Abstract
Fluorescent microscopy experiments show that when 2′-O-methyl-modified molecular beacons (MBs) are introduced into NIH/3T3 cells, they elicit a nonspecific signal in the nucleus. This false-positive signal can be avoided by conjugating MBs to macromolecules (e.g. NeutrAvidin) that prevent nuclear sequestration, but the presence of a macromolecule makes efficient cytosolic delivery of these probes challenging. In this study, we explored various methods including TAT peptide, Streptolysin O and microporation for delivering NeutrAvidin-conjugates into the cytosol of living cells. Surprisingly, all of these strategies led to entrapment of the conjugates within lysosomes within 24 h. When the conjugates were pegylated, to help prevent intracellular recognition, only microporation led to a uniform cytosolic distribution. Microporation also yielded a transfection efficiency of 93% and an average viability of 86%. When cells microporated with MB–NeutrAvidin conjugates were examined via flow cytometry, the signal-to-background was found to be more than 3 times higher and the sensitivity nearly five times higher than unconjugated MBs. Overall, the present study introduces an improved methodology for the high-throughput detection of RNA at the single cell level.
INTRODUCTION
The ability to measure gene expression, using techniques such as DNA microarrays and reverse transcriptase PCR, has proven to be invaluable in molecular biology, drug development and clinical diagnostics. A major limitation of these approaches, however, is they generally report only the relative change in gene expression for a population of cells. In many cases, it is the aberrant RNA expression of only a small percentage of cells that may be of interest. Therefore, there is still a need for high-throughput methods that allow RNA levels to be measured at the single cell level.
Over the past decade, much effort has been devoted to developing fluorescent probes that are capable of imaging RNA at the single-cell level. Of these, perhaps molecular beacons (MBs) have garnered the most attention. MBs are antisense oligonucleotide probes labeled with a ‘reporter’ fluorophore at one end and with a quencher at the other end (1). In the absence of complementary nucleic acid targets, the MBs assume a stem–loop configuration. As a result, the fluorophore and quencher are brought into close proximity and fluorescence is quenched. When MBs hybridize to complementary targets, the fluorophore is separated from the quencher and fluorescence is restored. MBs have already been used to study gene expression in a variety of live cell applications, ranging from monitoring the transport and the distribution of β-actin mRNAs in motile fibroblasts to the detection of specific RNAs in living cancer cells (2–13).
Although MBs clearly hold promise as a tool for the analysis of RNA at the single cell level, we have previously shown that when nuclease-vulnerable MBs are introduced into living cells via microinjection, they are quickly sequestered into the nucleus and generate false-positive signals (5). Presumably, these false-positive signals arise when MBs interact with nucleases and/or nucleic acid-binding proteins. One strategy to overcome nuclear sequestration involves attaching MBs to macromolecules such as quantum dots or streptavidin, whereby MB conjugates cannot pass through the nuclear pores and are effectively retained in the cytoplasm (2,5). This eliminates the nonspecific signal observed in the nucleus.
It is likely that before MB–macromolecular conjugates become widely adopted as a tool for RNA imaging, strategies that allow for the efficient delivery of these probes into the cytosol of living cells must be identified. Although microinjection can be used to direct the probes into the cytosol, this method is tedious, inefficient and impractical for gene expression analysis of large numbers of cells. Alternative delivery methods that have recently been shown to be effective in delivering unmodified MBs into living cells include cell penetrating peptides (CPPs) and Streptolysin O (SLO), which is a pore-forming bacterial exotoxin that binds to cell membranes (4,9,14). The specific CPP used was the 11 amino-acid peptide of the human immunodeficiency virus type I TAT protein. To date, TAT peptide has been demonstrated to direct the internalization of numerous cargoes including small peptides (15), proteins and polymers (16–18), oligonucleotides including molecular beacons (9,19), phage vectors (16), liposomes and polymersomes (20,21) and nanoparticles (22,23). Internalization of TAT peptide is thought to be through micropinocytosis although the exact mechanism is still under debate (24–28). SLO, on the other hand, permeabilizes adherent and nonadherent cells by reversibly forming pores on the cell surface (14,29–34). It has been shown that SLO can be used to deliver oligonucleotides (including MBs), dextran and proteins up to 100 kDa in mass into the cytosol (29,30,34). Although not previously utilized for delivering MBs, electroporation provides yet another possibility for delivering MB–macromolecular conjugates into living cells. The transient permeabilization of the plasma membrane during application of an electric field is a well-established technique for transfecting RNA, DNA, fluorescent dyes, dextrans, peptides and proteins into various cell lines (35–39). A common problem often experienced with electroporation is low cell viability; however, recent advances in electroporation technology such as the ability to perform electroporation in microliter volume spaces (e.g. pipette tips) has led to a reduction in the many harmful events associated with this process, including heat generation, metal ion dissolution, pH variation and oxide formation (www.microporator.com). This microliter volume electroporation process is known as microporation (Digital Bio Technology).
In this study, we compared CPPs, SLO and microporation as methods to deliver NeutrAvidins and MB–NeutrAvidin conjugates into living cells. NeutrAvidin, a deglycosylated variant of avidin, was utilized because its biotin-binding capabilities allows for facile conjugation to biotinylated MBs, it is of similar size as streptavidin and thus should prevent nuclear localization of MB conjugates, and it reportedly exhibits less nonspecific interactions than streptavidin (http://probes.invitrogen.com). Cells microporated with MB–NeutrAvidin conjugates were further examined via flow cytometry to confirm MB hybridization with cytoplasmic RNA. The intracellular signal-to-background of MB–NeutrAvidin conjugates was also measured and compared with unconjugated MBs. Overall, this study provides a strategy for the efficient, high-throughput analysis of specific RNA expression in single living cells.
