Abstract
Cation-induced folding into quadruplex structures for three model human telomeric oligonucleotides, d[AGGG(TTAGGG)3], d[TTGGG(TTAGGG)3A] and d[TTGGG(TTAGGG)3], was characterized by equilibrium titrations with KCl and NaCl and by multiwavelength stopped flow kinetics. Cation binding was cooperative with Hill coefficients of 1.5–2.2 in K+ and 2.4–2.9 in Na+ with half-saturation concentrations of 0.5–1 mM for K+ and 4–13 mM for Na+ depending on the oligonucleotide sequence. Oligonucleotide folding in 50 mM KCl at 25°C consisted of single exponential processes with relaxation times τ of 20–60 ms depending on the sequence. In contrast, folding in100 mM NaCl consisted of three exponentials with τ-values of 40–85 ms, 250–950 ms and 1.5–10.5 s. The folding rate constants approached limiting values with increasing cation concentration; in addition, the rates of folding decreased with increasing temperature over the range 15–45°C. Taken together, these results suggest that folding of G-rich oligonucleotides into quadruplex structures proceeds via kinetically significant intermediates. These intermediates may consist of antiparallel hairpins in rapid equilibrium with less ordered structures. The hairpins may subsequently form nascent G-quartets stabilized by H-bonding and cation binding followed by relatively slow strand rearrangements to form the final completely folded topologies. Fewer kinetic intermediates were evident with K+ than Na+, suggesting a simpler folding pathway in K+ solutions.
INTRODUCTION
Guanine-rich segments of DNA can associate into four-stranded structures known as G-quadruplexes that are characterized by stacked arrays of four guanine bases (1–5). Interest in G-quadruplexes has been stimulated by an emerging appreciation of their role in biological regulation as well as their potential to serve as novel drugs and drug targets. G-rich sequences have been found in eukaryotic telomeres, where they help maintain chromosomal integrity by preventing erosion of chromosomal ends during DNA replication (6). G-rich sequences are also present in the promoter regions of a number of genes, including oncogenes such as c-myc where they may regulate transcription (7,8). In addition, G-quadruplexes may also regulate immunoglobin switching (9). Synthetic G-quadruplex oligonucleotides (aptamers) currently are candidate drugs for treating cancer (3,10–17), prevention of thrombosis (18) and inhibition of HIV replication (19). In addition, drugs that bind to specific quadruplex DNAs may be useful in treating a variety of pathological conditions including cancer (20).
Structural determinations by X-ray crystallography and NMR have shown that the fundamental building block of G-quadruplexes is the G-tetrad, a planar arrangement of four guanines H-bonded to each other that form a cyclic structure with a central cavity (3,5,21). In addition to H-bonding forces, G-quadruplexes are stabilized by nonpolar interactions between stacked tetrads, electrostatic interactions involving monovalent cation binding to the O6 carbonyls of guanines within a quartet and by hydration effects (22). In addition to specific cation binding, nonspecific polyelectrolyte cation interactions (23) are also likely to be important for quadruplex stabilization.
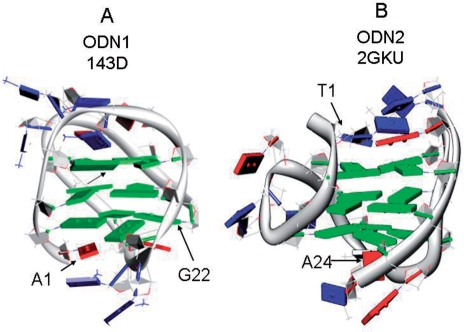
A variety of molecular topologies have been found for G-quadruplex structures owing to the conformational plasticity of oligonucleotides (3,5,21). The stereochemistry of the glycosidic bonds (anti or syn), strand polarity (parallel or antiparallel), molecularity (mono-, bi- or tetra-molecular), the sequence and topology of the loops connecting the G-quartets and the nature of the sequences flanking the G-stacks combine to generate many different 3D shapes. For example, one of the oligonucleotides in this study, d[AGGG(TTAGGG)3] (ODN1), a model of the human telomere sequence, forms a unimolecular G-quadruplex in solutions containing K+ or Na+. NMR studies (24) have shown that ODN1 in Na+ forms a basket structure characterized by antiparallel strands, two lateral loops and one diagonal loop (see the schematic diagram in Figure 1A).
Figure 1.
Cartoon showing the folding topology of ODN1 in Na+ [G3(T2AG3)3, PDB id 143D, (A)] and ODN1 [T2G3(T2AG3)3A, PDB id 2GKU] in K+ (B). Bases are shown as slabs in which blue, T; red, A; green, G. The figures were drawn using the molecular graphics program Chimera (65) from NMR-derived coordinates for the first conformer listed in pdb files 143D (24) and 2GKU (28).
Quadruplex conformations in K+ are of particular interest because the relatively higher intracellular concentration of this cation suggests that K+-bound structures predominate under physiological conditions. A novel structure for ODN1 described as a parallel-stranded propeller characterized by three double-chain reversal loops was found in the presence of K+ ions by X-ray crystallography (25). However, biophysical studies showed that the propeller structure is not the predominant form in K+ solution and that this structure may have been preferentially stabilized by the macromolecular crowding conditions necessary for crystal growth (26). NMR data indicate that ODN1 in K+ solutions probably exists as an equilibrium mixture of more than one conformer (27).
Luu et al. (28) recently published an NMR-derived solution structure of d[TTGGG(TTAGGG)3A] (ODN2) in K+. This structure (shown in Figure 1B) exhibits a ‘hybrid’ fold consisting of a mixture of three parallel strands and one antiparallel strand connected by loops located on edges (28). In addition, Yang and colleagues have recently characterized G-quadruplex structures in K+ solution (29–31). A similar structure based on circular dichroism using oligonucleotides containing brominated bases has been proposed (32).
A characteristic feature of biological polymers is their ability under physiological conditions to fold into specific 3D structures that are functionally definitive. Characterization of the kinetics of folding is essential for providing insight into potential pathways of folding (33). For example, kinetic analysis can reveal the presence of folding intermediates as well as define the number and nature of rate-limiting, high-energy intermediates (e.g. the transition state). Considerable progress has been made in understanding mechanisms of polypeptide folding; however, delineating pathways of oligonucleotide folding has received less attention (34) possibly because of the richness of the folding landscape for DNA has not been appreciated as functionally important until recently. For example, recent kinetic studies have provided insight into the mechanism of DNA and RNA hairpin formation (35,36) as well as folding pathways for complex structures such as ribozymes (37,38) and riboswitches (39).
The kinetics of formation of monomolecular G-quadruplexes has not been extensively characterized (40). In one recent study, Zhao et al. (15) investigated the folding and unfolding kinetics of telomeric oligonucleotides using surface plasmon resonance to detect unfolded structures by trapping them with a complementary oligonucleotide. The kinetic constants resulting from this study did not agree well with thermodynamic analyses of quadruplex folding, possibly because of difficulties in interpreting solid/liquid phase kinetics (40) as well as daunting numerical problems associated with fitting complex coupled rate equations to obtain rate constants. Defining the unfolding kinetics of telomeric structures has however been more amenable to experimental analysis. Balasubramanian et al. for example utilized fluorescence resonance energy transfer to determine unfolding rates of single molecules of human telomeric G quadruplexes (41–43). In addition, the kinetics of multimolecular strand association has also been investigated; as expected of multibody collisional reactions, tetramer formation is extremely slow, with half times of days at 37°C (40,44–47).
