individual R- and S-CrA-PdG diastereomers (data not shown).
Figure 3.
Induction of new Top1-mediated DNA cleavage sites on the DNA strand opposite to the R/S-CrA-PdG adduct. (A) Double-stranded (ds) control (WT) or R/S-CrA-PdG (R/S) adducted oligonucleotides were 3′-end labeled on the scissile strand (upper) with α-32P ddATP (shown as 32P-A in B). DNA was then reacted with Top1 and camptothecin in the presence or absence of NaCNBH3 (50 mM). The size of the fragments generated by Top1 is indicated to the right. Asterisks indicate the positions of new Top1-mediated DNA cleavage sites induced by the CrA-PdG adduct. (B) Summary of Top1-mediated DNA cleavage sites from (A).
The cleavage reactions were also carried out under reducing conditions so as to monitor the effects of DNA–Top1 crosslink formation by CrA-PdG adducts on Top1-mediated DNA cleavage. The comparison of lanes 1–4 and lanes 5–8 in Figure 3A demonstrates that the presence of NaCNBH3 results in only a minor reduction of the CPT-induced Top1 cleavage product (site 13) in the unmodified DNA substrate. In contrast, NaCNBH3 completely suppresses the Top1-mediated DNA cleavage induced in the CrA-PdG adducted DNA substrate (Figure 3A, compare lanes 9–12 and lanes 13–16). It is likely that this inhibition is attributable to the formation of a stable DNA–protein crosslink between Top1 and the ring open form of the CrA-PdG adduct (previously shown in Figure 1A). Taken together, the results presented in Figure 3 demonstrate that CrA-PdG adduct formation can both inhibit Top1-mediated DNA cleavage at adduct sites and induce the formation of new high-affinity Top1 cleavage sites in the near vicinity. Moreover, CrA-PdG adducts are able to sequester the DNA cleaving activity of Top1 by trapping the enzyme in a cleavage incompetent complex via the formation of a stable DNA–Top1 crosslink.
Stability of the CrA-PdG adduct-induced Top1 cleavage complexes
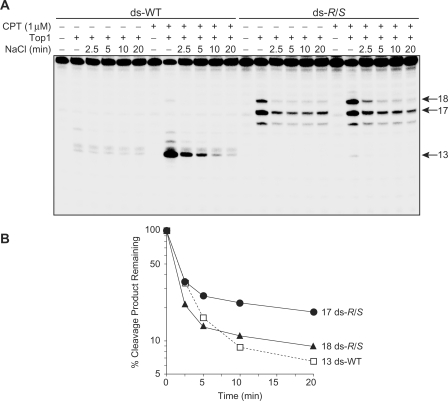
To further characterize the Top1 cleavage complexes induced by the CrA-PdG adduct, we investigated whether these cleavage complexes were reversible using a salt reversal assay. Indeed, one characteristic of the Top1 cleavage reaction is that in the presence of high salt concentrations the cleavage–religation equilibrium is shifted toward religation (34). Religation of the CPT-induced DNA cleavage site (13-mer) in the unmodified DNA substrate was previously demonstrated to be complete within 10 min following the addition of 0.35 M NaCl (Figure 4A and B) (35). More interestingly, whereas the CPT-sensitive cleavage site at position 18 in the DNA substrate containing the CrA-PdG adduct had similar reversal kinetics to that of the complexes trapped by CPT in the unmodified DNA substrate (site 13), the cleavage at the CPT-independent site at position 17, albeit partially reversible, was much more stable (Figure 4A and B). This slowing of the religation step generated by the CrA-PdG adduct is possibly caused by distortion of the DNA structure and consequent inhibition of the nucleophilic attack on the tyrosylphophodiester bond by the cleaved 5′-hydroxyl DNA strand (see Discussion section).
Figure 4.