MATERIALS AND METHODS
MB design
Antisense firefly luciferase 2′-O-methyl RNA MBs possessing a TEX 613 fluorophore (Ex: 596 nm, Em: 613 nm) at the 5′ end, an Iowa Black RQ quencher, IBRQ, at the 3′ end and a biotin-dT group incorporated in the 3′ stem was synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA). The antisense sequence of firefly luciferase (pGL3-Luc 235-252, Promega, Madison, Wisconsin, USA) was chosen because it is not complementary to any known endogenous RNA target in NIH/3T3 cells. Specifically, the antiluciferase MB employed was TEX613-mGmUmCmAmCmCmUmCmAmGmCmGmUmAmAmGmUmGmAmUmGmTmCmG(bio-dT) mGmAmC-IBRQ. A complementary Luciferase RNA oligonucleotide target was also synthesized, with the sequence rUrGrGrArCrArUrCrArCrUrUrArCrGrCrUrGrArGrUrA.
Synthesis of NeutrAvidin/MB conjugates
NeutrAvidin (Pierce, Rockford, IL, USA) was dissolved in 50 mM Sodium Borate buffer, pH 8 at a concentration of 10 mg/ml and reacted with Cy5-NHS ester (Amersham Biosciences, Piscataway, NJ, USA) or Alexa750-NHS ester (Invitrogen, Carlsbad, CA, USA) at dye to NeutrAvidin molar ratio of 2.5:1. Similarly, an additional sample of NeutrAvidin was labeled with FITC (Sigma, St. Louis, MO, USA) at a dye to NeutrAvidin molar ratio of 10:1. The fluorescent conjugates were purified on NAP-5 gel chromatography columns (Amersham Biosciences) in Phosphate Buffer (48 mM K2HPO4, 4.5 mM KH2PO4, 14 mM NaH2PO4), pH 7.2. The number of fluorophores per NeutrAvidin was determined using a Cary100 spectrophotometer (Varian). It was determined that there were 3.6 FITC, 1.5 Cy5 and 1.7 Alexa750 per NeutrAvidin, respectively. Pegylated NeutrAvidin conjugates were synthesized by further reacting fluorescently labeled NeutrAvidins with Methyl PEOn-NHS ester (n = 8, Pierce) at various PEG to NeutrAvidin molar ratios (200:1, 100:1, 50:1 and 25:1), and purified by repeated filtration and dilution on Microcon YM-30 centrifugal devices (30 000 MW cutoff; Millipore, Billerica, MA, USA). Initial experiments indicated that when the pegylated NeutrAvidins were microporated into NIH/3T3 cells, those with 200:1 and 100:1 PEG labeling ratios yielded a diffuse cytoplasmic distribution of NeutrAvidin as determined by fluorescence microscopy. Microporation and fluorescence microscopy protocols are described below. NIH/3T3 cells that were microporated with NeutrAvidin that was pegylated at a labeling ratio of 50:1 and 25:1 exhibited punctate cytoplasmic fluorescence. As a result, a 100:1 labeling ratio was used for the subsequent studies. TAT-NeutrAvidin conjugates were synthesized by reacting fluorescently labeled NeutrAvidins, pegylated and unpegylated, with biotin-TAT (YGRKKRRQRRR-biotin) at a molar ratio of 4:1 TAT to NeutrAvidin. MB–NeutrAvidin conjugates were synthesized by reacting fluorescently labeled NeutrAvidins with biotinylated antisense MBs at a molar ratio of 1.5:1 MB to NeutrAvidin, prior to pegylation. All conjugation reactions were allowed to react overnight at room temperature. All conjugates were purified on YM-30 centrifugal devices and concentrations were determined spectrophotometrically.
Cell culture and transfection
NIH/3T3 cells (ATCC, Manassas, VA, USA) were cultured in DMEM supplemented with 10% fetal bovine serum and incubated in 5% CO2 at 37°C. To generate cells that express Firefly luciferase, the NIH/3T3 cells were infected with adenovirus, H4′ 040CMVffLuciferase (Penn Genomic Center, Philadelphia, PA, USA), at a multiplicity of infection of 104 particles per cell. Infection was carried out 24 h prior to delivery of NeutrAvidin conjugates without any apparent loss of viability. Firefly activity was confirmed by making bioluminescent measurements on a Glomax 20/20 luminometer (Promega) following the administration of SteadyGlo (Promega).
Microinjection
Microinjection was carried out using a Femtojet and Injectman NI 2 (Eppendorf, Westbury, NY, USA) microinjection system fitted with Femtotips I (Eppendorf). Prior to use, Femtoptips were treated with Hexamethyldisilazane (Fluka) for 10 min, followed by repeated washes in phosphate buffer. Cells were microinjected with samples containing 2.5 or 10 μM antisense 2′-O-methyl MBs in phosphate buffer, pH 7.2.
TAT-mediated delivery
NIH/3T3 cells were seeded in 48-well plates in DMEM-FBS with no antibiotics 1 day prior to experimentation such that they were 50–70% confluent on the second day. Pegylated and unpegylated TAT-Cy5-NeutrAvidin were added to the wells at a final NeutrAvidin concentration of 200 nM in 100 μl media. Fusogenic peptides were also added to the wells at final concentrations ranging from 0 to 100 μM. The specific fusogenic peptides tested were INF-7 (GLFEAIEGFIENGWEGMIDGWYG-(PEG)6-NH2), GALA-INF3 (GLFEAIEGFIENGWEGLAEALAEALEALAA-(PEG)6-NH2) and HA2 (GLFGAIAGFIENGWEGMIDGWYG-(PEG)6-NH2). The (PEG)6 linker was added to each peptide to increase solubility. All of the peptides were synthesized by Biopeptide Co., Inc., San Diego, CA, USA. Fluorescent images of the cells were acquired over the course of 24 h as described below.