The purpose of the present study was to characterize the kinetics of folding of simple monomolecular G-quadruplexes in homogeneous solution and to use the kinetic information to suggest folding pathways consistent with the kinetic data. To accomplish this aim, we used stopped flow mixing coupled with rapid wavelength scanning to detect quadruplex formation from its spectroscopic signature: an increase in absorbance at ∼295 nm (48). G-quadruplex folding provides an especially convenient system for studying polynucleotide folding because folding to a well-defined state can be driven simply by addition of a suitable cation. In the current study, we investigated K+ and Na+-driven folding of three oligonucleotides models of the human telomere, two of which (ODN1 and ODN2) have well-defined solution structures. ODN1 is a 22-nt model of the human telomere sequence d[AGGG(TTAGGG)3]; ODN2 is a 24-nt oligonucleotide in which the 5′A of 143D is replaced with TT along with the addition of a 3′A. We also characterized a truncated 23-nt version of ODN2 (designated ODN3) in which A24 located at the 3′-terminus was deleted. The NMR structure of ODN2 shows that access to the cation binding channel may be hindered by A24. Removal of this base might be expected to influence cation binding.
The results of our investigation show that cation-driven G-quadruplex formation in these model systems is a multistep process in which a rapidly formed cation–oligonucleotide complex converts by one or more slower rearrangements to give the final equilibrium product. Our results indicate that K+-driven folding requires two kinetic steps to arrive at the final folded product, while Na+-dependent folding occurs in four kinetic steps. We also found that removal of the 3′A from ODN2 results in both an increase in cation affinity compared to ODN2 as well an increase in folding rate, suggesting that accessibility of cations to the interior of partially folded intermediates may be an important determinant of folding rate.
MATERIALS AND METHODS
Oligonucleotides
Desalted oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA, USA). They were dissolved in folding buffer (10 mM tetrabutylammonium phosphate, 1 mM EDTA, pH 7.0) as 500–900 µM stock solutions and stored at 4°C. This buffer system was chosen because the relatively large size of the tetrabutylammonium cation precludes its ability to stabilize quadruplex structures. Oligonucleotide concentrations were estimated from the absorbance at 260 nm determined at room temperature with a JASCO V-550 spectrophotometer using ε = 228.5 mM−1 cm−1, 230.6 mM−1 cm−1 and 244.3 mM−1 cm−1 for the unfolded forms of ODN1, ODN2 and ODN3, respectively (49).
Other reagents
Monobasic tetrabutylammonium phosphate (Fluka), ethylenediamine tetraacetic acid (ACS reagent) and KCl (Fluka) were obtained from Sigma Chemical Co. (St Louis, MO, USA). NaCl (ACS reagent) was from J.T. Baker (Phillipsburg, NJ, USA). Folding buffer was prepared by titration to the desired pH with tetrabutylammonium hydroxide (40% w:v in water, Sigma).
Equilibrium titrations
The extent of folding was assessed by measuring changes in oligonucletodide UV absorbance as a function of added KCl or NaCl. Not only are these experiments of intrinsic interest, but also are required as a prelude to the stopped flow mixing experiments to establish the expected changes in absorption and to determine minimal cation concentrations necessary to induce complete folding. Absorption spectra were measured at 1 nm intervals from 220 to 340 nm after serial additions of either KCl or NaCl with a JASCO V-550 spectrophotometer equipped with a magnetic stirrer and a peltier-thermostatted cuvette holder maintained at 25°C. Equal volumes of KCl or NaCl in folding buffer were added to sample and reference cuvettes that contained either the oligonucleotide in folding buffer or folding buffer alone. Final oligonucleotide concentrations were in the range 6–10 µM. The spectra were corrected for oligonucleotide dilution (<6%) resulting from the addition of the cation solution. Difference spectra 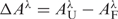 were generated by subtracting the absorption spectrum obtained after each addition of cation from the spectrum of the fully unfolded oligonucleotide. The resulting titration data thus consist of a matrix of elements ▵Ai,j of cation concentration i and wavelength j.
were generated by subtracting the absorption spectrum obtained after each addition of cation from the spectrum of the fully unfolded oligonucleotide. The resulting titration data thus consist of a matrix of elements ▵Ai,j of cation concentration i and wavelength j.
Analysis of titration data
The titration curves were initially assumed to fit a two-state cation-induced folding mechanism:
| 1 |
where U and F represent the unfolded and folded states of the oligonucleotide and M is a cation. Preliminary analysis of the titration data indicated that cation binding is cooperative. We therefore fit the data to the Equation 2 (the classical Hill equation) using the nonlinear least squares module of the program Origin 7.0 (OriginLab Corp., Northampton, MA, USA) to optimize values of the adjustable constants n and K0.5:
| 2 |
where αi = ▵Ai/▵Amax, [M]i is the cation concentration at αi, K0.5 = [M] at αi = 0.5 and n is the Hill coefficient, which can be interpreted as the average number of bound cations at αi = 0.5.
Kinetic experiments
The kinetics of oligonucleotide folding was assessed by measuring time-dependent changes in UV absorbance with a stopped flow mixing device equipped with a rapid scanning monochromator (On-Line Instrument Systems, Bogart, GA, USA). Absorption spectra consisting of 200 equally spaced data points covering the wavelength range of 255–364 nm were collected with a time resolution of up to 1 ms/spectrum. Data points collected at wavelengths <270 nm and at ‘zero’ time (the instant of flow stop) were discarded prior to analysis because of low signal to noise ratio at the shorter wavelengths and because the ‘zero’ time point was unreliable due to mixing artifacts. A circulating water bath was used to maintain a constant reaction temperature. The kinetic experiments were carried out under pseudo first-order conditions (e.g. [cation] >> [oligonucleotide]). Data sets for three to five successive mixing experiments were averaged for analysis. Control experiments consisting of mixing oligonucleotide with cation-free buffer and mixing cation-free buffer with buffer containing high salt indicated the absence of absorbance changes due to oligonucleotide dilution or artifacts due to changes in refractive index resulting from mixing low and high salt solutions.
The reaction progress curves were analyzed in terms of difference spectra (▵Aλ versus time) generated by subtracting the absorption spectrum determined at time t from the absorption spectrum of the completely folded oligonucleotide, e.g. 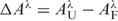 . Thus, the data sets consisted of a matrix A of elements Ai,j = ▵A at λi and tj. The data matrices were analyzed by singular value decomposition (SVD) (see below) and nonlinear least squares fitting to rate equations corresponding to first-order coupled reactions. The program Specfit/32 (version 3.0.39, Spectrum Software Associates, Marlborough, MA, USA) was used for these analyses. The criterion for an acceptable fit to a mechanism was minimal systematic deviations of the fitted progress curves from the experimental data points. For convenience in discussion, the optimized pseudo first-order rate constants thus obtained have been converted to relaxation times τ from the relationship τi = 1/ki.
. Thus, the data sets consisted of a matrix A of elements Ai,j = ▵A at λi and tj. The data matrices were analyzed by singular value decomposition (SVD) (see below) and nonlinear least squares fitting to rate equations corresponding to first-order coupled reactions. The program Specfit/32 (version 3.0.39, Spectrum Software Associates, Marlborough, MA, USA) was used for these analyses. The criterion for an acceptable fit to a mechanism was minimal systematic deviations of the fitted progress curves from the experimental data points. For convenience in discussion, the optimized pseudo first-order rate constants thus obtained have been converted to relaxation times τ from the relationship τi = 1/ki.
