Abstract
Changing a highly conserved amino acid in motif A of any of the four yeast family B DNA polymerases, DNA polymerase α, δ, ɛ or ζ, results in yeast strains with elevated mutation rates. In order to better understand this phenotype, we have performed structure–function studies of homologous mutants of RB69 DNA polymerase (RB69 pol), a structural model for family B members. When Leu415 in RB69 pol is replaced with phenylalanine or glycine, the mutant polymerases retain high-catalytic efficiency for correct nucleotide incorporation, yet have increased error rates due to increased misinsertion, increased mismatch extension and inefficient proofreading. The Leu415Phe mutant also has increased dNTP insertion efficiency opposite a template 8-oxoG and opposite an abasic site. The 2.5 Å crystal structure of a ternary complex of RB69 L415F pol with a correctly base-paired incoming dTTP reveals that the phenylalanine ring is accommodated within a cavity seen in the wild-type enzyme, without steric clash or major change in active site geometry, consistent with retention of high-catalytic efficiency for correct incorporation. In addition, slight structural differences were observed that could be relevant to the reduced fidelity of L415F RB69 pol.
INTRODUCTION
The study of DNA polymerases with amino acid replacements introduced on the basis of structural information has been useful for understanding polymerase catalytic mechanism, their abilities to discriminate against mismatches, rNTPs and nonnatural dNTPs, and their abilities to use (or not) various primer templates, including those containing lesions. Among numerous studies of mutant enzymes in polymerase families A, B, X, Y and RT, replacements for conserved amino acids often result in reduced catalytic activity. In other, less common instances, amino acid replacements for conserved residues do not strongly reduce polymerization activity but do alter substrate specificity, sometimes in a highly informative manner. Examples under study in several laboratories involve B family polymerases, whose members include the polymerases for bacteriophage T4 and its close relative RB69, and four eukaryotic enzymes, pol α, pol δ, pol ɛ and pol ζ. The active sites of these enzymes, as well as those of members of other polymerase families, are comprised of highly conserved amino acid sequence motifs designated A, B and C. Motif A contains the conserved hydrophobic residue of interest in this study (see alignments in (3,7,9,10–11). This is a methionine in yeast pol ɛ and a leucine in the other five polymerases mentioned above. In the crystal structure of RB69 pol (1), this leucine (Leu415) is immediately adjacent to invariant Tyr416, which interacts with the sugar of the incoming dNTP in the polymerase active site and has an important role in preventing incorporation of NTPs (2).
The consequences of replacing the conserved leucine/methionine with other amino acids have now been examined with five different B family members. In a seminal study of T4 pol, substituting Leu412 with methionine yielded bacteriophage that replicated efficiently but had an elevated mutation rate (3). Subsequent biochemical studies indicated that this mutator effect results from inefficient proofreading due to defective movement of mismatches generated by the polymerase into the exonuclease active site (4,5). In yeast pol α, the homologous Leu868 was replaced with several different amino acids (6,7). Particularly informative were the L868F and L868M mutants, which have normal polymerase activity, increased mismatch extension efficiency (L868M) and reduced DNA synthesis fidelity in vitro. Yeast strains harboring L868F and L868M pol α alleles had elevated spontaneous mutation rates that were enhanced by a defect in mismatch repair, indicating reduced replication fidelity in vivo. In addition, the mutator effect of the pol α L868M mutant allele was strongly elevated by inactivating the 3′ exonuclease activity of pol δ (6), implying that the 3′ exonuclease activity of pol δ may proofread errors made by pol α, a type of extrinsic proofreading (8).
Three different studies (9–11) have shown that replacing Leu612 in yeast pol δ with other amino acids also yields yeast strains with mutator phenotypes that were further increased when mismatch repair is inactivated, again suggesting reduced replication fidelity in vivo. Indeed, yeast L612M pol δ has reduced fidelity in vitro, despite retaining 3′ exonuclease activity (10). This suggests that, like L412M T4 pol (5), L612M pol δ sometimes fails to partition mismatched termini to the 3′ exonuclease active site. By analogy to the promiscuous mismatch extension ability of L868M pol α (6), the partitioning defect of L612M pol δ could also be due to promiscuous mismatch extension. Supporting and extending this interpretation is the fact that M644F (12) and M644G (13) mutants of yeast pol ɛ share similar biochemical properties with those seen for the T4 pol, pol α and pol δ mutants, i.e. robust catalytic efficiency, reduced fidelity, promiscuous mismatch extension and reduced proofreading efficiency.
Two sets of observations indicate that replacing the conserved leucine or methionine in motif A of B family polymerases with other amino acids may also affect synthesis when copying damaged templates. Firstly, the corresponding motif A residue in Y family pol η is a phenylalanine (Phe34). Because pol η can efficiently bypass a cis-syn cyclobutane pyrimidine dimer (CPD), the translesion synthesis (TLS) ability of a L868F pol α mutant was examined with a CPD, a 6-4 UV photoproduct and an abasic site. L868F pol α bypassed all three lesions more efficiently than did wild-type pol α, and conversely, the Phe34 to Leu replacement in pol η decreased bypass (7). Second, replacing Leu979 in motif A of yeast pol ζ, the B family polymerase that performs mutagenic DNA synthesis in vivo, with any of six different amino acids resulted in yeast strains with UV-induced mutant frequencies that were higher than that of strain with wild-type pol ζ (14). This suggests the L979F pol ζ has reduced fidelity during bypass of UV photoproducts. Interestingly, while four of the replacements resulted in reduced survival following UV irradiation, the rev3-L979F and rev3-L979M strains had normal survival, suggesting that they retained robust pol ζ catalytic activity.
The above studies indicate that the conserved Leu/Met in motif A is critical for substrate discrimination and possibly the TLS capacity of B family polymerases. Moreover, these mutator polymerases are proving to be very useful for probing biological functions of polymerases in vivo, including the roles of pol ɛ (13) and pol δ (15) in leading and lagging strand DNA replication and the contribution of pol δ-dependent replication errors to cancer susceptibility in mice (16). Thus, it is of interest to obtain a better understanding of the nucleotide selectivity, mismatch extension, proofreading efficiency and TLS efficiency and fidelity of these B family enzymes. This is currently hampered somewhat by the lack of structural information on pol α, pol δ, pol ɛ or pol ζ. Fortunately, however, a number of studies demonstrated that RB69 pol serves as an excellent structural surrogate for its eukaryotic homologs (1,17,18). On that basis, here we construct and characterize the biochemical properties of RB69 pol mutants containing Phe and Gly replacements for Leu415, and then present the crystal structure of L415F RB69 pol with a dideoxy-terminated primer template and correctly base-paired incoming dTTP.