SLO delivery
The method used to delivery MB conjugates into NIH/3T3 cells by reversible membrane permeabilization with SLO was modified from previously established methods described by Walev et al. (29) and Faria et al. (34). Briefly, 110 U of SLO (Sigma) was activated in 100 μl of PBS containing 5 mM dithiothreitol (Pierce) and 0.05% BSA for 2 h at 37°C. NIH/3T3 cells plated in 48-well plates were washed once with PBS and were incubated in 100 μl of HBSS containing 30 mM HEPES, pH 7.2 for 20 min at 37°C. NeutrAvidin (5, 15, 25 or 50 μl) and an optimized amount (50 μl) of activated SLO was then added to the cells yielding a final NeutrAvidin concentration of 0.8, 2.3, 3.6 or 6.25 μM, respectively. After incubating the cells for 20 min at 37°C, 300 μl of ice-cold resealing media (DMEM–FBS containing 2 mM Ca+2) was added to each well. Cells were then allowed to reseal at 4°C for 60 min (30). After the resealing step, the media was replaced with fresh resealing media and images were acquired at various time points over the course of 24 h.
Microporation
Microliter volume electroporation was performed with a OneDrop MicroPorator (MP-100, BTX Harvard Apparatus). Specifically, NIH/3T3 cells were seeded in 48-well plates in DMEM–FBS with no phenol red and no antibiotics 1 day prior to experimentation. The cells were then trypsinized and suspended in media without phenol red, washed with 1× PBS, and finally resuspended in resuspension buffer R (BTX Harvard Apparatus) at a concentration of 120 000 cells per 11 μl. NeutrAvidin conjugates (1 μl) were added to the cells such that the final concentration of NeutrAvidin was 1, 2 or 3 μM probe. The 10 μl of the cells (i.e. 100 000 cells) were then microporated at 1250 V, 1500 V or 1700 V with a 10 ms pulse width, 3 pulses total. Following microporation the cells were either seeded in 24-well plates and imaged at various time points over the course of 24 h or immediately suspended in 1× PBS for flow cytometric analysis.
Lysosomal labeling
Twenty-four hours after the NeutrAvidin conjugates were delivered into NIH/3T3 cells with CPPs, SLO or microporation, the cells were washed with 1× PBS, and LysoTracker Green DND-153 (Invitrogen) was added at a final concentration of 1 μM in 1× PBS. Cells were incubated with the LysoTracker loading solution for 2 min and then washed and incubated with fresh PBS. The cells were immediately imaged via fluorescence microscopy.
Cytotoxicity assay
The effect of microporation voltage and NeutrAvidin concentration on the viability of NIH/3T3 cells was assessed via an MTT assay (ATCC) according to the manufacture's instructions. To determine the effect of voltage, cells were microporated at 0, 1250, 1500 or 1740 V in the presence of 3 μM FITC-PEG-NeutrAvidin conjugates. The number of pulses was kept constant at 3 and the pulse width was held constant at 10 ms. To determine the effect of NeutrAvidin concentration, cells were microporated at 1740 V in the presence of 0, 1, 2 or 3 μM of FITC-PEG-NeutrAvidin conjugates. FITC-labeled, pegylated NeutrAvidins were used for the MTT assay to ensure that there was no significant spectral overlap between the FITC absorbance (490 nm) and the MTT absorbance (570 nm). Absorbances were measured on a Cary100 spectrophotometer (Varian). Viability of cells was determined by dividing the absorbance measurements of the electroporated cells by the absorbance of cells that were not electroporated (0 V).
Fluorescent imaging
All microscopy images were acquired with an Olympus IX 81 motorized inverted fluorescence microscope equipped with a back-illuminated EMCCD camera (Andor), an X-cite 120 excitation source (EXFO) and Sutter excitation and emission filter wheels. Images of LysoTracker Green, Texas Red, Cy5 and Alexa750 were acquired using the filter sets (HQ480/40, HQ535/50, Q505LP), (HQ560/55, HQ645/75, Q595LP), (HQ620/60, HQ700/75, Q660LP) and (HQ710/75, HQ810/90, Q750LP) (Chroma, Rockingham, VT 05101), respectively. A LUC PLAN FLN 40× objective (NA 0.6) was used for all cell imaging studies. Results were analyzed with NIH Image J.
Flow cytometry analysis
Transfection efficiency
Following microporation of NIH/3T3 cells in the presence of FITC-labeled, pegylated NeutrAvidin conjugates (as described in the Cytotoxicity assay section), the cells were washed once in PBS and analyzed on a Guava Excite flow cytometer fitted with a 488 nm excitation laser. Flow cytometry data was analyzed with FLOWJO (Version 7.2.2; Tree Star, Ashland, OR, USA). Fluorescence signal intensity was defined as the mean intensity of cells lying within a predefined gate. The appropriate gate was defined on an FSC versus SSC dot plot of NIH/3T3 cells that were not electroporated (0 V).
Luciferase RNA detection
Wild-type or adenovirus-infected NIH/3T3 cells were microporated at 1740 V in the presence of 700 nM MB–NeutrAvidin conjugates (NeutrAvidin was labeled with Alexa750 and pegylated) or 700 nM unconjugated MBs. The cells were washed in 1× PBS before being analyzed using an LSR II flow cytometer (Becton Dickinson) equipped with a 532 nm laser, a 600 nm long pass dichroic filter and a 610 nm PE-Texas Red emission filter (610/20). Flow cytometry data were analyzed as described above.
RESULTS AND DISCUSSION
2′-O-methyl RNA MBs elicit a false-positive signal in the nucleus of living cells
Previously, we have shown that when nuclease-vulnerable molecular beacons are introduced into living cells, they are quickly sequestered into the nucleus and elicit a false-positive signal (5). To determine whether this was a result of nucleus degradation and/or nonspecific interactions, we investigated whether MBs composed of 2-O′-methyl RNA backbones, which are generally considered resistant to nuclease degradation also elicit a false-positive signal in the nucleus of living cells. Specifically, we microinjected antisense luciferase 2′-O′-methyl RNA MBs that were not complementary to any known endogenous RNA into NIH/3T3 cells. It was hypothesized that these MBs would remain quenched unless opened due to non-specific interactions. Consequently, an increase in fluorescence would indicate that nonspecific interactions could cause false-positive events.