Analysis of equilibrium and kinetic data by SVD
As stated above, major goal of this study was to suggest plausible pathways for the cation-induced formation of G-quadruplexes and to investigate selected structural variables that might influence these pathways. An important tool in this analysis is enumeration of significant kinetic and equilibrium intermediates present as the unfolded state progresses to the fully folded state. SVD analysis of the data facilitates definition of the minimum number of absorbing species required to fit the multiwavelength data sets and thereby rigorously test the validity of the two-state folding model.
In brief, SVD is a method of analyzing large data sets that aids in distinguishing significant signals from noise, provides a relative measure of the contribution of each signal to the total, and for spectroscopic data, defines basis spectra that show the contribution of each species to the experimental spectra. If the data are fit to an analytical expression describing an explicit reaction mechanism SVD provides the actual absorption spectra of the reactants, products and any intermediates, their concentration profiles as a function of an experimental variable such as ligand concentration or time, the best fitting constants for the mechanism and an estimate of the quality of the fit. Details of SVD data analysis as applied to biochemical equilibria and kinetics are available in a number of comprehensive reviews (50–52); a summary of the SVD analyses used in these studies is available in the Supplementary Data of this article.
RESULTS
Folding equilibria
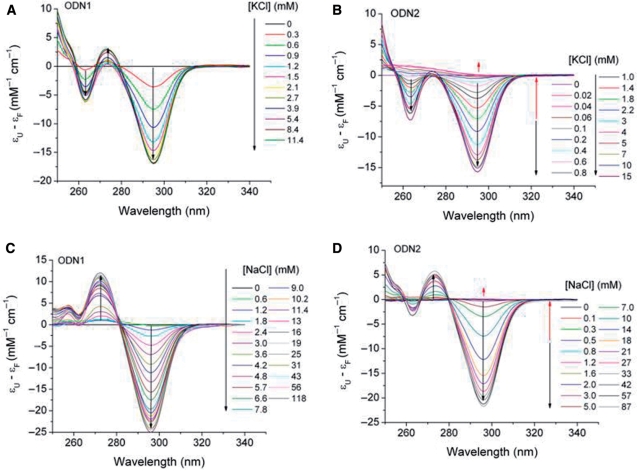
It has been previously established that cation-dependent formation of G-quadruplex structures is accompanied by characteristic changes in the UV absorption spectra of folded compared to unfolded oligonucleotides (48,53). These changes generally involve an increase in absorbance at ∼295 nm for the folded state. To determine the expected absorbance changes and their dependence on cation concentration for the oligonucleotides in this study, we measured the UV spectra of ODN1, ODN2 and ODN3 as a function of KCl and NaCl concentrations. The results of representative experiments are shown in Figure 2 for ODN1 and ODN2 and for ODN3 in Supplementary Figure S1. The difference spectra show that K+-induced folding of ODN1 and ODN2 is accompanied by relatively small absorbance changes between 250 and 275 nm and somewhat larger changes between 275 and 300 nm, with maxima and minima at ∼265 and ∼295 nm, respectively (Figure 2A and B). Na+-induced folding of both oligonucleotides showed a similar absorbance increase at ∼295 nm; however, in contrast to folding in KCl, a decrease in absorbance was noted in the 275 nm spectral region (Figure 2C and D). The spectral changes in the wavelength range 250–340 nm shown in Figure 2 were achieved within a few minutes after mixing and were stable for at least 1 h; however, we observed a small spectral drift of unknown origin at wavelengths below ∼250 nm for folding ODN1 and ODN3. Thus, data points at wavelengths <250 nm were removed from the data sets prior to their evaluation.
Figure 2.
Spectrophotometric titrations of oligonucleotide models of the human telomeric sequence X-G3(T2AG3)3-Y. In (A and C), ODN1 (X = T, Y = 0) was titrated with KCl or NaCl, respectively. In (B and D), ODN2 (X = TT, Y = A) was titrated with KCl or NaCl, respectively. The arrows in the figures indicate decreasing or increasing absorbance changes with successive additions of cation. The final cation concentrations after each addition are shown at the right in each panel. Difference spectra are presented as the difference between the absorption coefficients ε for the unfolded (U) and the folded (F) structures. Oligonucleotide strand concentrations were 4–6 µM in folding buffer (10 mM tetrabutylammonium phosphate, 1 mM EDTA, pH 6.97). The temperature was 25°C.
Additional notable features characterizing the spectra in Figure 2 include a lack of isosbestic wavelengths and a biphasic absorbance change at ∼295 nm when ODN2 was titrated with either KCl or NaCl. As indicated by the direction of the arrows in Figure 2, the absorbance in the 295 nm range decreased with increasing cation concentration; however, above 0.2–0.3 mM cation, the absorbance at this wavelength increased with increasing cation concentration. The lack of isosbestic points and the biphasic absorbance changes suggest the presence of appreciable concentrations of one or more spectroscopically distinct intermediates. This conclusion is supported by the SVD analysis given in the Supplementary Data, where the basis spectra and concentration profiles of the spectroscopically significant titration intermediates are summarized.
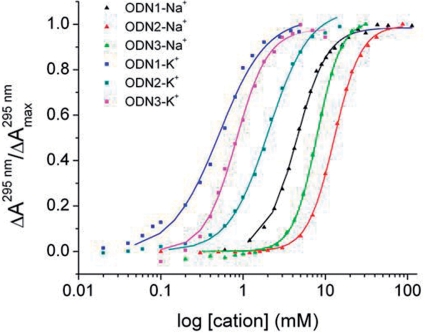
Titration curves for K+ and Na+-induced folding of ODN1, ODN2 and ODN3 derived from the absorbance changes at 295 nm are shown in Figure 3. The lines superimposed on the experimental data points represent the best fit of the data to Equation 2. The optimal values of the parameters K0.5 (the ligand concentration for half saturation) and n (the index of cooperativity) are summarized in Table 1. Hill coefficients for K+ binding were between 1.5 and 2.2, whereas for Na+ binding, n ranged from 2.4 to 2.9 depending on the oligonucleotide sequence. Hill coefficients >1 indicate that oligonucleotide folding is positively cooperative with respect to cation concentrations; the observed values of n are consistent with the predicted binding stoichiometry of two K+ and three Na+ bound per oligonucleotide, which can accommodate a K+ ion between each of the three quartets or a Na+ ion within the central cavity of each quartet.
Figure 3.
Fraction of folded oligonucleotide (α) as a function of added cation. The points were derived from the absorbance changes at 295 nm shown in Figure 2 for ODN1 and ODN2. For ODN3, the points are from spectra in Figure S1. The lines were derived by fitting the points to the Hill equation as described in the text. The resulting optimal fitting parameters n (Hill coefficient) and K0.5 (cation concentration at half the saturation) are summarized in Table 1.
Table 1.