MATERIALS AND METHODS
Constructing and purifying RB69 pol mutants
RB69 polymerase mutants were constructed by the QuickChange™ site-directed mutagenesis method (Stratagene, La Jolla, CA, USA) using the plasmid pCW19R (pRB.43Pol+ Exo+) for expressing wild-type RB69 pol and pCW50 (pRB.43Pol+Exo−) for expressing exonuclease-deficient (D222A/D327A) RB69 pol (kindly provided by John W. Drake, NIEHS) (19). Plasmids containing the desired changes and no other changes (confirmed by sequence analysis) were introduced into BL21(DE3) RIL cells (Stratagene) and expressed overnight at 15°C. Frozen cells from 1.5 l cultures were suspended in 50 mM Tris–HCl (pH 7.8), 5% glycerol, 500 mM NaCl, 1 mM DTT, 1 mM EDTA and 1 mM PMSF. The suspensions were sonicated for 2 min, centrifuged and clear lysates were passed through a 5 ml Q column (GE Healthcare Biosciences, Piscataway, NJ, USA) to remove DNA. The flow through was diluted 6-fold with 50 mM Tris–HCl (pH 7.8), 5% glycerol and 1 mM DTT to reduce the salt concentration, and loaded onto a second 5 ml Q column equilibrated with 50 mM Tris–HCl (pH 7.8), 5% glycerol and 1 mM DTT, and eluted with the same buffer plus 200 mM NaCl. Peak fractions were concentrated and the buffer was exchanged with 50 mM sodium phosphate (pH 6.8), 10% glycerol and 3 mM DTT. The sample was loaded onto a 5 ml heparin column (GE Healthcare Biosciences) and eluted with a gradient of 100 mM to 1 M NaCl. Peak fractions were dialyzed with 1 l of buffer [25 mM K2HPO4, 25 mM KH2PO4 (pH 6.85), 10% glycerol, 3 mM DTT, 1 mM Na2S2O5 and 0.01% NaN3] and further purified on a hydroxyapatite column (Biorad, Hercules, CA, USA). A final purification step with a Q column (GE Healthcare Biosciences) was used to obtain samples without phosphate and with high purity. Samples for crystallization were concentrated to ∼17 mg/ml in 10 mM Tris–HCl (pH 7.5), 2.5% glycerol, 10 mM KCl and 3 mM DTT, and then frozen in small aliquots using liquid nitrogen.
Measuring polymerase-specific activity
Polymerase-specific activity was measured by incorporation α-P32-dATP into activated calf-thymus DNA (GE Healthcare Biosciences). A 25 μl reaction mixture contained 66 mM Tris–HCl (pH 8.8), 17 mM (NH4)SO4, 5 mM DTT, 6.5 mM MgCl2, 10% glycerol, 100 μg BSA, 6.25 μg activated calf-thymus DNA, 13 nM α-P32 dATP and 100 μM each of dNTP. Reactions were started by adding 5 nM RB69 pol, the mixtures were incubated at 37°C for 30 min, and then reactions were quenched with 12 mM EDTA. The products were passed through two consecutive G-25 spin columns (GE Healthcare Biosciences) to remove α-P32 dATP. The relative specific activities were calculated based on the relative radioactivity counted with a scintillation counter. To ensure linearity, <20% of the substrate was incorporated into product by all enzymes.
Gap-filling reactions and product analysis to determine fidelity
Reaction mixtures (25 μl) contained 25 mM Tris–acetate (pH 7.5 at 37°C), 10 mM MgCl2, 150 mM KOAc, 1 mM of each dNTP, 2 mM dithiothreitol, 0.2 nM gapped M13mp2 DNA and 10 nM RB69 pol. Reactions were incubated at 37°C for 15 min. Under these conditions, when DNA products were analyzed by agarose gel electrophoresis as described in ref. (20), all reactions filled the gap without obvious strand displacement [data not shown, but for typical result, see Figure 3 in (20)]. DNA products of reactions were introduced into Escherichia coli cells and plated as described in ref. (20) to score blue M13 plaques (correct synthesis) and light blue and colorless plaques (containing errors). The types of errors were determined by sequencing the lacZ α-complementation gene in single-stranded DNA isolated from independent mutant M13 plaques, allowing calculation of error rates as previously described (21). Error rates for wild type are recalculated from raw data published in ref. (22) without background correction for direct comparison. All materials for the fidelity assay were from previously described sources (20,23). The statistical significance of differences in pairs of error rates was calculated using Fisher's exact test.
Figure 3.
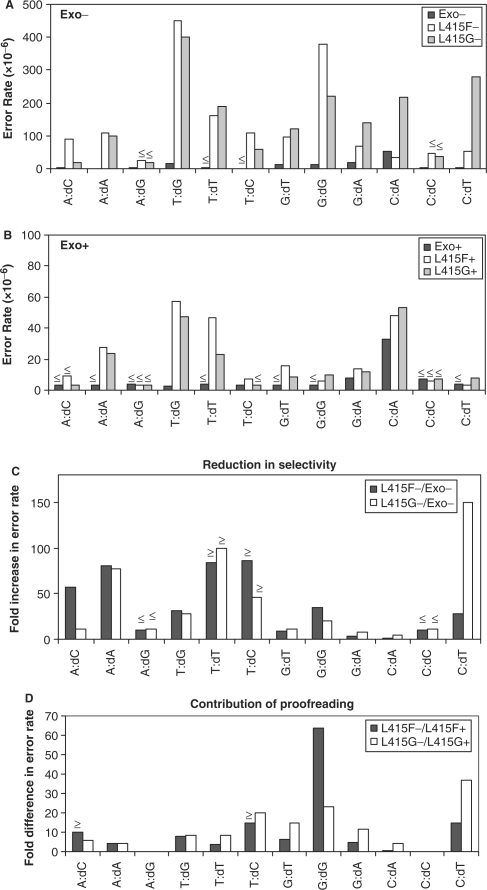
Error rates of RB69 pol derivatives for the 12 different single base mismatches. (A) Error rates for exonuclease deficient RB69 pols. (B) Error rates for exonuclease proficient RB69 pols. The less than or equal to symbol in (A) and (B) indicates that errors involving these mismatches were not observed and therefore error rates do not exceed the rate calculated assuming that one event had occurred. (C) Reduction in nucleotide selectivity of F− and G− pols compared to L− pol. (D) Contribution of proofreading by comparing error rates of the F− and G− pols to F+ and G+ pols.