Following cytoplasmic injection of 2-O′-methyl RNA MBs, there was a gradual increase in fluorescence observed in the nucleus (Figure 1a). To determine whether the increase in signal was simply the result of MB accumulation or nonspecific opening of MBs, the total fluorescence of the cell was quantified over time. It was determined that the total MB fluorescence increased by ∼82% over the course of 30 min, which suggests that nonspecific opening of MBs was creating false-positive signals. To further explore whether the increase in fluorescence was in response to undesirable MB hybridization or due to nonspecific protein interactions, live cell competitive inhibitions studies were performed. Specifically, a 10-fold excess of unlabeled 2′-O-methyl RNAs with the same targeting sequence as the MBs were coinjected with MBs into NIH/3T3 cells. Despite the presence of a competitive inhibitor, we continued to observe an increase in MB fluorescence in the nucleus, suggesting that protein interactions are likely responsible for the false-positive signals.
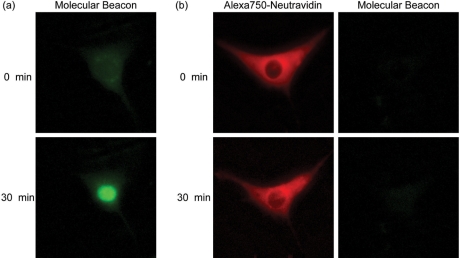
Figure 1.
Intracellular fluorescence of MBs and NeutrAvidin–MB conjugates. (a) Fluorescent microscopy images of NIH/3T3 cells injected with 2′-O-methyl RNA MBs. The MBs were not perfectly complementary to any known endogenous RNA in mice. (b) NIH/3T3 cells were injected with MB–NeutrAvidin conjugates. NeutrAvidin was labeled with the fluorescent dye, Alexa750, to facilitate imaging of their intracellular distribution.
Previously, we and others have demonstrated that by linking MBs to macromolecules such as quantum dots or streptavidin they can be effectively retained in the cytoplasm (2,5). In the present study, we conjugated 2′-O-methyl RNA MBs to fluorescently labeled NeutrAvidins to prevent nuclear sequestration. Immediately following cytoplasmic injection, fluorescent signals from both the MBs and NeutrAvidins were only observed in the cytoplasm of the cells (Figure 1b). While NeutrAvidin signals were clearly distinguished from the background, the MB fluorescence signal was extremely faint and barely detectable above the background. Thirty minutes after the injection, the total MB fluorescence within the entire cell increased by only ∼6%. This small increase in signal likely results from a small amount of unbound MB that localized to the nucleus (i.e. a faint fluorescent signal was observed in the nucleus). As a result, conjugation of NeutrAvidins to MBs not only helps overcome nuclear sequestration of the MBs, but also effectively prevents the generation of false-positive signals.
NeutrAvidin conjugates are directed towards lysosomes when TAT-peptide, SLO or microporation are used for intracellular delivery
To investigate whether TAT peptides can be used to deliver NeutrAvidin conjugates into the cytosol of living cells, NIH/3T3 cells were treated with Cy5-labeled NeutrAvidins conjugated to TAT peptides and imaged at various time points. Shortly after the addition of the TAT–NeutrAvidin conjugates a uniform fluorescent signal could be observed on the surface of the cells. By 4 h, punctate fluorescent staining could be observed within the cytoplasm of the cells, while no fluorescence was observed in the nucleus. Similar patterns persisted over the course of 24 h (Figure 2a). Live-cell staining with the lysosomal labeling dye, LysoTracker, indicated that the majority of the fluorescent TAT–NeutrAvidin conjugates were localized in lysosomes (Figure 2d). We further explored the possibility of enhancing endosomal release of TAT-NeutrAvidins by employing pH-sensitive fusogenic peptides, i.e. INF-7, GALA-INF3 and HA2. These fusogenic peptides have previously been reported to disrupt endosomes, causing them to release their contents (40,41). When NIH/3T3 cells were co-incubated with TAT–NeutrAvidin and various concentrations of each of the fusogenic peptides, a punctate fluorescent staining pattern was still observed in the cytoplasms. This signal was largely colocalized with the lysosomal labeling dye, LysoTracker (data not shown). It should be noted that these results do not completely rule out the possibility that some endosomes were disrupted by the fusogenic peptides. For example, it is possible that the cytoplasmic signal was too weak to be observed relative to the bright fluorescent signal emanating from the lysosomes; however, it does appear that endosomal disruption is inefficient. Therefore, based on our observations, TAT peptides are not effective in delivering fluorescently labeled NeutrAvidin conjugates into the cytosol of living cells.
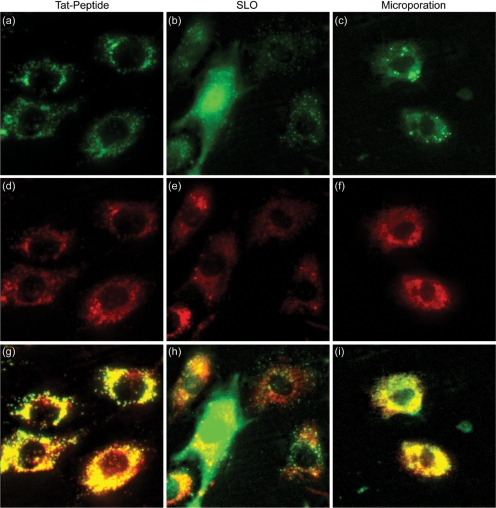
Figure 2.
Delivery of NeutrAvidin conjugates into single living cells. Cy5-labeled NeutrAvidins were delivered into NIH/3T3 cells either by (a) TAT-peptide, (b) SLO or (c) microporation. Images corresponding to TAT-peptide-based delivery and microporation were acquired 24 h following delivery. Images corresponding to delivery via SLO were acquired immediately after the permeabilized membranes were resealed, ∼1.5 h. Subsequent staining of the cells with LysoTracker was used to determine whether the pegylated NeutrAvidin was localized within lysosomes for each of the delivery methods, (d) TAT-peptide, (e) SLO and (f) microporation. The merged NeutrAvidin and lysosome images for (g) TAT peptide, (h) SLO, and (i) microporation show all colocalized fluorescent signals as yellow in color. The contrast in the yellow channel of the merged images was increased to more clearly distinguish areas of colocalization from the pure red and green signals.