Reversibility of the CrA-PdG adduct-induced Top1 cleavage complexes. (A) Double-stranded (ds) control (WT) or R/S-CrA-PdG (R/S) adducted oligonucleotides were reacted with Top1 in the presence or absence of 1 µM camtothecin at 25°C for 20 min. DNA cleavage was reversed by adding 0.35 M NaCl and monitored over time. (B) Reactions carried out as in (A) were quantified and the percentages of Top1-mediated cleavage products remaining after salt reversal are represented as semi-log plots.
Detection of DNA–Top1 covalent complexes by crotonaldehyde in vivo
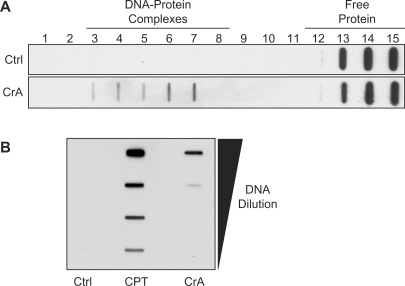
Finally, to investigate whether Top1 can be trapped by CrA-PdG adducts in cellular DNA, we employed the immunocomplex of enzyme (ICE) bioassay. This technique permits the detection of Top1 covalently linked to genomic DNA through the used of cesium chloride (CsCl) gradient fractionation and subsequent western blot analysis (27,28). MCF7 cells were incubated with 100 µM of crotonaldehyde for 6 h, and the cell extracts were fractionated by centrifugation with a CsCl gradient. In untreated control samples, Top1 was found exclusively in the upper fractions (12–15) of the CsCl gradient as free protein (Figure 5A). In contrast, in the cells treated with crotonaldehyde, Top1 was found in the upper fractions as well as in the fractions near the bottom of the gradient or the DNA-containing fractions (3–7) (Figure 5A), which is indicative of the formation of DNA–Top1 covalent complexes. DNA–Top1 complexes could also be detected as early as 1 h after treatment with 10 mM crotonaldehyde (Figure 5B). As a positive control, CPT was shown to trap DNA–Top1 complexes as expected (Figure 5B). These results suggests that crotonaldehyde can produce stable cellular DNA adducts, which can subsequently form DNA–Top1 covalent complexes in vivo, albeit much less efficiently than CPT.
Figure 5.
Detection of DNA–Top1 covalent complexes by crotonaldehyde in cultured mammalian cells. (A) MCF7 cells were treated with 100 μM of crotonaldehyde (CrA) for 6 h. Equal numbers of cells were lyzed and separated by CsCl gradients centrifugation. Fractions were collected from the bottom of the tubes (numbered 1–15). The presence of Top1 was assayed in each of the gradient fractions by western blotting with a Top1 monoclonal antibody. Cellular DNA was contained in fractions 3–7, which corresponds to the DNA–Top1 complexes. (B) MCF7 cells were treated with 0.1 μM CPT or 10 mM CrA for 1 h. All of the DNA-containing fractions (3–7) were collected from the CsCl gradient, pooled together, serial diluted and then blotted with the Top1 monoclonal antibody.
DISCUSSION
Crotonaldehyde is a bifunctional electrophile. An interesting feature of crotonaldehyde is its ability to form primary DNA adducts that are capable of participating in secondary reactions with cellular macromolecules. In this article, we report the induction of DNA–Top1 crosslinks both directly and indirectly by crotonaldehyde-derived DNA adducts. In addition, the formation of stable DNA–Top1 covalent complexes is observed in cells treated with crotonaldehyde (Figure 5). Thus, the results reported here suggest that these types of DNA lesions, if unrepaired, may contribute to some degree to the mutagenic and carcinogenic consequences of crotonaldehyde exposure.