Kinetics analysis
Substrates for insertion and extension assays are listed subsequently. The template sequence for both primer extension and insertion assays was 5′-ACGTCGTGACTGAGAAAACCCTGGCGTTACCA. The primer sequence for T-T mismatch primer extension was 5′-GTAACGCCAGGGTTTTCTCT. The primer sequence for A-T primer extension was 5′-GTAACGCCAGGGTTTTCTCA. The correct nucleotide inserted with these substrates was dGTP. The primer for correct insertion of dATP and mininsertion of dTTP opposite template T was 5′-GTAACGCCAGGGTTTTCTC. Primers were 5′ end-labeled with [γ-32P] ATP and T4 polynucleotide kinase, and then annealed with the template (50% excess). Reaction mixtures contained 25 mM Tris–acetate (pH 7.5 at 37°C), 10 mM MgCl2, 150 mM KOAc, 2 mM dithiothreitol, 100 nM P32-labeled primer/template, varying concentrations of dNTP and a fixed concentration of RB69 pol that when used with each enzyme resulted in <20% product formation. Reactions were initiated by mixing 5 μl premixed enzyme–DNA complex with 5 μl Mg-dNTP, and incubated at 37°C for 10 min before quenching with 10 μl of formamide loading buffer (95% deionized formamide, 25 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol). DNA products were separated by 12% denaturing polyacrylamide gel electrophoresis and product bands were quantified by phosphoimagery. Data were fit to the Michaelis–Menten equation by curve fitting with the KaleidaGraph program to obtain kcat and Km.
Measuring TLS efficiency
Oligonucleotides for TLS studies were from previously described sources and primer templates were prepared as described (24–26). Reaction mixtures (30 μl) contained 25 mM Tris–HCl (pH 7.5 at 37°C), 5 mM MgCl2, 150 mM KOAc, 50 μM of each dNTP, 2 mM DTT and 4 pmol of primer template. Reaction mixtures were preheated at 37°C for 1 min and reactions were initiated by adding 4 fmol of RB69 pol. This creates a 1000:1 ratio of primer template to polymerase, thereby generating DNA product molecules that largely result from a single cycle of processive synthesis (using criteria described in ref. (25). Incubation was at 37°C and aliquots were removed after 15 min and terminated by adding an equal volume of formamide loading buffer containing 95% deionized formamide, 25 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol. DNA products were separated by 12% denaturing polyacrylamide gel electrophoresis and product bands were quantified by phosphoimagery. Termination probabilities and insertion, extension and bypass efficiencies were calculated as described earlier (25,27).
Measuring 8-oxo-G bypass fidelity
Reactions were performed as described earlier, but with an unlabeled primer, 8 pmol of primer template, 0.8 pmol of RB69 pol and 50 μCi of α-32P-dCTP, and incubation was for 5 min. Newly synthesized, internally labeled 32-mer to 36-mer (−) strand DNA products of complete bypass that contain polymerization errors were purified, hybridized to gapped circular M13 DNA and used to infect E. coli host cells, score plaque colors and quantify insertion specificity, all as described previously (25,27). Light blue plaques resulted from incorporation of dCMP opposite 8-oxo-G and blue plaques resulted from misincorporation opposite 8-oxo-G, with the identity of the error established by DNA sequence analysis.
Determining the structure of L415F RB69 pol
Crystals of the ternary complex of exonuclease-deficient L415F RB69 pol were prepared as described (1) for the ternary complex of the RB69 pol (L−), using the same substrate. The sequence of the primer strand was 5′-GCGGACTGCTTACC* (where C* is ddC) and the sequence of the template strand was 3′-GCGCCTGACGAATGGACA. The polymerase (17 mg/ml) was mixed in an equimolar ratio with annealed primer-template duplex, and dTTP was then added to 1.6 mM. Using the hanging-drop method, an equal volume of 220 mM CaCl2, 25% (w/v) PEG 350 monomethyl ether and 50 mM Tris–HCl (pH 7.0) was mixed with the protein–DNA solution and allowed to equilibrate by vapor diffusion against 0.5 ml of well solution. Crystals typically grew in 1–2 days at room temperature, and were then transferred to the cryoprotectant [220 mM CaCl2, 30% (w/v) PEG 350 monomethyl ether and 50 mM Tri–HCl (pH 7)] and flash frozen in liquid nitrogen. X-ray data were collected with an in-house X-ray source using a 007HF generator, varimaxHF mirrors and a Saturn92 CCD detector from Rigaku, Woodlands, TX, USA. The initial structure was solved by molecular replacement using the ternary complex structure of RB69 pol (1) as the starting model. The structure was refined through several iterative cycles of annealing with CNS (28), followed by manual refinement of the structure with coot (29) and O (30). The PDB ID code is 3CQ8. The fo(L415F)−fo(wt) difference map between the L415F mutant and the wild-type ternary complex structure was calculated with the model phases and experimental structure factors in CNS (28).
RESULTS
We purified RB69 pol derivatives containing Leu (wild-type), Phe or Gly at amino acid position 415, each of which either retained 3′ exonuclease activity or lacked exonuclease activity due to two changes (D222A/D327A) in the exonuclease active site. These six polymerases, which for brevity are referred to here as L+, L−, F+, F−, G+ and G−, were compared for specific activity, for single base substitution and deletion error rates, for catalytic efficiency of insertion and mismatch extension using single dNTPs, and for TLS efficiency with three different lesions.
Specific activity of RB69 L415F/G mutants
To assess the effect of the amino acid replacements on polymerization activity, we measured polymerase-specific activity using activated DNA as a substrate. The polymerase-specific activity of F+ and G+ were 93 and 63%, respectively, of wild-type, L+ RB69 pol. The polymerase-specific activities of F− and G− were 84 and 40%, respectively, of L−.
Fidelity of RB69 L415F mutant polymerases
The fidelity of the six polymerases was compared for mistakes made when copying an undamaged LacZ template sequence in gapped M13mp2 DNA (see Materials and methods section). The M13mp2 DNA products of gap-filling DNA synthesis by wild-type L+ RB69 pol had a lacZ mutant frequency of 0.0008 (Table 1), a value similar to that of the uncopied DNA (typically 0.0005–0.0007). As seen in an earlier study of RB69 pol (19), synthesis by the L− RB69 pol generated a higher mutant frequency (0.0029), as expected from loss of proofreading activity. The F− and G− mutants generated products with mutant frequencies that were 9-fold higher, indicating that both exonuclease-deficient polymerases had reduced nucleotide selectivity. The lacZ mutant frequencies of the F+ and G+ mutant polymerases were also higher than that of their L+ parent (Table 1, compare 0.0037 and 0.0032 to 0.0008), suggesting that the exonuclease proficient mutant enzymes proofread some errors less efficiently than normal.
Table 1.
LacZ mutant frequencies generated by RB69 pols
| RB69 pol | Plaques | lacZ mutants | Mutant frequency | Sequenced | Substi- tutions | – 1 |
|---|---|---|---|---|---|---|
| Exonuclease-proficient RB69 pols | ||||||
| Wild-type | 54 411 | 52 | 0.0008 | 22 | 18 | 2 |
| L415F | 35 729 | 133 | 0.0037 | 108 | 93 | 12 |
| L415G | 27 120 | 88 | 0.0032 | 88 | 71 | 13 |
| Exonuclease-deficient RB69 pols | ||||||
| Wild-type | 91 799 | 276 | 0.0029 | 159 | 93 | 40 |
| L415F | 13 252 | 333 | 0.0250 | 97 | 80 | 20 |
| L415G | 4 881 | 128 | 0.0260 | 128 | 113 | 20 |
A few mutants contained a single base addition or a change involving more than one base.