To investigate whether SLO could be used to efficiently deliver NeutrAvidin conjugates into the cytosol of living cells, NIH/3T3 cells were treated with SLO and varying amounts of fluorescently labeled NeutrAvidins (0.8, 2.3, 3.6 and 6.25 μM). Immediately after the permeabilized membranes were resealed, intracellular fluorescent signals were detectable but only when relatively high concentrations of NeutrAvidin were utilized (>3 μM) (Figure 2b). Furthermore, there was a large cell-to-cell variation in total cellular fluorescence, and also variability in the intracellular pattern of fluorescence. Specifically, some cells appeared to exhibit fluorescent signals throughout the entire cell, including both cytoplasm and the nucleus, while other cells exhibited a more punctate pattern. After 24 h, nearly all of the cells exhibited a punctate fluorescent pattern (data not shown). Colocalization of the punctate fluorescent signals with the LysoTracker dye confirmed that the majority of the NeutrAvidins were localized in lysosomes.
To investigate whether microporation could be used to efficiently deliver NeutrAvidin into the cytosol of living cells, 3 μM of Cy5-labeled NeutrAvidins were microporated into NIH/3T3 cells. Bright fluorescent signals were observed in the cells immediately following microporation, while the cells were still in suspension; however, after 24 h the fluorescent signal became punctate (Figure 2c). Colocalization of the NeutrAvidin signal with the LysoTracker dye indicated that the NeutrAvidins were localized in lysosomes (Figure 2f).
In order to identify whether the Cy5 dye was responsible for targeting the Cy5–NeutrAvidin conjugates to lysosomes, we delivered just the Cy5 dye into living cells via SLO and microporation. In both cases, the dye was initially diffused throughout the cell, but over time the fluorescent signal in the cytoplasm dissipated suggesting leakage of the dye from the cytoplasmic compartment. After 24 h there was still some signal remaining in the lysosomes, but the fluorescent signal was extremely faint. Based on these findings, we do not believe that Cy5 targeted the NeutrAvidin to lysosomes. To further confirm this point, we also delivered Alexa750-labeled NeutrAvidin into cells via SLO and microporation. In each case, we observed similar intracellular localization as with Cy5–NeutrAvidin. This suggests that the fluorescent label is not responsible for targeting NeutrAvidin to lysosomes.
Pegylation improves cytosolic delivery of NeutrAvidin conjugates by microporation
Considering the uniform distribution of fluorescently labeled NeutrAvidin observed following microinjection, it was surprising to find that CPPs, SLO and microporation all resulted in punctate fluorescent staining. In particular, both SLO and microporation directly porate the cellular membrane and will allow direct passage of extracellular proteins into the cytoplasm. In response to these puzzling results, we hypothesized that coating the fluorescently labeled NeutrAvidin with polyethylene glycol (PEG) may improve cytosolic delivery. PEG coatings are commonly used to improve solubility, diminish the potential for aggregation, improve biocompatibility, and minimize nonspecific interactions.
When pegylated NeutrAvidin conjugates were delivered into the cells by TAT peptide, a punctate fluorescent pattern was again observed in the cytoplasm of NIH/3T3 cells (Figure 3a). Colocalization of the NeutrAvidin-signal with the LysoTracker dye again indicated that the majority of the proteins were localized in the lysosomes (Figure 3d). Therefore, pegylation did not seem to improve the cytosolic delivery of NeutrAvidin, as the majority of the probes were still localized in lysosomes. Overall, we conclude that TAT peptides can effectively be used to internalize macromolecules into cells, but these macromolecules predominantly reside in lysosomes.
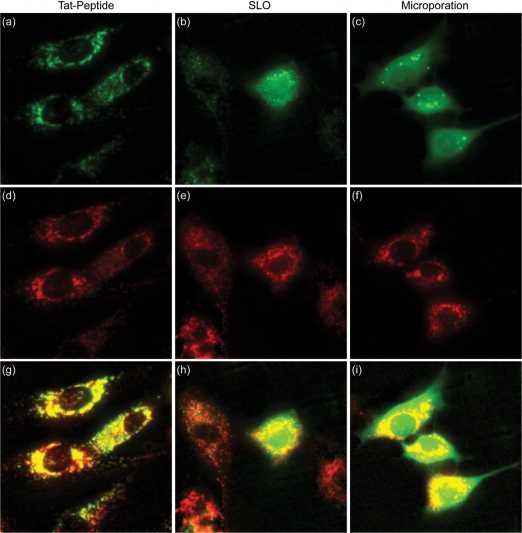
Figure 3.
Delivery of pegylated NeutrAvidin into single living cells. Cy5-labeled NeutrAvidins colabeled with PEG were delivered into NIH/3T3 cells either by (a) TAT peptide, (b) SLO or (c) microporation. Images corresponding to TAT-peptide-based delivery and microporation were acquired 24 h following delivery. Images corresponding to delivery via SLO were acquired immediately after the permeabilized membranes were resealed, ∼1.5 h. Subsequent staining of the cells with LysoTracker was used to determine whether the pegylated NeutrAvidin was localized within lysosomes for each of the delivery methods, (d) TAT-peptide, (e) SLO, and (f) microporation. The merged NeutrAvidin and lysosme images for (g) TAT peptide, (h) SLO and (i) microporation show all colocalized fluorescent signals as yellow in color. The contrast in the yellow channel of the merged images was increased to more clearly distinguish areas of colocalization from the pure red and green signals.
Pegylated NeutrAvidins were also delivered into the cells by SLO (Figure 3b). Again, pegylation did not seem to significantly improve the cytosolic delivery of fluorescently labeled NeutrAvidin. Specifically, immediately after the permeabilized membranes were resealed, a few cells appeared to exhibit fluorescent signals throughout the cell but most cells exhibited punctate fluorescent signals. Furthermore, there was still a large cell-to-cell variation in fluorescence between cells. After 24 h, nearly all of the cells exhibited a punctate fluorescent pattern (data not shown). Colocalization of the punctate fluorescent signals with the LysoTracker signal confirmed that the majority of the NeutrAvidins were localized in the lysosomes.