Initial studies by Kurtz and Lloyd (23) established the predisposition of CrA-PdG adducts to take part in secondary reactions with proteins by demonstrating the formation of crosslinks between CrA-PdG adducts and the tetrapeptide KWKK. However, further analysis revealed that the bond formed between the CrA-PdG adduct and the peptide occurred at the terminal amino group and not the ε-amino groups of the lysine residues. The rationalization for this selectivity was based on the intrinsic lower pKa of the α-amine relative to the ε-amino groups. Nevertheless, the specific microenvironment in enzyme active sites can decrease the pKa of lysyl side chains and effectively render them more nucleophilic. The related acrolein-dG adduct has been reductively trapped with histones and the base-excision repair protein T4 pyrimidine dimer glycosylase (T4 PDG) (36). Additionally, DNA–protein crosslinks between the aldehydic form of an abasic site with a catalytic N-terminal threonine of T4 PDG have also been trapped by NaBCNH3 treatment (37,38). In the case of Top1, it has recently been shown that pyridoxal 5′-phosphate can act as a catalytic inhibitor of Top1 through the formation of a Schiff base between its aldehydic moiety and the ɛ-amino group of a critical active site lysine residue (39,40). Indeed, examination of the available crystal structures of Top1 bound to DNA reveals the presence of several potentially nucleophilic lysine sidechains within the DNA-binding pocket capable of forming Schiff base-mediated crosslinks between CrA-PdG adducts and Top1 (41). It is likely that there are several different populations of crosslinking residues in Top1. Moreover, besides the ɛ-amino group of lysine, additional reactive amino acid residues may include the sulfhydryl group of cysteine and the imidazole group of histidine.
The propensity of primary CrA-PdG adducts to participate in secondary reactions is derived from its ability to undergo a ring-opening reaction from a cyclic, ring-closed form in single-stranded DNA or nucleosides into a reactive ring-open aldehydic moiety when paired opposite dC in a duplex environment (20). Similar ring-opening reactions have been observed with the related adducts γ-OH-PdG and M1dG (42,43). DNA–Top1 crosslinks only appeared in the presence of the CrA-PdG adducted duplex DNA substrate (Figure 1B), which indirectly confirms this structural transformation by CrA-PdG adducts in duplex DNA. That observation is consistent with previous findings that demonstrated a markedly slower rate of DNA–peptide crosslink formation for a single-stranded CrA-PdG adducted DNA substrate compared to a duplex substrate (23). Thus, it is plausible to envisage that CrA-PdG adducts exist largely as the ring-open form in vivo by reason of the native structure of the DNA within the cell is double stranded.
CrA-PdG adducts can also redirect the DNA-cleaving activity of Top1 to alternative sites (Figure 6B). In this study, the presence of a single CrA-PdG adduct at the −3(G) position on the nonscissile strand of the DNA substrate (Figure 1A) completely suppresses the cleavage of the high-affinity CPT-dependent Top1 cleavage site (Figure 3A). This result may partly be explained by steric clashes between the presence of adducts in the DNA minor groove and the binding of Top1 (32,33,35). Recent NMR studies have shown that the CrA-PdG adduct in the ring-open form occupies the minor groove and extends toward the 5′-end of the adducted strand, overlaying roughly two adjacent base pairs (26), which correspond to A(−2) and A(−1) in the Top1 substrate used in these studies (Figure 1A). This minor groove interference by the CrA-PdG adduct thus blocks specific interactions needed for proper nucleophilic attack by the catalytic tyrosine of Top1. A similar site-specific inhibition has been observed for other minor groove adducts derived from polycyclic aromatic diol epoxides or acetaldehyde adducts (32,33,35). Alternatively, the CrA-PdG adduct may restrict CPT from accessing and stabilizing the Top1 cleavage complex, and as a result inhibit the formation of the observed DNA cleavage product (13-mer).
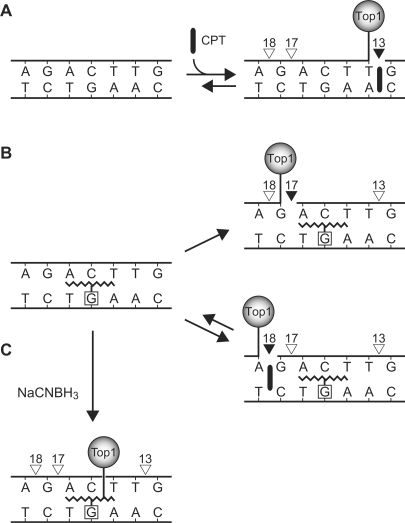
Figure 6.