Error specificity
Sequence analysis of collections of independent lacZ mutants (Table 1, columns on right) indicates that the vast majority of errors resulting from in vitro DNA synthesis are either single base deletions (open triangles in Figure 1) or single base substitutions. These were distributed nonrandomly throughout the LacZ template (Figure 1). From the number of occurrences, the mutant frequencies (Table 1) and knowledge of the number of template nucleotides at which these events result in a detectable change in plaque color, we calculated error rates for a variety of different single base changes generated by each of the six RB69 pols.
Figure 1.
Spectra of single base changes by mutant derivatives of RB69 pol. (A) Single base change by the F− (above the sequence) and F+ (below the sequence) polymerases. (B) Single base changes by the G− (above the sequence) and G+ (below the sequence) polymerases. Only errors resulting in detectable phenotypes are shown. Position 1 is the first transcribed nucleotide of the lacZ gene.
Single base deletions
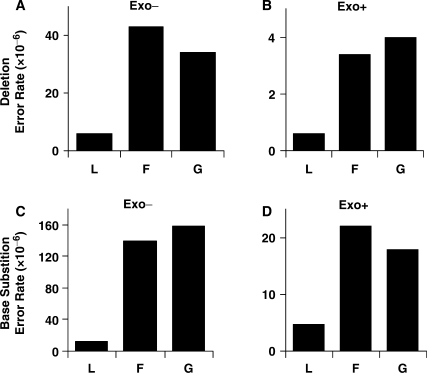
Error rates for single base deletions made by the F− and G− mutant polymerases are 6- to 7-fold higher than for the L− polymerase (Figure 2A). Among 20 single base deletions generated by the F− pol (Figure 1A), 14 were deleted from a homonucleotide run (error rate 5.9 × 10−7) and six were loss of a noniterated base (error rate of 2.7 × 10−7). Similarly, among 20 single base deletions generated by the G− pol (Figure 1B), nine were deleted from a homopolymeric run (error rate 3.0 × 10−7) and 11 were loss of a noniterated base (error rate of 3.9 × 10−7).
Figure 2.
Error rates of RB69 pol derivatives. (A and B) Single base deletions. (C and D) Single base substitutions.
Single base deletion error rates for all three exonuclease-proficient enzymes were about 10-fold lower than for their corresponding exonuclease-deficient derivatives (compare Figure 2B with 2A), indicating that ∼90% of misaligned intermediates are corrected by the exonuclease. Nonetheless, error rates for single base deletions by the F+ and G+ polymerases were 5- and 7-fold higher, respectively, than for the L+ polymerase (Figure 2B), indicating that at least some misaligned intermediates do escape proofreading. All 12 single base deletion errors by the F+ polymerase (Figure 1A), and 11 of 13 single base deletion errors by the G+ polymerase (Figure 1B), were within homonucleotide runs. The relative paucity of single nucleotide deletions of noniterated nucleotides clearly indicates that the misaligned intermediates for this type of event, which involves misaligned nucleotides close to the primer terminus, are more efficiently proofread than are misalignments in repetitive sequences, wherein misaligned nucleotides can be further upstream (see Discussion section).
Single base substitutions
Single base substitution errors were more common than single base deletions (Table 1, Figure 1). When all possible single base substitutions are considered collectively, average base substitution error rates for the F− and G− polymerases are >10-fold higher than for the L− polymerase (Figure 2C). When considering the 12 possible single base substitutions individually (Figure 3A), each of which is detectable at multiple sites in the LacZ template, average error rates for the F− and G− polymerases are higher than for the L− polymerase by factors ranging from ≥85-fold for T-dCMP and T-dTMP to ≤2-fold for C-dAMP (Figure 3C). The small increases for some mismatches likely reflect some degree of ‘correct’ insertion opposite rare cryptic lesions in the template, e.g. insertion of dAMP opposite uracil arising from cytosine deamination (see more below). These results reveal that both active site amino acid replacements reduce RB69 pol's discrimination against stable incorporation of incorrect dNTPs.
Error rate comparisons between the three exonuclease-proficient enzymes (Figures 2D and 3B) and their exonuclease-deficient counterparts (Figures 2C and 3A) reveal that the majority of single base mismatches are proofread. This is the case for the overall average error rate (Figure 2) and for the rates of many of the individual mismatches (Figure 3D). Nonetheless, the error rates of the F+ and G+ polymerases are higher than the L+ polymerase (Figures 2D and 3B), indicating that some misinsertions are extended by the mutant polymerases rather than excised, despite the fact that they retain 3′ exonuclease activity. To determine if escape from proofreading could be explained by promiscuous mismatch extension that reduces partitioning of a mismatch to the exonuclease active site, we performed steady-state kinetic analysis of single nucleotide incorporation by the three exonuclease-deficient polymerases.
Steady-state kinetic analysis of correct dNTP insertion
We first measured the efficiency of correct incorporation during extension of a correctly paired primer template. For insertion of dATP opposite template T (Table 2, first set of experiments), the kcat, Km and catalytic efficiency (kcat/Km) values for F− and G− polymerases were similar to those of the L− polymerase. For insertion of dGTP opposite template C (Table 2, third set of experiments), the kcat, Km and catalytic efficiency values for the F− enzyme were similar to those of L− polymerase, while the Km for the G− enzyme was 3-fold higher, reducing catalytic efficiency by 3-fold. These data and the specific activity measurements with activated DNA described above demonstrate that the two amino acid replacements at the polymerase active site have little effect on correct nucleotide incorporation.
Table 2.
Steady-state kinetics results for exo- RB69 polymerases
| Terminus | dNTP: Template | Enzyme | kcat (s−1) | Km (μM) | kcat/Km (s−1 M−1) | f efficiency | kcat/Km F,G/L | f F,G/L |
|---|---|---|---|---|---|---|---|---|
| G-C | dATP:T | L− | 0.12 ± 0.04 | 3.4 (±0.5) | 3.5 ± 0.7 × 104 | 1 | ||
| F− | 0.19 ± 0.13 | 3.8 (±0.8) | 6.5 ± 3.0 × 104 | 1 | 1.9 | |||
| G− | 0.20 ± 0.07 | 4.2 (±0.7) | 4.6 ± 1.0 × 104 | 1 | 1.3 | |||
| G-C | dTTP:T | L− | 0.0031 ± 0.001 | 1000 (±310) | 3.1 ± 0.5 | 9.0 × 10−5 | ||
| F− | 0.19 ± 0.13 | 1500 (±700) | 1.2 ± 0.3 × 102 | 1.8 × 10−3 | 37 | 20 | ||
| G− | 0.16 ± 0.01 | 910 (±110) | 1.8 ± 0.1 × 102 | 3.8 × 10−3 | 56 | 43 | ||
| T-A | dGTP:C | L− | 0.11 ± 0.01 | 0.65 (±0.08) | 17 ± 4 × 104 | 1 | ||
| F− | 0.24 ± 0.01 | 0.74 (±0.09) | 33 ± 3 × 104 | 1 | 1.9 | |||
| G− | 0.080 ± 0.007 | 1.8 (±1.0) | 5.6 ± 3.5 × 104 | 1 | 0.32 | |||
| T-T | dGTP:C | L− | 0.0038 ± 0.001 | 880 (±230) | 4.4 ± 1 | 2.6 × 10−5 | ||
| F− | 0.27 ± 0.10 | 420 (±90) | 6.6 ± 1 × 102 | 2.0 × 10−3 | 150 | 77 | ||
| G− | 0.10 ± 0.01 | 860 (±210) | 1.2 ± 0.5 × 102 | 3.7 × 10−3 | 27 | 14 |
The analyses were performed as described in Materials and methods section.