Finally, when microporation experiments were performed with pegylated NeutrAvidin conjugates, the majority of the fluorescent signal was observed in the cytoplasm of NIH/3T3 cells (Figure 3c). There were still a small number of bright fluorescent spots within each cell, but otherwise the intracellular distribution closely resembled that of NeutrAvidin following microinjection. The diffuse fluorescent signal persisted in the cytoplasm for at least 24 h. This could be beneficial for temporal measurements of gene expression in living cells. It should also be noted that aside from the desirable intracellular distribution of NeutrAvidin conjugates, another exciting benefit of using microporation for delivery is the small amount of sample needed. Only 10 μl of 3 μM sample was needed per 100 000 cells compared with 175 μl of 3.6 μM sample for SLO. This is an important benefit when only a limited amount of sample is available.
Microporation offers high cell viability and transfection efficiency
In general, electroporation is often considered an invasive procedure that results in poor viability. To specifically test the effect of ‘microporation’ on cell viability, an MTT cell proliferation assay was performed on cells that were microporated using various voltages and probe concentrations. In general, viability decreased as the microporation voltage was increased (Figure 4a). Specifically, the viability was 86 ± 5%, 78 ± 11% and 66 ± 10% (n = 3) for 1250, 1500 and 1740 V, respectively. Increasing the concentration of the pegylated NeutrAvidin conjugates from 0 to 3 μM did not seem to have any significant effect on cell viability (one-way ANOVA, F = 0.245, P = 0.79) (data not shown).
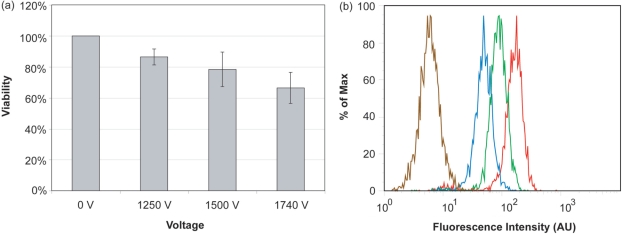
Figure 4.
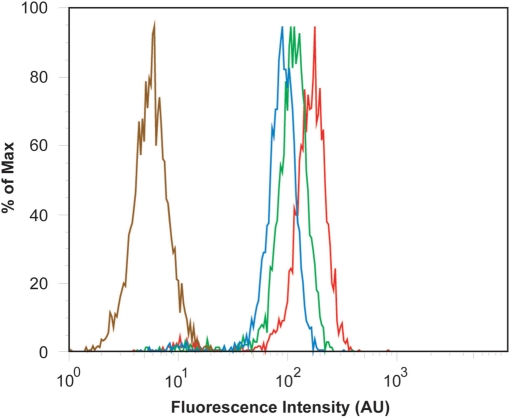
The effect of microporation voltage settings on cell viability and transfection efficiency. (a) NIH/3T3 cells were microporated in the presence of 3 μM FITC-labeled, pegylated NeutrAvidins at 0, 1250, 1500 or 1740 V. The corresponding viability was determined by MTT assay. Viability was determined relative to cells that were not microporated (0 V). (b) Flow cytometry histograms for NIH/3T3 cells microporated at different voltage levels 0 V (brown), 1250 V (blue), 1500 V (green) and 1740 V (red).
To determine the effect of voltage on transfection efficiency, cells were microporated with 3 μM of fluorescently labeled and pegylated NeutrAvidin at 0, 1250, 1500 and 1740 V before being analyzed by flow cytometry. It was found that the transfection efficiency increased with increasing voltage settings (Figure 4b). Specifically, transfection efficiencies at 1250, 1500 and 1740 V were 88, 93 and 96%, respectively. It was further shown that microporation itself (at 1740 V) does not affect cell autofluorescence. Overall, microporation appears to yield a high transfection efficiency of the NeutrAvidin conjugates. These results, combined with the high viability of NIH/3T3 cells after microporation, particularly at 1250 V, suggest that microporation provides a viable approach for the high-throughput delivery of MB–NeutrAvidin conjugates into living cells.
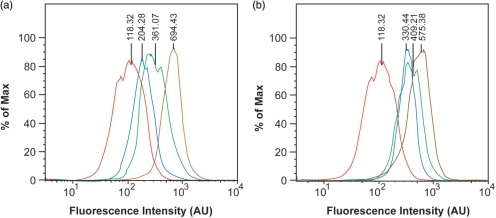
To evaluate the effect of probe concentration on transfection efficiency, cells were microporated with different concentrations of pegylated NeutrAvidin conjugates before being analyzed by flow cytometry (Figure 5). As expected, the average fluorescent signal per cell and the percentage of transfected cells increased as the concentration of NeutrAvidin was increased. Specifically, ∼93% of the cells exhibited a distinct fluorescent signal when microporated with 1 μM of NeutrAvidin and ∼96% of the cells exhibited a distinct fluorescent signal when microporated with 3 μM of NeutrAvidin. Our results are consistent with the general notion that higher voltage settings and higher extracellular protein concentrations can both lead to improved cytosolic delivery of proteins when applying electroporation.
Figure 5.
The effect of probe concentration on transfection efficiency. NIH/3T3 cells were microporated in the presence of 0 μM (brown), 1 μM (blue), 2 μM (green) and 3 μM (red) of FITC-labeled and pegylated NeutrAvidin at 1740 V before being analyzed by flow cytometry.