Summary of DNA–Top1 complex formation by (A) unmodified DNA substrate and (B–C) CrA-PdG adducted DNA substrate. Boxed nucleotide, site of adduct; open triangles, no cleavage; closed triangles, Top1 cleavage site. (A) In the unmodified substrate, Top1 cleavage complexes are trapped by CPT at site 13. Sites 17 and 18 are undetectable. (B) In the CrA-PdG adducted substrate, site 13 is suppressed and two cleavage sites are induced; site 17 is CPT-independent and site 18 is trapped by CPT. (C) Upon reduction by NaCNBH3, Top1 forms an indirect crosslink with the DNA via the CrA-PdG adduct and the formation of Top1 cleavage complexes is inhibited.
In addition to inhibiting the preferential Top1 cleavage site, the CrA-PdG adduct also induced the formation of two new Top1-mediate cleavage sites (17- and 18-mer) in the absence of CPT upstream of the preferential cleavage site (13-mer) (Figures 3A and 6). We believe that the formation of these novel DNA-cleavage sites is due to the interference of the CrA-PdG adduct with the Top1 binding at the preferential site (13-mer). On the basis of our previous studies, we presume that Top1 can bind and interact at several different positions on the oligonucleotides (32,33,35), and cleavage at the alternative sites only becomes kinetically favored when cleavage at the preferential site is inhibited, which is the case when the CrA-PdG adduct is present. Interestingly, both the 18-mer cleavage product in the adducted DNA substrate and the expected 13-mer cleavage product in the unmodified substrate contain similar features. First, both cleavage sites are enhanced by the addition of CPT. Second, each contain a guanine immediately 3′ to the cleaved bond (Figures 3B and 6), which is consistent with previous studies showing that CPT predominantly traps Top1 cleavage sites of this sequence context (44). Lastly, both have comparable religation kinetics (Figure 4B). Of greater interest is the auxiliary cleavage site (17-mer) that is induced by the CrA-PdG adduct. This cleavage site is unaffected by CPT treatment and exhibits slowed religation kinetics in comparison (Figure 4B) to the previously mentioned Top1 cleavage sites. The first characteristic is almost certainly due to the identity of the DNA bases around the cleavage site, whereas the second is most likely due to the distortion of the DNA structure after Top1 has cleaved the DNA backbone. Overall, our interpretation is that the factors effecting the induction of the two different populations of Top1 cleavage are based on: (i) geometrical changes in the DNA caused by the CrA-PdG adduct and (ii) on the selective trapping by CPT of the site bearing a guanine +1 (site producing the 18-mer) (44), which provides preferential binding for the drug (Figure 6B).
The presence of a reducing agent in the cleavage reactions presented here results in complete inhibition of the CrA-PdG induced cleavage sites, which suggests that the induction of Top1 cleavage complexes by CrA-PdG adducts and formation of a stable bona fide DNA–Top1 crosslinks at the site of the CrA-PdG adduct are mutually exclusive (Figure 6B and C). In other words, we propose that the Top1 cannot be bound to the DNA more than once in the presence of the CrA-PdG adduct. More specifically, Top1 is either bound at the alternative cleavage site on the DNA through its active site tyrosine to form a cleavage complex or bound to the CrA-PdG adduct through a given lysine residue to form a DNA–protein crosslink.