Optimal parameters for fitting cation-binding curves in Figure 2
| ODN | Sequence | Cation | K0.5 (mM) | Hill coefficient, n |
|---|---|---|---|---|
| 1 | d[AGGG(TTAGGG)3] | K+ | 0.51 ± 0.03 | 1.5 ± 0.1 |
| Na+ | 4.40 ± 0.04 | 2.38 ± 0.05 | ||
| 2 | d[TTGGG(TTAGGG)3A] | K+ | 2.05 ± 0.08 | 1.76 ± 0.07 |
| Na+ | 12.8 ± 0.1 | 2.38 ± 0.05 | ||
| 3 | d[TTGGG(TTAGGG)3] | K+ | 0.82 ± 0.03 | 2.2 ± 0.1 |
| Na+ | 7.9 ± 0.1 | 2.9 ± 0.1 |
Detection of equilibrium folding intermediates by SVD analysis
As mentioned earlier, the lack of isosbestic points in the cation-dependent absorption spectra, the biphasic changes in absorbance at ∼295 nm and the less than maximal Hill coefficients suggest significant concentrations of stable intermediates during the equilibrium titrations. Support for this conclusion is in the Supplementary Data. Briefly, we conclude from SVD analysis of the data in Figure 2 and Figure S1 that two or three distinct spectroscopic species contribute to the absorbance changes during the titration of all three oligonucleotides by either K+ or Na+ ions. The basis spectra for the intermediates and their concentration profiles during the titration are given in Figures S2–S4 in Supplementary Data. The major basis spectrum is contributed by the absorbance difference between the unfolded and folded species (e.g. AU – AF) while the minor basis spectra reflect the presence of partially saturated and/or partially folded intermediates (e.g. AI – AF). The concentration of the intermediates never constitutes more than a few percent of the total; at any point in the titration, the major species present are either the unfolded or the fully folded structures.
Folding kinetics
Cation-dependent folding kinetics of model telomeric oligonucleotides
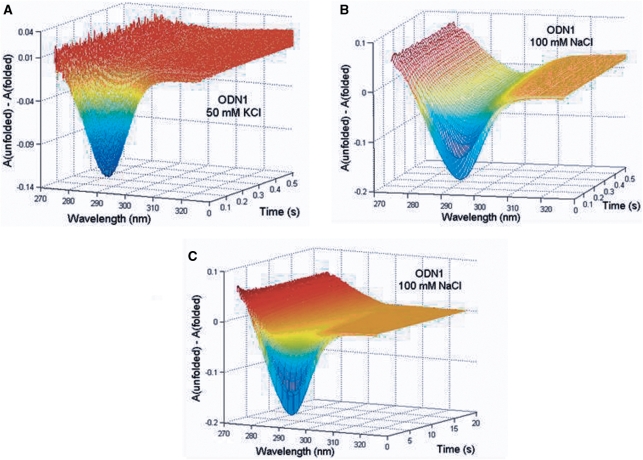
Figure 4 shows representative 3D plots of time-dependent changes in the UV difference spectra 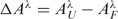 for folding of ∼5 µM ODN1 induced by 50 mM KCl (Panel A) and by 100 mM NaCl (Panels B and C). Limitations in signal-to-noise ratio prevented investigating the kinetics of folding over a large range of oligonucleotide concentrations; however, the kinetics of ODN1 folding was similar at both 3 and 8 µM strand concentrations, indicating that the observed folding kinetics is unimolecular with respect to oligonucleotide.
for folding of ∼5 µM ODN1 induced by 50 mM KCl (Panel A) and by 100 mM NaCl (Panels B and C). Limitations in signal-to-noise ratio prevented investigating the kinetics of folding over a large range of oligonucleotide concentrations; however, the kinetics of ODN1 folding was similar at both 3 and 8 µM strand concentrations, indicating that the observed folding kinetics is unimolecular with respect to oligonucleotide.
Figure 4.
Representative 3D plots of the wavelength and time dependence of spectral changes accompanying cation-induced folding of ODN1. (A) Shows the time-dependent evolution of absorbance changes between 270 and 340 nm for K+-induced folding of ODN1. (B and C) Illustrate resolution of absorbance changes into rapid (B) and slower (C) time domains for Na+-induced folding of ODN1. ODN1 (8 µM) was mixed with 100 mM KCl or 200 mM NaCl (all reagents in folding buffer). The temperature was 25°C. The reactants were diluted 2-fold and the path length of the observation cuvette was 2.0 cm.
Both data sets in Figure 4 clearly show the kinetic evolution of absorption changes in the 295 region that constitute the spectroscopic signature of a G-quadruplex. K+-induced folding is characterized by rapid monophasic changes in the oligonucleotide absorption spectrum that were complete within 100–200 ms, while Na+-induced folding exhibited a multiphasic time course characterized by rapid absorbance changes that were complete within a few hundred ms followed by relatively slower changes that required 20–25 s to complete. The multiphasic nature of the Na+-dependent progress curves are clearly shown by comparing panels B and C, which depict the absorbance changes on two different time scales. Similar kinetic data were obtained for folding ODN2 and ODN3.
Analysis of cation-induced folding kinetics
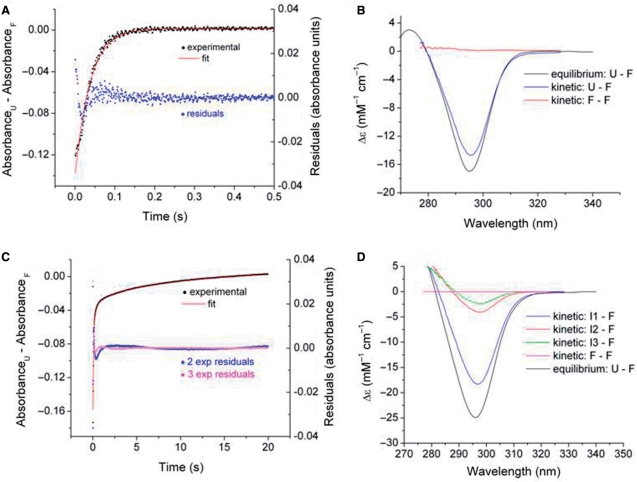
Kinetic data matrices consisting of rows of time points and columns of ▵A values at specific wavelengths as shown in Figure 4 were analyzed by SVD and nonlinear least squares fitting procedures to establish (i) the minimum kinetic mechanism that fit the data; (ii) the kinetic difference spectra of the species involved and (iii) the associated rate constants and their standard errors. Figure 5 shows the results of an analysis of the data set for K+-induced folding data for ODN1 in Figure 4A. SVD analysis revealed that the time–wavelength profile could be adequately represented by a two-state reaction U → F. Fitting the data matrix to a single exponential process resulted in an adequate representation of the progress curve for the reaction. This is illustrated by the congruence of the experimental and calculated progress curves at the 295 nm and the residual plot shown in Figure 5A. The relatively large residual for the initial data points can be attributed to a mixing artifact. For the data set shown, the best fitting value of the observed pseudo first-order rate constant kobs is 26.7 ± 0.2 s−1. Similar monophasic kinetics results observed for K+-induced folding of ODN2 and ODN3 (kobs values of 4.6 ± 0.2 s−1 and 15.8 ± 0.3 s−1, respectively). Complete results for K+-induced folding of ODN2 and ODN3 are given in the Supplementary Data (Figures S5–S7).
Figure 5.