Steady-state kinetic analysis of misinsertion and mismatch extension
Among 12 possibilities, the error rates of the F− and G− polymerases were most strongly increased for the A-dATP, T-dCTP and T-dTTP mismatches (Figure 3C). The T-dTTP mismatch is a signature error made by the homologous yeast pol ɛ mutant containing a glycine substitution, and the resulting T-A to A-T transversions in yeast have been informative regarding the function of pol ɛ in leading strand replication (13). For these reasons, we chose to measure the steady-state kinetic parameters for misinsertion of dTTP opposite template T, and for correct incorporation of dGTP opposite template C during extension of a primer template containing a T-T mismatch. For misinsertion of dTTP opposite T (Table 2, second set of experiments), all three polymerases had a similar apparent Km. However, the F− and G− polymerases had a significantly increased kcat (0.19 s−1 and 0.16 s−1, respectively) relative to L− polymerase (0.0031 s−1), such that their catalytic efficiency for misinsertion was 20- and 43-fold higher, respectively, than that of L− polymerase. These data are consistent with the increased error rates for this mismatch seen in complete gap-filling synthesis reactions, and clearly demonstrate that the two mutant polymerases have reduced nucleotide selectivity. For correct extension of a T-T mismatch (Table 2, fourth set of experiments), all three polymerases again had a similar apparent Km, while the F− and G− polymerases again had significantly increased kcat values (0.27 s−1 and 0.10 s−1, respectively) relative to wild-type (0.0038 s−1). Accordingly, their catalytic efficiencies for mismatch extension were 77- and 14-fold higher, respectively, than that of L− polymerase. This promiscuous T-T mismatch extension likely suppresses partitioning of the mismatch to the exonuclease active site for excision, thus accounting for the higher error rates for T to A substitutions by the F+ and G+ mutants enzymes as compared the wild-type (Figure 3B).
TLS efficiency
Several published observations indicate that homologous L868F pol α (7) and L979F pol ζ (14) mutants have increased TLS capacity. For this reason, we compared the TLS efficiency of the F+ and F− mutant polymerases to their wild-type counterparts, using three lesions with different coding capacity: 8-oxo-guanine, which can ambiguously pair with dCTP or dATP, a synthetic abasic site (tetrahydrofuran) lacking base coding potential and a cis-syn cyclobutane thymine-thymine dimer (TTD) that blocks synthesis by accurate replicative polymerases. Reactions were performed using 1000-fold more primer template than polymerase, such that DNA products primarily result from one cycle of processive DNA synthesis.
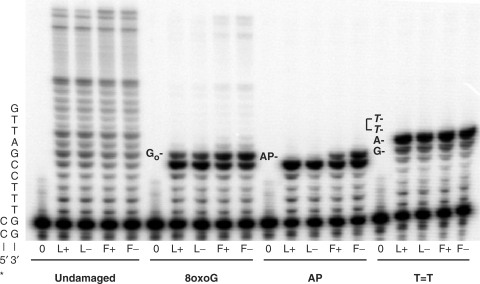
The four polymerases copied the undamaged DNA template to similar extents (Figure 4). With both L+ and L− polymerases, synthesis was strongly blocked by 8-oxo-guanine in the template (Figure 4). The strongest block occurred following incorporation opposite the template base preceding the lesion, and to a slightly lesser extent following incorporation opposite 8-oxo-G. Faint product bands were observed that reflected incorporation of seven or more nucleotides (these are more obvious upon long exposure, data not shown), indicating that at least some bypass by the L+ and L− enzymes does occur. However, quantification of band intensities and subsequent calculations (25) indicate that 8-oxo-G bypass efficiency relative to the undamaged template was only 1.1 and 1.2%, respectively, for the L+ and L− polymerases. Synthesis by the F+ and F− polymerases differed in two ways. First, the efficiency of insertion opposite 8-oxo-G increased from ∼10% for the L+ and L− enzymes to 33 and 44% for the F+ and F− polymerases, respectively. Second, replacing L415 with Phe increased the relative 8-oxo-G bypass efficiency to 2.7 and 3.8% for the F+ and F− enzymes, respectively. Interestingly, when the probability of extension following insertion opposite the lesion was quantified, similar low values (8–10%) were obtained for all four polymerases. Thus, the increased bypass efficiencies of the F+ and F− polymerases are largely explained by the observed increase in the probability of insertion opposite the lesion.
Figure 4.
TLS by RB69 polymerases. The sequence of the template primer is shown on the left. Go, 8-oxoG; AP, Abasic site analog; T=T, cyclobutane dimer.
Results were somewhat different for synthesis using a template with a synthetic abasic site. Both L+ and L− polymerases were more strongly blocked by this lesion than by 8-oxo-G, and as observed with 8-oxo-G, strong blockage occurred whether the exonuclease was intact or not (Figure 4). Essentially, complete blockage occurred following incorporation opposite the template base preceding the lesion, and no bypass synthesis was detected. For the F+ and F− polymerases, the efficiency of insertion opposite the abasic site was increased, and slightly more so with the F− as compared to the F+ polymerase. Complete lesion bypass was not observed under these conditions, which allow a single encounter of the polymerases with the lesion. However, in reactions allowing multiple encounters (excess polymerase, longer incubation times), a small amount of complete AP site bypass by the L415F mutant pols was observed (data not shown). Interestingly, the results with the TTD differed yet again. Following insertion opposite the template base preceding the 3′ T of the TTD, no subsequent incorporation was detected for any of the four RB69 pols (Figure 4).