Alterations in Top1 enzymatic activities are not limited to the occurrence of CrA-PdG adducts. Other similar covalent DNA adducts that lie within the minor groove, such as the benzo[a]pyrene or benzo[c]phenanthrene diol epoxide (BaP and BcPh, respectively) adducts (32,33), or the acetaldehyde adduct (35), have been shown to both enhance as well as suppress Top1 cleavage depending on the position of the adduct relative to the cleavage site. Analogous to CrA-PdG adducts, BaP and BcPh adducts are dominant inducers of new Top1 cleavages (32,33). In addition, the noncovalent minor groove binding drug, ecteinascidin 743, can also influence the catalytic cycle of Top1 by inhibiting the DNA religation step (i.e. poisoning Top1) (45). Together with the present results obtained with the CrA-PdG adduct, these studies highlight the importance of the contacts between the DNA minor groove and the catalytic active site of Top1.
Although we are unable to discriminate between what types of DNA–Top1 covalent complexes are induced by crotonaldehyde in mammalian cells (Figure 5), both lesions have the potential to be damaging in vivo. Of interest is the recent study showing that crotonaldehyde treated mice exhibit an increase in the frequency of chromosomal aberrations (46). These cytogenetic effects of crotonaldehyde are reminiscent of the chromosomal abnormalities observed after the stabilization of DNA–Top1 cleavage complexes with Top1 poisons, such as CPT. On the basis of the results of the present investigation, it might be concluded that the harmful effects of crotonaldehyde exposure may to a degree stem from the direct and indirect formation of DNA–Top1 covalent complexes, which can lead to DNA damage if unrepaired.
ACKNOWLEDGEMENTS
The National Institutes of Health supported part of this work through a program project grant (ES05355) and center grant (ES00267) to M.P.S. and C.J.R. The work done in the Laboratory of Molecular Pharmacology was supported by the NIH Intramural Program, Center for Cancer Research, National Cancer Institute. Funding to pay the Open Access publication charges for this article was provided by the Center for Cancer Research, NCI, NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Dry Cleaning, Some Chlorinated Solvents and Other Industrial Chemical. Lyon, France: International Agency for Research on Cancer; 1995. International Agency for Research on Cancer. [PMC free article] [PubMed] [Google Scholar]
- 2.Chung FL, Chen HJ, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–2111. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 3.Wang MY, Chung FL, Hecht SS. Identification of crotonaldehyde as a hepatic microsomal metabolite formed by α-hydroxylation of the carcinogen N-nitrosopyrrolidine. Chem. Res. Toxicol. 1988;1:28–31. doi: 10.1021/tx00001a005. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS, Upadhyaya P, Wang M. Reactions of α-acetoxy-N-nitrosopyrrolidine and crotonaldehyde with DNA. IARC Sci. Publ. 1999;125:147–154. [PubMed] [Google Scholar]
- 5.Neudecker T, Eder E, Deininger C, Henschler D. Crotonaldehyde is mutagenic in Salmonella typhimurium TA100. Environ. Mol. Mutagen. 1989;14:146–148. doi: 10.1002/em.2850140303. [DOI] [PubMed] [Google Scholar]
- 6.Neudecker T, Lutz D, Eder E, Henschler D. Crotonaldehyde is mutagenic in a modified Salmonella typhimurium mutagenicity testing system. Mutat. Res. 1981;91:27–31. doi: 10.1016/0165-7992(81)90065-8. [DOI] [PubMed] [Google Scholar]
- 7.Czerny C, Eder E, Runger TM. Genotoxicity and mutagenicity of the α, β-unsaturated carbonyl compound crotonaldehyde (butenal) on a plasmid shuttle vector. Mutat. Res. 1998;407:125–134. doi: 10.1016/s0921-8777(97)00069-4. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes PH, Kanuri M, Nechev LV, Harris TM, Lloyd RS. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ. Mol. Mutagen. 2005;45:455–459. doi: 10.1002/em.20117. [DOI] [PubMed] [Google Scholar]