SVD and nonlinear least squares analysis for K+- and Na+-induced folding of ODN1 (data sets of Figure 4). Panels A anc C compare experimental progress curves at 295 nm to simulated curves generated by global fitting of the wavelength–time courses data matrix to a single exponential process for K+ (A) folding and triple exponential processes Na+ folding of ODN1 (C). The optimal rate constants are given in the text. The residual plots (A and C) indicate the deviations of the experimental and fitted absorbance changes. The kinetic and static difference spectra are compared in (B) for K+-driven folding and in (D) for Na+-driven folding. The equilibrium difference spectra are taken from the titration data of Figure 2 and the kinetic difference spectra were derived by SVD and least square fitting procedures as described in the text. The Δɛ values are derived with reference to the fully folded structure.
SVD analysis of multiwavelength kinetic data coupled with fitting to a specific mechanism allows determination of the kinetic spectra of the kinetically significant species. Since our analysis of the K+-induced folding data can be described by a single exponential process, the significant spectra will be those of the unfolded species U and the folded species F. Since the difference spectral data were analyzed, the basis spectra are represented by ▵A1 = AU – AF and ▵A2 = AU – AU. Obviously, the latter difference spectrum will consist of a null spectrum if only two species, U and F, are present during the course of the reaction. As shown in Figure 5B, this expectation is fulfilled for K+-induced folding of ODN1. The kinetic difference spectrum for ODN1-KCl that represents the difference in absorbance between U and F, overlays to within ∼10% with the difference spectrum determined by equilibrium titration. This result indicates that ∼90% of the expected absorbance change was observable in the stopped flow apparatus; the unobserved change most likely can be accounted for by the dead time of the apparatus. The second significant difference spectrum is essentially a null spectrum as expected for the two-species reaction pathway. Similar results were obtained for K+-induced folding of ODN2 and ODN3 (Figure S8).
The kinetics of Na+-dependent oligonucleotide folding was analyzed in a similar fashion as described above for K+-induced folding. SVD and curve fitting analysis showed that three sequential exponential processes were required to adequately reproduce the data sets for folding of all of the oligonucleotides in Na+ solutions. The results of a representative experiment are shown in Figure 5C and D for the data matrix for Na+-induced folding of ODN1 depicted in Figure 4B and C. Fitting to single and double exponential rate expressions resulted in large systematic deviations of the fitted and experimental progress curves. Figure 5C shows that three sequential exponentials (modeling the U → I1 → I2 → F reaction pathway) were required to match the experimentally observed progress curve at 294 nm. The residual plots show that the three-exponential fit (magenta points) gives a significantly better fit of the progress curve than two exponential fit (blue points). The optimal rate constants for ODN1 folding in 100 mM NaCl (Figure 4B and C) are 11.9 ± 0.1, 1.07 ± 0.05, 0.0095 ± 0.0020 s−1. The corresponding constants for ODN2 and ODN3 in 100 mM NaCl are 14.5 ± 0.6, 3.0 ± 0.2 and 0.7 ± 0.1 s−1 for ODN2 and 26.2 ± 1.4, 4.0 ± 0.3 and 0.67 ± 0.03 s−1 for ODN3, respectively.
The kinetic difference spectra associated with the ODN1/100 mM NaCl data set are illustrated in Figure 5D. As with K+, the most significant difference spectrum is that for U–F with the less intense spectra attributed to intermediates U − I1 and U − I2. An overlay of the kinetic and static difference spectra are in reasonable agreement, which indicates that the kinetics account for the expected absorbance change as determined over the relevant wavelength range. The corresponding fits for ODN2 and ODN3 are given in the Supplementary Data section (Figure 5B).
Dependence of the observed rate constant kobs on [KCl] and [NaCl]
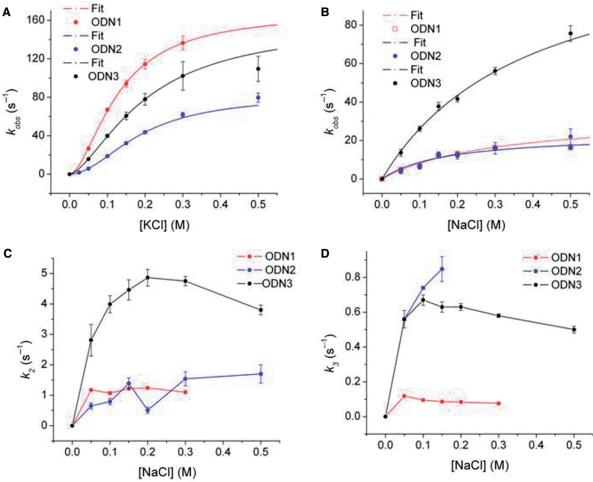
Previous studies have shown that cation exchange in G-quadruplexes occurs on a microsecond time scale (6,54–56). Based on this, we expected that the folding rates might be independent of cation concentration on the millisecond time scale accessible to stopped-flow mixing. However, as shown in Figure 6A and B, the observed folding rate constants (kobs) increased with cation concentration in a nonlinear fashion, approaching maximal values at higher concentrations. The functional relationship between kobs and [M+] differed for the two cations in that kobs for K+-induced folding exhibited a sigmoidal dependence on cation concentration, while the relationship between kobs and [Na+] was hyperbolic. This type of saturation behavior can be mechanistically interpreted as a cation-dependent preequilibration step, e.g. a rate-limiting isomerization of an intermediate that is stabilized by bound cations. The sigmoidal dependence of kobs on [K+] (Figure 6A) suggests that the intermediate contains more than one bound cation (mechanism 3):
| 3 |
Equation 4 below describes the dependence of such a rate-limiting isomerization mechanism on ligand binding as a function of ligand concentration [M+]:
| 4 |
where k2 is the value of kobs at saturating cation concentration, K = (k−1 + k2)/k1 and n is related to the number of M+ ions involved in formation of the complex. To obtain optimal values of the parameters n, K and k2, the data in Figure 6A, were fit by nonlinear least squares to Equation 4; the resulting parameter values are summarized in Table 2. The values of n for K+-driven folding of ODN1, ODN1 and ODN3 are 1.6–1.9, suggesting that formation of the preequilibrium complex requires the participation of two K+ ions in keeping with the known stoichiometry of two K+ per three quartets. Values for k2 (which can be interpreted as the rate of formation of the final folded form from the preequilibrium complex) are ∼160 s−1 for ODN1 and ODN3 and ∼80 s−1 for ODN2. These rate constants convert to relaxation times of about 6 and 12 ms for the rate-limiting step of K+-dependent folding of these structures.
Figure 6.
Dependence of the rate constants for oligonucleotide folding on cation concentration. (A) Shows sigmoidal dependence of kobs on [KCl] for ODN1, ODN2 and ODN3. The lines illustrate the best fit of the data to Equation 4 determined by nonlinear least squares analysis. The line was calculated using the optimal values for n, k2 and K in Table 2. (B) Shows a similar fit on [NaCl] of the fastest rate constant for ODN1, ODN2 and ODN3 folding. The lines represent the dependence of kobs on [NaCl] calculated using the optimal values of k2 and K in Table 2. (C and D) Show the dependence of the rate constants for the slower relaxations (k3 and k4) on [NaCl]. The lines connect the data points but have no theoretical basis. The error bars represent the SD of the fitted parameter.
Table 2.