Flow cytometric analysis of RNAs in single living cells
In order to evaluate whether pegylated NeutrAvidin–MB conjugates could be used to detect gene expression in single living cells, microporation experiments were performed using antisense luciferase 2-O′-methyl RNA MB conjugates. Specifically, MB–NeutrAvidin conjugates were microporated into NIH/3T3 cells that express the luciferase protein (luc+). Two negative control experiments were conducted to examine whether the measured fluorescent signal reflected MB hybridization or false-positive events. The first control experiment consisted of microporating MB–NeutrAvidin conjugates into NIH/3T3 cells that do not express luciferase (luc−). The second negative control consisted of microporating luc+ cells in the absence of MB–NeutrAvidin conjugates (i.e. no probe). This experiment was performed to determine the level of cellular autofluorescence. A positive control experiment was also conducted to determine the maximum attainable signal in the cells. This experiment consisted of microporating MB conjugates prehybridized to synthetic luciferase RNA targets into the luc+ cells. Representative flow cytometry results are shown in Figure 6a. In order to calculate the signal-to-background (S:B) of the MB–NeutrAvidin conjugates, first the mean fluorescent signal of cells microporated in the absence of probe (i.e. autofluorescence) was subtracted from the other histograms. Then, the mean fluorescent signal of luc+ cells microporated with prehybridized MB–NeutrAvidin conjugates was divided by the mean fluorescent signal of luc− cells microporated with MB–NeutrAvidin conjugates, giving an S:B of 6.7. When MB–NeutrAvidin conjugates were microporated into Luc+ cells, the mean fluorescent signal was calculated to be 182% higher than luc− cells microporated with MB–NeutrAvidin conjugates.
Figure 6.
Flow cytometric measurements of MB fluorescence in NIH/3T3 cells. NIH/3T3 cells were microporated in the presence of (a) MB–NeutrAvidin conjugates or (b) unconjugated MBs. The flow cytometry histograms correspond to cells that do not contain luciferase RNA targets and were microporated in the absence of probe (negative control, red); cells that do not contain luciferase RNA targets and were microporated in the presence of probes (negative control, blue); cells that express luciferase RNA targets and were microporated in the presence of probes (green); and cells that express luciferase and were microporated in the presence of pre-hybridized probes (positive control, brown).
To highlight the importance of designing MBs that are not sequestered into the nucleus, analogous flow cytometry experiments were conducted with NIH/3T3 cells that were microporated with unconjugated antisense 2′-O-methyl RNA MBs (i.e. not conjugated to NeutrAvidin) (Figure 6b). It was found that unconjugated MBs exhibit significantly higher background fluorescence in Luc− cells compared with cells microporated with the MB–NeutrAvidin conjugates. Specifically, the S:B of cells microporated with unconjugated MBs was calculated to be only 2.15. This S:B is more than three times lower than the S:B achieved with MB–NeutrAvidin conjugates. The low S:B observed with unconjugated MBs likely stems from the high background signal present in the nucleus (Figure 1a). This high background also has a significant negative effect on the MB's ability to sensitively detect RNA. Specifically, when unconjugated MBs were microporated into Luc+ cells, the mean fluorescent signal was calculated to be only 37% higher than luc− cells microporated with unconjugated MBs. Therefore, the sensitivity for detecting luciferase RNA targets was nearly five times lower than with MB–NeutrAvidin conjugates. These results clearly highlight the advantages of retaining MBs in the cytoplasm for accurate RNA detection and may present an opportunity for quantification of RNA in living cells.
In summary, we demonstrate that MBs composed of 2′-O-methyl RNA backbones only elicit a nonspecific signal in the nucleus. When MBs are retained in the cytoplasmic compartment, by conjugation to NeutrAvidins, false-positive signals are reduced to marginal levels. Delivery of these MB–NeutrAvidin conjugates can be achieved by microporation, which allows for high transfection efficiency and high viability. Utilizing flow cytometry for high-throughput analysis of gene expression, MB–NeutrAvidin conjugates allowed for a higher sensitivity and improved dynamic range in RNA detection compared with conventional MBs. As a result, MB–NeutrAvidin conjugates can be used as an effective tool for detecting the expression of specific RNAs in living cells.
ACKNOWLEDGEMENTS
This material is based upon work supported in part by the National Institute of Health (NCI) R21 CA125088, the National Science Foundation BES-0616031 and the American Cancer Society RSG-07-005-01. Funding to pay the Open Access publication charges for this article was also provided by the grants mentioned above.
Conflict of interest statement. I, Dr Mark Behlke, am employed by Integrated DNA Technologies, Inc. (IDT), which offers oligonucleotides for sale similar to some of the compounds described in the article. IDT is however not a publicly traded company and I personally do not own any shares/equity in IDT.