- 9.Chung FL, Tanaka T, Hecht SS. Induction of liver tumors in F344 rats by crotonaldehyde. Cancer Res. 1986;46:1285–1289. [PubMed] [Google Scholar]
- 10.Chung FL, Hecht SS. Formation of cyclic 1,N2-adducts by reaction of deoxyguanosine with α-acetoxy-N-nitrosopyrrolidine, 4-(carbethoxynitrosamino)butanal, or crotonaldehyde. Cancer Res. 1983;43:1230–1235. [PubMed] [Google Scholar]
- 11.Chung FL, Young R, Hecht SS. Formation of cyclic 1, N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 12.Winter CK, Segall HJ, Haddon WF. Formation of cyclic adducts of deoxyguanosine with the aldehydes trans-4-hydroxy-2-hexenal and trans-4-hydroxy-2-nonenal in vitro. Cancer Res. 1986;46:5682–5686. [PubMed] [Google Scholar]
- 13.Budiawan, Eder E. Detection of 1,N2-propanodeoxyguanosine adducts in DNA of Fischer 344 rats by an adapted 32P-post-labeling technique after per os application of crotonaldehyde. Carcinogenesis. 2000;21:1191–1196. [PubMed] [Google Scholar]
- 14.Chung FL, Nath RG, Nagao M, Nishikawa A, Zhou GD, Randerath K. Endogenous formation and significance of 1,N2-propanodeoxyguanosine adducts. Mutat. Res. 1999;424:71–81. doi: 10.1016/s0027-5107(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 15.Nath RG, Chung FL. Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Natl Acad. Sci. USA. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nath RG, Ocando JE, Chung FL. Detection of 1,N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- 17.Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1,N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lao Y, Hecht SS. Synthesis and properties of an acetaldehyde-derived oligonucleotide interstrand cross-link. Chem. Res. Toxicol. 2005;18:711–721. doi: 10.1021/tx0497292. [DOI] [PubMed] [Google Scholar]
- 19.Nechev LV, Kozekov I, Harris CM, Harris TM. Stereospecific synthesis of oligonucleotides containing crotonaldehyde adducts of deoxyguanosine. Chem. Res. Toxicol. 2001;14:1506–1512. doi: 10.1021/tx0100690. [DOI] [PubMed] [Google Scholar]
- 20.Cho YJ, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, et al. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 2000;13:1149–1157. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- 23.Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 24.Kuykendall JR, Bogdanffy MS. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 1992;283:131–136. doi: 10.1016/0165-7992(92)90145-8. [DOI] [PubMed] [Google Scholar]
- 25.Cho YJ, Kozekov ID, Harris TM, Rizzo CJ, Stone MP. Stereochemistry modulates the stability of reduced interstrand cross-links arising from R- and S-α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine in the 5′-CpG-3′ DNA sequence. Biochemistry. 2007;46:2608–2621. doi: 10.1021/bi061381h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho YJ, Wang H, Kozekov ID, Kozekova A, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Rizzo CJ, et al. Orientation of the crotonaldehyde-derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA adduct hinders interstrand cross-link formation in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 2006;19:1019–1029. doi: 10.1021/tx0600604. [DOI] [PubMed] [Google Scholar]
- 27.Shaw JL, Blanco J, Mueller GC. A simple procedure for isolation of DNA, RNA and protein fractions from cultured animal cells. Anal. Biochem. 1975;65:125–131. doi: 10.1016/0003-2697(75)90498-4. [DOI] [PubMed] [Google Scholar]
- 28.Takagi K, Dexheimer TS, Redon C, Sordet O, Agama K, Lavielle G, Pierre A, Bates SE, Pommier Y. Novel E-ring camptothecin keto analogues (S38809 and S39625) are stable, potent, and selective topoisomerase I inhibitors without being substrates of drug efflux transporters. Mol. Cancer. Ther. 2007;6:3229–3238. doi: 10.1158/1535-7163.MCT-07-0441. [DOI] [PubMed] [Google Scholar]