Optimimal parameters for fitting cation dependence of kobs in Figure 6A and B
| ODN | Cation | K (mM) | k1max (s-1) | n |
|---|---|---|---|---|
| 1 | K+ | 0.129 ± 0.004 | 167.9 ± 4.1 | 1.73 ± 0.09a |
| Na+ | 0.07 ± 0.03 | 20.6 ± 3.5 | 1b | |
| 2 | K+ | 0.188 ± 0.009 | 82.4 ± 4.1 | 1.93 ± 0.05 |
| Na+ | 0.17 ± 0.05 | 23.4 ± 2.8 | 1b | |
| 3 | K+ | 0.201 ± 0.005 | 156.9 ± 2.9 | 1.56 ± 0.02 |
| Na+ | 0.40 ± 0.08 | 129 ± 17 | 1b |
aKCl data fit to Equation 5 (Hill equation).
bNaCl data fit to Equation 7 (hyperbola).
As shown earlier, the kinetics of Na+-driven oligonucleotide folding required three exponential processes with time constants differing by ∼10-fold. This suggests that Na+-induced folding requires at least three intermediates:
| 5 |
As observed with K+-induced folding, the rate of the most rapid relaxation in Na+ approached a limiting value as the concentration of the ion concentration increased. However, in contrast to the folding kinetics in K+ solution, the relationship between kobs and [M+] was hyperbolic rather than sigmoidal (i.e. n = 1 in Equation 4). Equation 6 thus describes the relationship between the rapid formation of I1 and the rate of its conversion to I2:
| 6 |
The hyperbolic dependence of kobs on [Na+] suggests that a single sodium ion is required for formation of intermediate I1. The rates of formation of I2 and I3 also were [Na+]-dependent, suggesting that formation of these structures also involves stabilization of intermediates I2 and I3 by binding a second and third cation (Figure 6B and C).
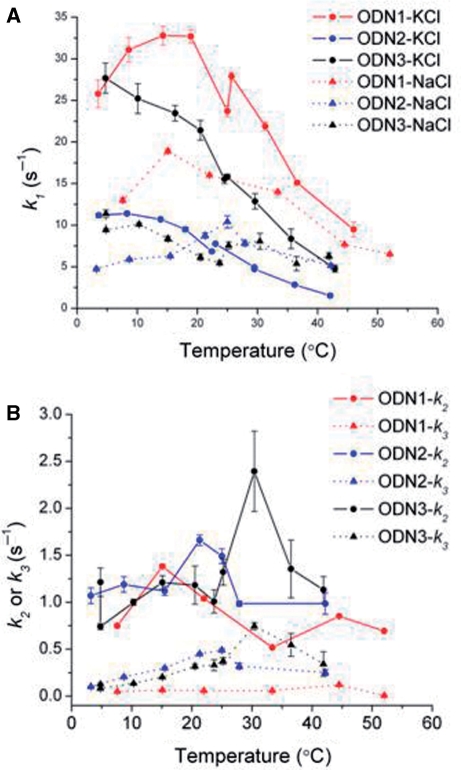
Temperature dependence of folding
Determination of the effect of temperature on the rate of biopolymer folding can provide useful information about the folding pathway that can suggest characteristics of the transition state ensemble, defined as high-energy conformations that lie on the pathway between the unfolded and folded states (33,57–59). In favorable cases, the thermodynamic activation parameters ▵G*, ▵H* and ▵S* associated with formation of the rate-limiting, high energy ensemble can be determined (33,58,60). Nonlinear Eyring plots (ln k/T versus 1/T) are frequently found with biopolymer folding reactions rather than the linear Arrhenius relationship characteristic of the temperature dependence of the rate of a simple chemical reaction. In the current study, anomalous Eyring plots were observed for cation-induced G-quadruplex folding. Figure 7A shows that kobs for ODN1 and ODN2 folding exhibited temperature optima at ∼10–20°C. The rate of K+-induced folding of ODN1 exhibited the greatest temperature dependence with kobs decreasing ∼5-fold between 15°C and 45°C. The rate of Na+-dependent folding of ODN2 was less sensitive to temperature, showing an approximate 2-fold decrease in the same temperature range. For ODN1 folding in Na+, kobs increased as temperature increased from 7°C to 15°C, but then decreased with further temperature increases. The slower rate constants k2 and k3 associated with the slower steps of Na+-induced folding also exhibited small variations with temperature. The mechanistic implications of these negative apparent activation energies are discussed below.
Figure 7.
Temperature-dependence of the rate constant kobs for K+- and Na+-induced folding of ODN1, ODN2 and ODN3 (A) and for the slower relaxations associated with folding in NaCl (B). The error bars represent the SD of the fitted parameter.
DISCUSSION
The primary purpose of the experiments presented here is to define the kinetics of intramolecular folding of G-quadruplex structures and from these data derive a self-consistent mechanism that fits with the known structures of the folded oligonucleotides. As indicated in the Introduction section, a search of the literature revealed few published studies that directly examine the folding kinetics of simple G-quadruplexes; to our knowledge, no kinetic study has been published that utilizes fold-specific changes in the UV spectrum to define the kinetics of quadruplex formation. We therefore undertook the current study using rapid multimixing stopped flow spectrophotometry to define the time-dependent changes in absorption spectra that accompany cation-driven quadruplex formation. The cations chosen were the major intracellular cation K+ along with Na+; the latter may play a regulatory role in conformational switching of G-quadruplexes by virtue of its ability to induce structural rearrangements in quadruplexes (22). The oligonucleotides chosen include two models of the human telomere, d[AG3(TTAGGG)3] (ODN1) and d[TTG3(TTAGGG)3A] (ODN2) for which solution structures have been defined by NMR. In addition, we determined the effect on the folding kinetics of deletion of the 3′-A of ODN2. This base stacks against the penultimate guanine and therefore could influence the access of ions to their binding sites within the quadruplex. Lastly, as an integral part of the kinetic analysis to define the spectroscopic endpoints of folding, we measured cation-induced folding isotherms as a means of assessing cation dependence of folding in which the data were analyzed in detail by SVD to suggest the presence of intermediate spectroscopic species. These studies provide new information on the kinetics and equilibria of cation-induced folding for three simple models of a human telomere.
The equilibrium titrations show that both K+ and Na+ induce human telomeric oligonucleotides to fold in a cooperative fashion with Hill coefficients consistent with the known stoichiometry of two K+-binding sites between G-quartets and three Na+-binding sites located within each quartet. The spectroscopic changes and well-defined stoichiometry of cation binding indicate that the titration and kinetic experiments reflect specific binding events rather than nonspecific electrostatic interactions between cations and backbone phosphates, which, for G-quadruplexes, has been shown not to exhibit a preference for either cation (61). In agreement with previous studies (61), apparent binding affinities for K+ were greater than that for Na+. However, even though cation binding is known to be necessary for stabilization of individual G-quartets, cooperativity has not been previously reported, nor has direct evidence of partially liganded quartets been presented. Analysis of the wavelength dependence of the titration curves suggests the presence of intermediates with less than a full stoichiometric complement of cations. This result implies a sequential mechanism of cation stabilization rather than a concerted one. This is suggested by the observation that the Hill coefficients associated with the binding reactions were generally less than the theoretical maxima for each of the oligonucleotides studied with the exception of ODN3 in Na+ solution, where the n-value approached the theoretical maximum of 3.0.