The fidelity TLS when bypassing template 8-oxo-G
Next, we measured how accurately the RB69 pols bypass template 8-oxo-G. The DNA products of complete bypass (dNTP insertion opposite the lesion, plus multiple extensions) were recovered and hybridized to gapped M13mp2 DNA containing a nonsense codon in the lacZ α-complementation gene. Resulting M13 plaque colors were then scored, with light blue plaques resulting from incorporation of dCMP opposite 8-oxo-G (present as one of the bases of the nonsense codon) and blue plaques resulting from misincorporation opposite 8-oxo-G. The identity of the error leading to blue plaque color was then established by DNA sequence analysis of DNA from independent plaques. The results (Table 3) indicate that all four forms of RB69 pol primarily incorporate either dCMP or dAMP opposite 8-oxo-G, with the proportion depending on the form of RB69 pol used. L+ prefers to incorporate dCMP, but dAMP incorporation is only 5-fold less likely. The proportion of dAMP incorporation increases very little when the 3′ exonuclease is inactivated (i.e. with L− pol), indicating that 8-oxo-dG·dAMP mismatches are not efficiently proofread. Results with the F+ and F− RB69 pols are similar to those with the L+ and L− pols (Table 3). This further implies inefficient proofreading of 8-oxo-dG·dAMP mismatches, and also demonstrates that substituting phenylalanine for Leu415 does not strongly affect misincorporation of dAMP opposite 8-oxo-G.
Table 3.
8-oxo-dG bypass fidelity by RB69 polymerases
| L+ | L− | F+ | F− | |
|---|---|---|---|---|
| Light blue plaques | 87.5% (1108) | 84.6% (1549) | 91% (1065) | 82.3% (1205) |
| Dark blue plaques | 10% (127) | 13.7% (251) | 8.2% (96) | 17.2% (252) |
| dATP:Go (%) | 17 | 23 | 14 | 29 |
| C:A ratio | 5.0 | 3.4 | 6.3 | 2.5 |
In parentheses are the numbers of plaques scored. A small percentage of plaques were colorless due to sequence changes within the chemically synthesized oligonucleotides and/or due to occasional polymerization errors located at positions other than at the nonsense codon.
Crystal structure of L415F RB69 pol
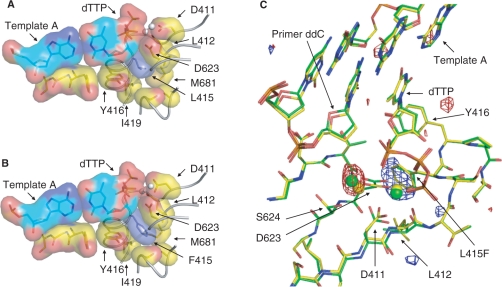
Next, we solved the 2.5 Å crystal structure of a closed ternary complex of the F− RB69 pol with a correct A-dTTP base pair at the active site (Table 4). This structure (Figure 5B) is similar to that of the L− polymerase [Figure 5A, adapted from ref. (1)] with a RMSD of 0.38 Å on 901 C-α atoms between the new structure and the L− polymerase structure (PDB ID code 1IG9). A comparison of the two structures reveals two clear difference peaks (Figure 5C). One is a strong positive peak in the (Fo − Fo) electron density map indicating the L to F change, with the phenylalanine side chain snugly accommodated in a pocket (Figure 5B). The other is a somewhat weaker negative peak at the catalytic metal-binding site suggesting a difference in Ca2+ occupancy. Other, even weaker difference peaks are also observed, but the differences are close to the noise level. Consistent with the weak density at the catalytic metal-binding site, the side chain of Ser624 (arrow in Figure 5C) moves away from the catalytic metal ion in the L415F mutant structure. There is also a small shift of the main chain backbone between residue 411 to residue 412 (arrows in Figure 5C), centered around C-α of D411 with the largest shift being 0.57 Å. This shift moves the D411 side-chain slightly closer to the nucleotide binding metal ion site. In addition, the sugar ring of the primer terminal nucleotide (labeled ‘Primer ddC’ in Figure 5C) is moved about 1.0 Å and its electron density is wider, suggesting that the sugar ring may adopt two conformations, one like that of wild-type RB69 pol and a second, slightly shifted conformation.
Table 4.
Data collection and refinement statistics
| Characteristics | Values |
|---|---|
| Data collection statistics | |
| Unit cell | a = 80.840, b = 117.006, c = 127.823 |
| Space Group | P212121 |
| Unique reflections | 42 387 |
| Resolution (highest shell) (Å) | 50–2.5 (2.59–2.5) |
| Rsym (highest shell) | 0.076 (0.514) |
| Completeness (highest shell) (%) | 98.6 (99.5) |
| Average Redundancy | 5.6 (4.5) |
| <I>/<σ(I)> (highest shell) | 17.6 (2.8) |
| Refinement statistics | |
| Final R (Rfree) (%) | 21.2 (28.1) |
| RMSD bonds (Å), angles (deg) | 0.007, 1.21 |
| Rotamer outliers (%) | 1.77 |
| Ramachandran outliers (%) | 0.78 |
| Ramachandran favored (%) | 90.4 |
| Average B factors | |
| All atoms | 46.9 |
| Protein atoms | 47.1 |
| DNA atoms | 50.4 |
| Incoming dTTP | 31.1 |
| Water atoms | 38.8 |
| Calcium atoms | 63.1 |
Figure 5.
Surface contact diagram of RB69 polymerase active site around residue 415. (A) L− ternary complex. (B) F− ternary complex. (C) Overlay of the active site of L− (green) and F− (yellow) pols, including the fo(L415F) − fo(wt) difference electron density map. The blue positive peak (4σ) indicates the additional density introduced by the L415F change. The red negative peak (4σ) indicates reduced density at the catalytic metal site in the F− structure compared to the density in the L− structure. In both structures shown, the metal is calcium rather than magnesium, the metal ion most likely to activate polymerization in vivo.
DISCUSSION
Amino acid replacements for the highly conserved Leu/Met in motif A of eukaryotic B family DNA polymerases result in mutator polymerases that are useful for probing biological functions, including proofreading of pol α-dependent replication errors by the 3′ exonuclease activity of pol δ (6), the role of pol ɛ (13) and pol δ (15) in leading and lagging strand DNA replication, the contribution of pol δ replication errors to cancer susceptibility in mice (16) and the contribution of pol ζ errors to spontaneous and UV-induced mutagenesis in yeast (14). Given the utility of these mutator alleles, it is of interest to obtain a better understanding of how amino acid replacements at this position affect the properties of these family B polymerases. Because there are as yet no structures of eukaryotic family B polymerases, the present study was undertaken to characterize the biochemical properties and obtain initial structural information on homologous mutants of RB69 pol. The results are informative regarding several properties of family B members.