- 29.Bonven BJ, Gocke E, Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985;41:541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- 30.Pourquier P, Ueng LM, Fertala J, Wang D, Park HJ, Essigmann JM, Bjornsti MA, Pommier Y. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. 7, 8-dihydro-8-oxoguanine and 5-hydroxycytosine. J. Biol. Chem. 1999;274:8516–8523. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
- 31.Pourquier P, Ueng LM, Kohlhagen G, Mazumder A, Gupta M, Kohn KW, Pommier Y. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J. Biol. Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 32.Pommier Y, Kohlhagen G, Pourquier P, Sayer JM, Kroth H, Jerina DM. Benzo[a]pyrene diol epoxide adducts in DNA are potent suppressors of a normal topoisomerase I cleavage site and powerful inducers of other topoisomerase I cleavages. Proc. Natl Acad. Sci. USA. 2000;97:2040–2045. doi: 10.1073/pnas.040397497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pommier Y, Kohlhagen G, Laco GS, Kroth H, Sayer JM, Jerina DM. Different effects on human topoisomerase I by minor groove and intercalated deoxyguanonsine adducts derived from two polycyclic aromatic hydrocarbon diol epoxides at or near a normal cleavage site. J. Biol. Chem. 2002;277:13666–13672. doi: 10.1074/jbc.M200209200. [DOI] [PubMed] [Google Scholar]
- 34.Pommier Y, Jenkins J, Kohlhagen G, Leteurtre F. DNA recombinase activity of eukaryotic DNA topoisomerase I; effects of camptothecin and other inhibitors. Mutat. Res. 1995;337:135–145. doi: 10.1016/0921-8777(95)00019-g. [DOI] [PubMed] [Google Scholar]
- 35.Antony S, Theruvathu JA, Brooks PJ, Lesher DT, Redinbo M, Pommier Y. Enhancement of camptothecin-induced topoisomerase I cleavage complexes by the acetaldehyde adduct N2-ethyl-2'-deoxyguanosine. Nucleic Acids Res. 2004;32:5685–5692. doi: 10.1093/nar/gkh902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 37.Golan G, Zharkov DO, Grollman AP, Dodson ML, McCullough AK, Lloyd RS, Shoham G. Structure of T4 pyrimidine dimer glycosylase in a reduced imine covalent complex with abasic site-containing DNA. J. Mol. Biol. 2006;362:241–258. doi: 10.1016/j.jmb.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 38.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl Acad. Sci. USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christmann-Franck S, Fermandjian S, Mirambeau G, Der Garabedian PA. Molecular modelling studies on the interactions of human DNA topoisomerase IB with pyridoxal-compounds. Biochimie. 2007;89:468–473. doi: 10.1016/j.biochi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Vermeersch JJ, Christmann-Franck S, Karabashyan LV, Fermandjian S, Mirambeau G, Der Garabedian PA. Pyridoxal 5'-phosphate inactivates DNA topoisomerase IB by modifying the lysine general acid. Nucleic Acids Res. 2004;32:5649–5657. doi: 10.1093/nar/gkh897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 42.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2′-deoxyguanosine. J. Biol. Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 43.Mao H, Schnetz-Boutaud NC, Weisenseel JP, Marnett LJ, Stone MP. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl Acad. Sci. USA. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaxel C, Capranico G, Kerrigan D, Kohn KW, Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem. 1991;266:20418–20423. [PubMed] [Google Scholar]
- 45.Takebayashi Y, Pourquier P, Yoshida A, Kohlhagen G, Pommier Y. Poisoning of human DNA topoisomerase I by ecteinascidin 743, an anticancer drug that selectively alkylates DNA in the minor groove. Proc. Natl Acad. Sci. USA. 1999;96:7196–7201. doi: 10.1073/pnas.96.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jha AM, Singh AC, Sinha U, Kumar M. Genotoxicity of crotonaldehyde in the bone marrow and germ cells of laboratory mice. Mutat. Res. 2007;632:69–77. doi: 10.1016/j.mrgentox.2007.04.008. [DOI] [PubMed] [Google Scholar]