The kinetic studies reveal that cation-induced folding of these monomeric model telomeric sequences probably proceeds through rapid formation of an intermediate cation–oligonucleotide complex that subsequently isomerizes in a slower step(s) to the fully folded structure. For K+-induced folding, a single kinetically significant isomerization was observed, while Na+-induced folding was more complex, requiring three steps subsequent to the initial rapid equilibrium step to achieve a final folded structure.
The apparent negative activation energy of folding is consistent with a variety of complex folding mechanisms, including a change in rate-limiting step with temperature, a temperature-induced change in stability of the ground states or a change in the heat capacity Cp of the activated complex (58) brought about by temperature-dependence of ▵H*. It is conceivable that all of these factors may contribute to the observed negative temperature dependence of quadruplex folding. First, saturation kinetics implies a multistep folding pathway in which different steps may become rate limiting at different temperatures. Second, recent studies demonstrate that G-quadruplex folding is accompanied by a significant ▵Cp (62). Changes in Cp during nucleic acid folding have been attributed both to a decrease in exposure of nonpolar (particularly guanine groups) and polar groups to solvent as well as base stacking (62).
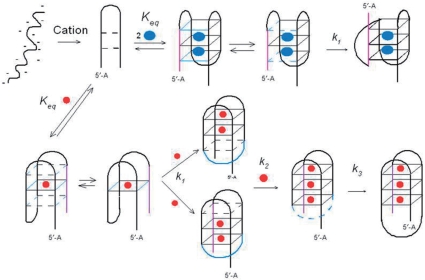
A hypothetical folding pathway that provides a structural framework for interpretation of the complex kinetics of cation-driven folding of G-quadruplexes is illustrated in Figure 8. This pathway, which is modified from one recently suggested by Mashimo et al. (63), incorporates the kinetic evidence in this article. At low ionic strength, oligonucleotides probably exist as an ensemble of extended structures as dictated by internal electrostatic repulsion of the backbone phosphate anions. Introduction of quadruplex-stabilizing cations will rapidly reduce this electrostatic repulsion in accordance with polyelectrolyte theory (23), which indicates that a shell of loosely bound cations is associated with polynucleotides. This charge neutralization will result in the collapse of the extended structures to more compact structures (the electrostatic collapse shown in Figure 8). Such a collapse has been discussed previously with respect to nucleotide folding and conceptually resembles the hydrophobic collapse that drives protein folding (6,34). For the specific experimental starting conditions of these studies (e.g. 10 mM tetrabutylammonium phosphate), the ionic strength is appreciable and the unfolded starting ensemble probably has already collapsed into compact structures.
Figure 8.
Schematic illustration of a hypothetical reaction scheme for cation-induced folding of oligonucleotide models of the human telomere. The orientation of the structures in the diagram is the same as in Figure 1. Blue spheres represent K+ ions and red spheres represent Na+ ions. For simplicity, single initial and intermediate conformers are illustrated, although these most likely consist of an ensemble of rapidly equilibrating, approximately isoenergetic structures. For example, identification of the central quartet in the lower pathway as the site of binding for the first Na+ is arbitrary. The equilibrium states of ODN1 in Na+ and ODN2 in K+ (shown in Figure 1) consist of single conformers. In contrast, ODN1 in K+ solution consists of a mixture of conformers.
In the second step, in the diagram, a small fraction of the collapsed structures may consist of hairpins with internal H-bonds between appropriate bases; e.g. hairpins in proper register for folding into stacked G-quartets could be stabilized by intrachain hydrogen bonding between A8 and T18 located in the loop regions in ODN2. Such correctly folded structures could then spontaneously fold back on themselves to generate H-bond stabilized quartets. Cation incorporation at specific binding sites either within or between quartets would then stabilize the stacked structures. The dependence of the folding rate on cation concentration (Figure 6) suggests that two K+ ions or one Na+ ion may stabilize these intermediates. For the K+-stabilized structures, the rate-limiting step is evidently folding of the thermodynamically stable structure, which for ODN2 involves formation of the transverse loop in Figure 8. As indicated in the diagram, the G2–G4 H-bonded structures may be in equilibrium with the H-bond free structures, thereby allowing rotation to occur about G3 of the H-bond free conformers, allowing formation of the antiparallel strand from the parallel orientation Fluctuations in polynucleotide H-bonding has been shown to take place on a millisecond time scale in homopolymers (64). We suggest that this conformational rearrangement may be the rate-limiting step characterized by k2 in reaction sequence 3.
Na+-stabilized quadruplexes are generally topographically more complex than the K+-bound structures. A kinetic consequence of this structural complexity is kinetic complexity, demonstrated here by the two additional relaxations required to achieve a stable folded state. The dependence of the intermediate relaxation k2 on a single Na+ ion (Figure 6B) suggests that this complex may consist of a double hairpin structure stabilized by one bound Na+ ion. This structure could undergo transient separation of the two arms of the double hairpin followed by a rotation about the loop connecting strands one and two of the initial chair structure. This conformational change resulting in formation of the transverse loop at the bottom of Figure 8 could be driven by H-bonding involving the first and fourth strands and also by incorporation of a Na+ ion within the quartet. The dependence of k2 and k3 on [Na+] suggests that these steps involve stepwise cation-dependent conformational changes.
CONCLUSIONS
Folding of cation-driven human telomeric model oligonucleotides is cooperative with respect to cation concentration in equilibrium titrations; however, the binding equilibria were not consistent with a two-state, all-or-none folding reaction as small concentrations of spectroscopically distinct intermediates were detected. These species presumable are partially folded species with less than a full complement of bound cations.
The kinetics of K+-induced folding occurs in two steps. The data are consistent with the folding pathway U + 2K+ ⇋ I − K + 2 → F − K + 2. Intermediate I may be a chair structure and the rate limiting step may involve a conformational change forming the lateral loop connecting strands one and two of the quadruplex. The folding process is relatively rapid at physiological concentrations of K+, requiring about 6 ms for formation of the fully folded structure.
For Na+-driven folding, three exponential relaxations are evident. As with K+, the cation dependence of the rapid relaxation suggests formation of a Na+-oligonucleotide complex that subsequently undergoes a series of kinetically significant conformational changes as the final folded structure is attained. A minimal pathway for Na+-induced folding that is consistent with the kinetic data is U + 3Na+ ⇋ I1 − Na+ → I2 − Na+ → I3 − Na+ → F − 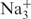 . The actual stoichiometry of Na+ binding to the intermediates remains to be determined, but may involve successive additions of ions to the partially folded structures rather than the concerted addition of K+ suggested by the dependence of kobs on [K+] (Figure 6). The multistep, Na+-driven folding kinetics may result from the more complex topography of the quadruplexes stabilized by this ion, which requires two loop rearrangements to give double-strand reversals. In the presence of Na+ as the sole cation, formation of the initial folding intermediate is nearly as fast as with K+, but the presence of the two additional conformational rearrangements (requiring formation of multiple new H-bonding partners) increases the time for complete folding to as much as 25 s for the model sequences studied.
. The actual stoichiometry of Na+ binding to the intermediates remains to be determined, but may involve successive additions of ions to the partially folded structures rather than the concerted addition of K+ suggested by the dependence of kobs on [K+] (Figure 6). The multistep, Na+-driven folding kinetics may result from the more complex topography of the quadruplexes stabilized by this ion, which requires two loop rearrangements to give double-strand reversals. In the presence of Na+ as the sole cation, formation of the initial folding intermediate is nearly as fast as with K+, but the presence of the two additional conformational rearrangements (requiring formation of multiple new H-bonding partners) increases the time for complete folding to as much as 25 s for the model sequences studied.