Catalytic efficiency for correct nucleotide incorporation
RB69 pols with nonconservative phenylalanine or glycine substitutions for Leu415 retain high-specific activities with activated DNA, and the exonuclease-deficient enzymes have steady-state kinetic constants for correct dNTP insertions that are similar to those of the exonuclease-deficient wild-type polymerase (Table 2). Relatively normal activity has also been reported for equivalent derivatives of other eukaryotic family B members, including human L864F and yeast Leu868F pol α (7) and exonuclease deficient yeast M644F and M644G pol ɛ (12,13). Genetic studies (14) imply that yeast pol ζ with the equivalent replacement (Leu979F) also retains high-polymerase activity. Retention of robust activity for correct incorporation is consistent with the fact that, in the ternary complex of F− RB69 pol with a correct base pair bound (Figure 5B), the phenylalanine side chain is accommodated within the space normally occupied by the Leu415 side chain plus adjacent open space (Figure 5A), without large changes in the position of surrounding residues or the geometry of the nascent base pair binding pocket. These observations are consistent with earlier studies that have used the structure of RB69 pol to model the effects of replacing leucine with other amino acid in pol α (7) and pol δ (11).
Misinsertion and mismatch extension efficiency
Despite retaining high-catalytic efficiency for correct incorporation, the F− and G− polymerases have elevated rates for a variety of different base substitution errors (Figure 2 and 3C). In fact, the identity of the amino acid at this position is a key determinant of the base substitution fidelity of several family B enzymes, including human L864F and yeast Leu868F pol α (7), yeast M644F and M644G pol ɛ (12,13) and yeast L612M pol δ (10). Beyond the general importance of this conserved motif A residue to fidelity of all these family B polymerases, two additional points are worth noting. First, different amino acid replacements at this position can have different effects, both on fidelity (12) and on several properties of yeast strains, such as mutation rates, cell cycling and DNA damage responses (11). Second, even the same amino acid replacement can differentially alter the error specificity of different members of family B. For example, we previously noted that phenylalanine replacement elevates yeast Leu868F pol α and M644F pol ɛ error rates to different extents for the same mispair [see Figure 5B in ref. (12)]. Comparing those data to the error specificity of L415F RB69 (Figure 3) reinforces this point, indicating that the consequences of the same amino acid replacement at this position are to some extent enzyme specific.
The elevated error rates for the F− and G− RB69 Pols clearly indicate that the first step towards stable misincorporation, discrimination against misinsertion of incorrect dNTPs, is reduced by the phenylalanine and glycine replacements. The conclusion is further supported by the kinetic data for the RB69 pols (Table 2). These data confirm that phenylalanine or glycine replacements reduce selectivity against dNTP misinsertion, and also reduce discrimination against correct incorporation onto a mismatched primer terminus. Promiscuity in mismatch extension is also conferred by glycine replacement in yeast pol ɛ (13) and by methionine replacement in yeast pol δ (10), and it is consistent with elevated UV-induced mutagenesis in yeast strains harboring rev3L979F and rev3L979M alleles encoding mutant pol ζ (14).
The selectivity of wild-type RB69 pol at both the insertion and mismatch extension steps results from large differences in both Km and kcat for correct and incorrect insertion (Table 2). Interestingly, while the F− and G− RB69 pols also exhibit large differences in the Km for correct and incorrect insertion, their kcat for misinsertion is similar to the kcat for correct insertion (Table 2). This suggests that when insertion of an incorrect dNTP is attempted, or when a mismatch is present at the primer terminus, assembly of the catalytic conformation may be easier to obtain when phenylalanine replaces leucine. This could be related to two of the slight differences seen when comparing the structure of wild-type (Leu415) and Phe415 RB69 pols: the 0.57 Å shift of D411 and the fact that the electron density of the primer terminus is more diffuse. These features are consistent with greater flexibility at the polymerase active site, which may allow the 3′ OH and the α phosphate to properly align for catalysis despite the presence of a mismatch.
Errors involving misalignments
Phenylalanine and glycine replacements for Leu415 increase the error rate of RB69 pol for single base deletions, which involve misaligned primer templates with an unpaired base in the template strand. Average error rates for the F and G mutant polymerases are similar for deleting noniterated bases and bases within homonucleotide runs (Figure 1 and see Results section), suggesting that these amino acid replacements may promote these errors via misaligned intermediates different from those predicted by the Streisinger hypothesis (31,32). Regardless of the mechanism or exact nature of the misaligned intermediate, the fact that phenylalanine and glycine replacements for Leu415 promote infidelity for single base deletions in repetitive as well as noniterated sequences implies that homologous changes in eukaryotic B family polymerases should be useful tools for probing the roles of these enzymes in determining the stability of repetitive DNA sequences that are abundant in eukaryotic genomes and are well-known hot spots for mutagenesis often associated with diseases.
Proofreading efficiency
Despite their 3′ exonuclease activities, the F+ and G+ polymerases generate single base substitution errors (Figures 2D and 3B) at higher rates than does wild-type RB69 pol. Thus, a small fraction of single base–base mismatches generated by the F and G mutant polymerases escape proofreading, most likely due to promiscuous mismatch extension (Table 2) at the expense of partitioning the mismatch to the exonuclease active site, as has been proposed for other B family pols (5,10,12,13). Consistent with the interpretation of reduced partitioning due to increased mismatch extension is the observation that L412M T4 DNA polymerase does not dissociate from DNA as readily as does wild-type T4 DNA polymerase (5). Proofreading is not limited to correcting single base-base mismatches, because the single base deletion error rates for all three exonuclease-proficient enzymes are about 10-fold lower than for their corresponding exonuclease-deficient derivatives (compare Figure 2B with 2A), indicating that ∼90% of misaligned intermediates are also proofread. Nonetheless, some misaligned intermediates generated by the F+ and G+ enzymes clearly do escape proofreading (Figure 2B), and these appear to preferentially arise within homonucleotide runs (Figure 1). The relative paucity of single base deletions of noniterated nucleotides clearly indicates that misaligned primer templates with an unpaired template strand base near the primer terminus are more efficiently proofread than are misalignments in repetitive sequences, just as has been reported before for other proofreading-proficient polymerases (33).
TLS parameters
Replacing L415 with phenylalanine affected the TLS properties of RB69 pol in a lesion-specific manner. Wild-type RB69 pol does not bypass a TTD or even insert a base opposite the 3′ T of the TTD, and inactivating its 3′ exonuclease did not increase TLS capacity (Figure 4). Similarly, neither wild-type nor exonuclease-deficient yeast pol δ is able to bypass a TTD or insert a base opposite the 3′ T (24). Moreover, replacing Leu415 in RB69 pol with phenylalanine does not enhance TTD bypass (Figure 4), which is interesting in light of an earlier study demonstrating that the homologous phenylalanine mutants of yeast and human pol α more efficiently bypass a TTD than do their wild type parents (7). One possible explanation for this difference among B family members is that our results with RB69 pol and pol δ are obtained using a large excess of primer template to permit a single encounter of the enzyme with the lesion, whereas the pol α studies (7), like most other TLS studies in the literature, use excess polymerase that allows a larger but unknown number of attempts at bypass. Thus, we cannot exclude that phenylalanine replacement might have a very small effect on the probability of TTD bypass per encounter by RB69 pol and pol δ. Alternatively, it is possible that the effect of phenylalanine replacement on TTD bypass efficiency is different among various B family members, just as is the case for error specificity when copying undamaged DNA (discussed above).