REFERENCES
- 1.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys. J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santangelo P, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen AK, Behlke MA, Tsourkas A. Avoiding false-positive signals with nuclease-vulnerable molecular beacons in single living cells. Nucleic Acids Res. 2007;35:e105. doi: 10.1093/nar/gkm593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake TJ, Medley CD, Sen A, Rogers RJ, Tan WH. Stochasticity of manganese superoxide dismutase mRNA expression in breast carcinoma cells by molecular beacon imaging. Chembiochem. 2005;6:2041–2047. doi: 10.1002/cbic.200500046. [DOI] [PubMed] [Google Scholar]
- 7.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan WH. Simultaneous monitoring of the expression of multiple genes inside of single breast carcinoma cells. Anal. Chem. 2005;77:4713–4718. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 8.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitin N, Santangelo PJ, Kim G, Nie SM, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, Wood WC, Yang L. Real-time detection of gene expression in cancer cells using molecular beacon imaging: new strategies for cancer research. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- 11.Perlette J, Tan WH. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 12.Santangelo P, Nitin N, LaConte L, Woolums A, Bao G. Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons. J. Virol. 2006;80:682–688. doi: 10.1128/JVI.80.2.682-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc. Natl Acad. Sci. USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhakdi S, Bayley H, Valeva A, Walev I, Walker B, Weller U, Kehoe M, Palmer M. Staphylococcal alpha-toxin, streptolysin-O, and Escherichia coli hemolysin: prototypes of pore-forming bacterial cytolysins. Arch. Microbiol. 1996;165:73–79. doi: 10.1007/s002030050300. [DOI] [PubMed] [Google Scholar]
- 15.Lindsay MA. Peptide-mediated cell delivery: application in protein target validation. Curr. Opin. Pharmacol. 2002;2:587–594. doi: 10.1016/s1471-4892(02)00199-6. [DOI] [PubMed] [Google Scholar]
- 16.Albarran B, To R, Stayton PS. A TAT-streptavidin fusion protein directs uptake of biotinylated cargo into mammalian cells. Protein Eng. Des. Sel. 2005;18:147–152. doi: 10.1093/protein/gzi014. [DOI] [PubMed] [Google Scholar]
- 17.Cao GD, Pei W, Ge HL, Liang QH, Luo YM, Sharp FR, Lu AG, Ran RQ, Graham SH, Chen J. In vivo delivery of a Bcl-xL fusion protein containing the TAT protein transduction domain protects against ischemic brain injury and neuronal apoptosis. J. Neurosci. 2002;22:5423–5431. doi: 10.1523/JNEUROSCI.22-13-05423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl Acad. Sci. USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abes R, Arzumanov AA, Moulton HM, Abes S, Lvanciva GD, Lversen PL, Gait MJ, Lebleu B. Cell-penetrating-peptide-based delivery of oligonucleotides: an overview. Biochem. Soc. Trans. 2007;35:775–779. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- 20.Christian NA, Milone MC, Ranka SS, Li GZ, Frail PR, Davis KP, Bates FS, Therien MJ, Ghoroghchian PP, June CH, et al. Tat-functionalized near-infrared emissive polymersomes for dendritic cell labeling. Bioconjugate Chem. 2007;18:31–40. doi: 10.1021/bc0601267. [DOI] [PubMed] [Google Scholar]
- 21.Torchilin VP, Levchenko TS, Rammohan R, Volodina N, Papahadjopoulos-Sternberg B, D'Souza GGM. Cell transfection in vitro and in vivo with nontoxic TAT peptide-liposome-DNA complexes. Proc. Natl Acad. Sci. USA. 2003;100:1972–1977. doi: 10.1073/pnas.0435906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT, Weissleder R. Tat peptide-derivatized magnetic nanoparticles allow in vivo tracking and recovery of progenitor cells. Nat. Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- 23.Ruan G, Agrawal A, Marcus AI, Nie S. Imaging and tracking of tat peptide-conjugated quantum dots in living cells: new insights into nanoparticle uptake, intracellular transport, and vesicle shedding. J. Am. Chem. Soc. 2007;129:14759–14766. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]
- 24.Ferrari A, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Caveolae-mediated internalization of extracellular HIV-1 tat fusion proteins visualized in real time. Mol. Ther. 2003;8:284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 25.Fittipaldi A, Ferrari A, Zoppe M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 26.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 27.Saalik P, Elmquist A, Hansen M, Padari K, Saar K, Viht K, Langel V, Pooga M. Protein cargo delivery properties of cell-penetrating peptides. A comparative study. Bioconjugate Chem. 2004;15:1246–1253. doi: 10.1021/bc049938y. [DOI] [PubMed] [Google Scholar]
- 28.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 29.Faria M, Spiller DG, Dubertret C, Nelson JS, White MRH, Scherman D, Helene C, Giovannangeli C. Phosphoramidate oligonucleotides as potent antisense molecules in cells and in vivo. Nat. Biotechnol. 2001;19:40–44. doi: 10.1038/83489. [DOI] [PubMed] [Google Scholar]
- 30.Fawcett JM, Harrison SM, Orchard CH. A method for reversible permeabilization of isolated rat ventricular myocytes. Exp. Physiol. 1998;83:293–303. doi: 10.1113/expphysiol.1998.sp004114. [DOI] [PubMed] [Google Scholar]
- 31.Giles RV, Grzybowski J, Spiller DG, Tidd DM. Enhanced antisense effects resulting from an improved streptolysin-O protocol for oligodeoxynucleotide delivery into human leukaemia cells. Nucleosides Nucleotides. 1997;16:1155–1163. [Google Scholar]
- 32.Giles RV, Spiller DG, Grzybowski J, Clark RE, Tidd DM. Selecting optimal oligonucleotide composition for maximal antisense effect following streptolysin O-mediated delivery into human leukaemia cells. Nucleic Acids Res. 1998;26:1567–1575. doi: 10.1093/nar/26.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer M, Vulicevic I, Saweljew P, Valeva A, Kehoe M, Bhakdi S. Streptolysin O: a proposed model of allosteric interaction between a pore-forming protein and its target lipid bilayer. Biochemistry. 1998;37:2378–2383. doi: 10.1021/bi9720890. [DOI] [PubMed] [Google Scholar]
- 34.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, Aktories K, Bhakdi S. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc. Natl Acad. Sci. USA. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oancea E, Teruel MN, Quest AFG, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J. Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen K, Meyer T. In vivo and in vitro characterization of the sequence requirement for oligomer formation of Ca2+/calmodulin-dependent protein kinase II alpha. J. Neurochem. 1998;70:96–104. doi: 10.1046/j.1471-4159.1998.70010096.x. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian K, Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89:963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- 38.Teruel MN, Blanpied TA, Shen K, Augustine GJ, Meyer T. A versatile microporation technique for the transfection of cultured CNS neurons. J. Neurosci. Methods. 1999;93:37–48. doi: 10.1016/s0165-0270(99)00112-0. [DOI] [PubMed] [Google Scholar]
- 39.Teruel MN, Meyer T. Electroporation-induced formation of individual calcium entry sites in the cell body and processes of adherent cells. Biophys. J. 1997;73:1785–1796. doi: 10.1016/S0006-3495(97)78209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The influence of endosome-disruptive peptides on gene-transfer using synthetic virus-like gene-transfer systems. J. Biol. Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 41.Prchla E, Plank C, Wagner E, Blaas D, Fuchs R. Virus-mediated release of endosomal content in-vitro – different behavior of adenovirus and rhinovirus serotype-2. J. Cell Biol. 1995;131:111–123. doi: 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]