As a final interesting observation, the number and identity of bases at the 5′ end of the quadruplex and addition of a base to the 3′ end of the quadruplex affects the rate of folding but does not alter the basic multistep mechanism. In particular, truncation of ODN2 by deletion of the 3′A markedly increases the limiting rate of the initial step in Na+-induced folding but has less influence on the limiting rate of K+-induced folding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank these colleagues for discussion and helpful comments: Andrew Lane, Nichola Garbett, John Trent, Luigi Petraccone and Chongkham Mekmaysy. This research was supported by National Cancer Institute grant CA35635. Funding to pay the Open Access publication charges for this article was provided by the James Graham Brown Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 2.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2001;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 5.Parkinson GN. Fundamentals of quadruplex structures. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 1–30. [Google Scholar]
- 6.Murthy VL, Rose GD. Is counterion delocalization responsible for collapse in RNA folding? Biochemistry. 2000;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dapic V, Bates PJ, Trent JO, Rodger A, Thomas SD, Miller DM. Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry. 2002;41:3676–3685. doi: 10.1021/bi0119520. [DOI] [PubMed] [Google Scholar]
- 11.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 12.Qi H, Lin CP, Fu X, Wood LM, Liu AA, Tsai YC, Chen Y, Barbieri CM, Pilch DS, Liu LF. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- 13.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 14.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Kan Z, Zeng Z, Hao Y, Chen H, Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J. Am. Chem. Soc. 2004;126:13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 16.Han H, Hurley LH. G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 17.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldrich E, O'Sullivan CK. Ability of thrombin to act as molecular chaperone, inducing formation of quadruplex structure of thrombin-binding aptamer. Anal. Biochem. 2005;341:194–197. doi: 10.1016/j.ab.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 19.Jing N, Rando RF, Pommier Y, Hogan ME. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 1997;36:12498–12505. doi: 10.1021/bi962798y. [DOI] [PubMed] [Google Scholar]
- 20.Olaussen KA, Dubrana K, Domont J, Spano JP, Sabatier L, Soria JC. Telomeres and telomerase as targets for anticancer drug development. Crit. Rev. Oncology/Hematology. 2006;57:191–214. doi: 10.1016/j.critrevonc.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Dai J, Carver M, Yang D. Biochimie. 2008. Polymorphism of human telomeric quadruplex structures. [Epub ahead of print, doi 10.1016/j.biochi.2008.02.026]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hud NV, Plavec J. The role of cations in determining quadruplex structure and stability. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 100–130. [Google Scholar]
- 23.Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Quart. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. Structure and Mechanism in Protein Science, W.H. Freeman and Co., New York. 1999. pp. 540–572. [Google Scholar]
- 34.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 35.Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA. 1998;95:8602–8606. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansari A, Kuznetsov SV, Shen Y. Configurational diffusion down a folding funnel describes the dynamics of DNA hairpins. Proc. Natl Acad. Sci. USA. 2001;98:7771–7776. doi: 10.1073/pnas.131477798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Downs WD, Cech TR. Kinetic pathway for folding of the Tetrahymena ribozyme revealed by three UV-inducible crosslinks. RNA. 1996;2:718–732. [PMC free article] [PubMed] [Google Scholar]
- 38.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 39.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mergny J-L, Gros J, De Cian A, Bourdoncle A, Rosu F, Saccà B, Guittat L, Amrane S, Mills M, Alberti P, et al. Energetics, kinetics and dynamics of quadruplex folding. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 31–80. [Google Scholar]
- 41.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green JJ, Ladame S, Ying L, Klenerman D, Balasubramanian S. Investigating a quadruplex-ligand interaction by unfolding kinetics. J. Am. Chem. Soc. 2006;128:9809–9812. doi: 10.1021/ja0615425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J. Am. Chem. Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 44.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 45.Petraccone L, Pagano B, Esposito V, Randazzo A, Piccialli G, Barone G, Mattia CA, Giancola C. Thermodynamics and kinetics of PNA-DNA quadruplex-forming chimeras. J. Am. Chem. Soc. 2005;127:16215–16223. doi: 10.1021/ja0545923. [DOI] [PubMed] [Google Scholar]
- 46.Mergny JL, De CA, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardin C, Leroy JL. The formation pathway of tetramolecular G-quadruplexes. Nucleic Acids Res. 2008;36:477–488. doi: 10.1093/nar/gkm1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 49.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henry ER, Hofrichter J. Singular value decomposition: application to analysis of experimental data. Methods Enzymol. 1992;210:129–192. [Google Scholar]
- 51.Hendler RW, Shrager RI. Deconvolutions based on singular value decomposition and the pseudoinverse: a guide for beginners. J. Biochem. Biophys. Methods. 1994;28:1–33. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 52.DeSa RJ, Matheson IB. A practical approach to interpretation of singular value decomposition results. Methods Enzymol. 2004;384:1–8. doi: 10.1016/S0076-6879(04)84001-1. [DOI] [PubMed] [Google Scholar]
- 53.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hud NV, Smith FW, Anet FAL, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 55.Li W, Miyoshi D, Nakano S, Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 56.Sheardy RD, Suh D, Kurzinsky R, Doktycz MJ, Benight AS, Chaires JB. Sequence dependence of the free energy of B-Z junction formation in deoxyoligonucleotides. J. Mol. Biol. 1993;231:475–488. doi: 10.1006/jmbi.1993.1295. [DOI] [PubMed] [Google Scholar]
- 57.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliveberg M, Tan YJ, Fersht AR. Negative activation enthalpies in the kinetics of protein folding. Proc. Natl Acad. Sci. USA. 1995;92:8926–8929. doi: 10.1073/pnas.92.19.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveberg M, Tan YJ, Silow M, Fersht AR. The changing nature of the protein folding transition state: implications for the shape of the free-energy profile for folding. J. Mol. Biol. 1998;277:933–943. doi: 10.1006/jmbi.1997.1612. [DOI] [PubMed] [Google Scholar]
- 60.Eyring H. The activated complex and the absolute rate of chemical reactions. Chem. Rev. 1935;17:65–77. [Google Scholar]
- 61.Xu Q, Deng H, Braunlin WH. Selective localization and rotational immobilization of univalent cations on quadruplex DNA. Biochemistry. 1993;32:13130–13137. doi: 10.1021/bi00211a023. [DOI] [PubMed] [Google Scholar]
- 62.Majhi PR, Qi J, Tang CF, Shafer RH. Heat capacity changes associated with guanine quadruplex formation: an isothermal titration calorimetry study. Biopolymers. 2008;89:302–309. doi: 10.1002/bip.20918. [DOI] [PubMed] [Google Scholar]
- 63.Mashimo T, Sugiyama H. Folding pathways of human telomeric hybrid G-quadruplex structure. Nucleic Acids Symp. Ser. 2007;51:239–240. doi: 10.1093/nass/nrm120. [DOI] [PubMed] [Google Scholar]
- 64.Basu HS, Shafer RH, Marton LJ. A stopped-flow H-D exchange kinetic study of spermine-polynucleotide interactions. Nucleic Acids Res. 1987;15:5873–5886. doi: 10.1093/nar/15.14.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.