Synthetic abasic sites strongly block DNA synthesis by most DNA polymerases [see ref. (34) and references therein]. Consistent with this fact and with elegant structural studies of exonuclease-deficient RB69 pol (17,18), none of the four RB69 pols examined here was able to completely bypass a synthetic abasic site (Figure 4). Moreover, while wild-type RB69 pol can insert dAMP opposite a synthetic abasic site under crystallization conditions (17), this insertion by L+ and L− RB69 pol is very inefficient when examined under the single hit reaction conditions used here (Figure 4). Compared to the L+ and L− polymerases, the F+ and F− polymerases had clearly increased dNTP insertion efficiency opposite the abasic site (Figure 4). Thus, the identity of the amino acid at this location in motif A does influence the efficiency of one key incorporation event required to bypass a synthetic abasic site. This may be relevant to the observation that the homologous phenylalanine mutants of yeast and human pol α more efficiently bypass a synthetic abasic site than do their wild-type parents (7). As discussed earlier for misinsertion, the phenylalanine replacement may promote greater flexibility at the polymerase active site, which may allow the 3′ OH and the α phosphate to properly align for dNTP insertion opposite the abasic site. However, further incorporation using the abasic site-containing template-primer terminus is not promoted by the phenylalanine replacement (Figure 4). This result differs from the increased efficiency for extending an undamaged but mismatched template-primer terminus (Table 2), i.e. the L415F replacement is a ‘separation-of-function’ mutant with regard to extension of these two different aberrant termini. The limited ability to extend the abasic site-containing template-primer terminus is likely related to the fact that insertion opposite the lesion is greater for F- RB69 pol than for F+ RB69 pol (Figure 4). This indicates a base inserted opposite the abasic site can be excised by the 3′ exonuclease of L415F RB69 pol, as has been described in earlier studies of wild-type RB69 pol (17,34).
Under the single-hit conditions used here, both the L+ and L− pols bypass template 8-oxo-G at about ∼1% of the efficiency that they bypass the corresponding undamaged G. This strong impediment to bypass results from inefficient insertion opposite 8-oxo-G (Figure 4, strong termination band preceding the lesion), plus inefficient extension of the resulting damaged template-primer terminus (Figure 4). Inefficient insertion opposite 8-oxo-G is consistent with kinetic analysis showing that L− RB69 pol inserts dCMP opposite 8-oxo-G 45-fold less efficiently than it inserts dCMP opposite undamaged G (18). The efficiency of insertion opposite 8-oxoG is enhanced 3-fold by the phenylalanine replacement, thereby increasing complete bypass efficiency by a similar amount. Just as seen with the abasic site, the efficiency of extending the 8-oxo-G-containing template-primer terminus is not enhanced by the phenylalanine replacement, again illustrating a differential consequence for extending undamaged mismatched termini and damaged termini. This separation of function concept also extends to the fidelity of DNA synthesis, because the phenylalanine replacement strongly affects the fidelity with which RB69 pol copies undamaged DNA, but has little effect on the fidelity of bypass of 8-oxo-G. Thus, RB69 pols primarily incorporate dCMP and dAMP opposite 8-oxo-G, in proportions that are generally consistent with previous kinetic data for L− RB69 pol (18). These proportions are not strongly influenced by proofreading, as is the case with T7 DNA polymerase (26) and they are not strongly influenced by substituting phenylalanine for Leu415 (Table 3).
This study was partly motivated by the desire to use mutant polymerases to probe the biological functions of eukaryotic B family members. In addition to the ‘spontaneous’ functions already mentioned in the Introduction section, the fact that phenylalanine replacement affects certain TLS bypass parameters for RB69 pol (this study) and for yeast and human pol α (7), suggests that mutants with amino acid replacements in motif A of pol α, δ, ɛ and ζ could be informative for understanding cytotoxic and mutagenic responses to DNA damaging agents. As one example, yeast strains encoding L979F pol ζ have normal survival but increased mutagenesis in response to exposure to UV light (14). Thus, studies to examine TLS parameters with homologous eukaryotic B family members should be informative, as should structural studies of mutant RB69 pols bound to lesions (17,18).
ACKNOWLEDGEMENTS
We thank John Drake for RB69exo+ and RB69exo− plasmids; Scott McCulloch for technical help with the TLS assay; Dinh Nguyen for assistance in DNA substrate preparation; Dinh Nguyen and the NIEHS DNA sequence core facility for sequence analysis of lacZ mutants; Joseph Krahn and Andrea Moon for assistance in data collection and crystal structure determination and Katarzyna Bebenek and Mercedes Arana for thoughtful suggestions on the article. This research was supported by Research Project Number Z01-ES065070 to TAK, in the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program, NIH, NIEHS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 2.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 3.Reha-Krantz LJ, Nonay RL. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. J. Biol. Chem. 1994;269:5635–5643. [PubMed] [Google Scholar]
- 4.Beechem JM, Otto MR, Bloom LB, Eritja R, Reha-Krantz LJ, Goodman MF. Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry. 1998;37:10144–10155. doi: 10.1021/bi980074b. [DOI] [PubMed] [Google Scholar]
- 5.Fidalgo da Silva E, Mandal SS, Reha-Krantz LJ. Using 2-aminopurine fluorescence to measure incorporation of incorrect nucleotides by wild type and mutant bacteriophage T4 DNA polymerases. J. Biol. Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- 6.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol. Cell. Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase delta in Saccharomyces cerevisiae. Genetics. 2005;170:569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 12.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase epsilon. Nucleic Acids Res. 2007;35:3076–3086. doi: 10.1093/nar/gkm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair. 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, Ladiges WC, Preston BD, Loeb LA. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol. Cell. Biol. 2007;27:7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bebenek A, Dressman HK, Carver GT, Ng S, Petrov V, Yang G, Konigsberg WH, Karam JD, Drake JW. Interacting fidelity defects in the replicative DNA polymerase of bacteriophage RB69. J. Biol. Chem. 2001;276:10387–10397. doi: 10.1074/jbc.M007707200. [DOI] [PubMed] [Google Scholar]
- 20.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 21.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bebenek A, Carver GT, Dressman HK, Kadyrov FA, Haseman JK, Petrov V, Konigsberg WH, Karam JD, Drake JW. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics. 2002;162:1003–1018. doi: 10.1093/genetics/162.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 24.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 25.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 26.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCulloch SD, Kunkel TA. Measuring the fidelity of translesion DNA synthesis. Methods Enzymol. 2006;408:341–355. doi: 10.1016/S0076-6879(06)08021-9. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb. Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Kroutil LC, Register K, Bebenek K, Kunkel TA. Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry. 1996;35:1046–1053. doi: 10.1021/bi952178h. [DOI] [PubMed] [Google Scholar]
- 34.Zahn KE, Belrhali H, Wallace SS, Doublie S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]