Abstract
Orthogonal ribosomes (o-ribosomes), also known as specialized ribosomes, are able to selectively translate mRNA not recognized by host ribosomes. As a result, they are powerful tools for investigating translational regulation and probing ribosome structure. To date, efforts directed towards engineering o-ribosomes have involved random mutagenesis-based approaches. As an alternative, we present here a computational method for rationally designing o-ribosomes in bacteria. Working under the assumption that base-pair interactions between the 16S rRNA and mRNA serve as the primary mode for ribosome binding and translational initiation, the algorithm enumerates all possible extended recognition sequences for 16S rRNA and then chooses those candidates that: (i) have a similar binding strength to their target mRNA as the canonical, wild-type ribosome/mRNA pair; (ii) do not bind mRNA with the wild-type, canonical Shine-Dalgarno (SD) sequence and (iii) minimally interact with host mRNA irrespective of whether a recognizable SD sequence is present. In order to test the algorithm, we experimentally characterized a number of computationally designed o-ribosomes in Escherichia coli.
INTRODUCTION
Gene expression involves two steps, transcription and translation. While a number of genetic tools exist for reprograming transcription in cells, far fewer tools exist for translation. Of the tools available in bacteria, the most popular are riboregulators, both cis- and trans-activating (1–4), and orthogonal ribosomes (o-ribosomes), also known as specialized ribosomes (5–9). In terms of reprograming translation, o-ribosomes are especially powerful as they enable one to partially decouple translation from the native protein synthesis machinery. In particular, o-ribosomes can translate genes with altered Shine-Dalgarno (SD) sequences not recognized by host ribosomes. Because of this fact, o-ribosomes can be used to explore translational regulatory mechanisms such as coupling (10,11) and to probe ribosome structure (6,12–15). Furthermore, o-ribosomes can be used to explore gene expression dynamics as they potentially provide a method for tuning translation rates. Finally, o-ribosomes may have application in synthetic biology as they introduce new functionality within cells (8,16–19).
O-ribosomes are duplicated ribosomes with mutations in the 3′ end of 16S rRNA that alter their specificity for mRNA (5–9) (Figure 1). In bacteria, translation initiation is primarily mediated by interactions between the 30S ribosomal subunit and the 5′ untranslated region of mRNA. Although many factors control this process, the most recognizable signal for translation is the SD sequence located approximately 6–12 bp upstream of the start codon (20–22). Complementary base-pair interactions between the SD sequence and the 3′ end of the 16S ribosomal RNA (rRNA), known as the anti-Shine-Dalgarno (ASD) sequence, serve to correctly position the 30S ribosomal subunit during the initiation process (22–24). The strength of this interaction is thought to influence translational efficiency as mutations in either the SD or ASD sequence that weaken the interaction reduce the amount of protein made (5,25). In the case of o-ribosomes, mutations are introduced into the ASD region such that they can base pair with complementary, noncanonical SD sequences not recognized by host ribosomes (5–9).
Figure 1.
Comparison of canonical and orthogonal translation. (a) Translation of canonical mRNAs is performed solely by the native ribosome and not the o-ribosome. (b) Orthogonal translation is specific only to cognate o-mRNAs. The native ribosomes are unable to translate the o-mRNA.
Initial efforts devoted towards engineering o-ribosomes in Escherichia coli involved changing two bases in the SD and ASD sequences (5,6). While these mutant ribosomes were sufficient for translating genes not recognized by host ribosomes, translation was inefficient (26). Furthermore, a number of researchers discovered that the o-ribosomes could be toxic to the cell (7,27). More recently, researchers have employed random mutagenesis and directed evolution to improve the functionality of o-ribosomes (7,8). Of notable significance is the recent work of Rackham and Chin, who proposed a novel dual-selection strategy for engineering o-ribosomes in E. coli. Unlike work in the past, their designs bypass many of the limitations associated with earlier ones, in particular toxicity.
To date, o-ribosome design has either involved ad hoc or random mutagenesis-based approaches. While these approaches have clearly been successful, one question is whether a rational, computational-based strategy could be employed in design. In particular, a computational approach would enable one to explicitly explore the different elements and associated hypotheses that factor into o-ribosome design. In this work, we propose a computational strategy for designing o-ribosomes in bacteria. The basic approach in our algorithm involves enumerating all possible ASD–SD pairs and then selecting those that minimally interfere with the translation of native mRNA. To demonstrate the utility of our algorithm, we experimentally tested a number of computationally designed o-ribosomes in E. coli. In the process, we were able to test a number of hypotheses regarding o-ribosome functionality. These findings should complement existing approaches based on random mutagenesis and screening.
MATERIALS AND METHODS
Bacterial strains, media and growth conditions
All cloning steps were performed in E. coli strain DH5αZ1 (F− deoR supE44 recA1 endA1 relA1 gyrA96 thi-1 Δ(lacZ-argF)U169 Φ80(lacZΔM15) hsdR17 attBλ::[PN25-tetR lacIq spcR]) (28). Subsequent experiments were conducted in E. coli strain LC100 (F− ilvG rfb-50 rph-1 attBλ::[PN25-tetR lacIq spcR]). LC100 was constructed by P1vir transduction of the chromosomally integrated TetR/LacI expression cassette from DH5αZ1 into strain MG1655 (29). Cultures were grown in Luria-Bertani (LB) liquid media for all experiments. All media were supplemented with 20 µg/ml chloramphenicol and 100 µg/ml ampicillin. Inducers anhydrotetracycline (aTc) and isopropyl-β-D-galactopyranoside (IPTG) were used at concentrations of 200 ng/ml and 1 mM, respectively, unless otherwise specified. All cultures were grown at 37°C.
O-ribosome expression systems
For ribosomal expression in E. coli, the rRNA operon rrnB was amplified by PCR using pKK3535 (a gift from H. Noller, UCSC) (30) using primers ATAGCGGGTACCGCCGCTGAGAAAAAGCGAAGC and ATACTGCAGTGTTCGTCTTCGGCACATAC bearing KpnI and PstI restriction sites (underlined). The resulting rrnB PCR fragment was cloned into the plasmid pZA31 (p15A origin, chloramphenicol resistance) under control of the aTc-inducible promoter PLtetO-1 (28), resulting in the plasmid pZA31-WT. Derivatives of pZA31-WT (pZA31-OR1 to pZA31-OR8) bearing mutations to the 3′ end of the 16S rRNA were made by enzymatic inverse PCR (EI-PCR) site-directed mutagenesis (31) using primers GGAAAGGTCTCAGGTTGGATCANNNNNNTACCTTAAAGAAGCGTACTTTGTAG and GAGTAGGTCTCAAACCGCAGGTTCCCCTACG bearing BsaI restriction sites (underlined), where the NNNNNN subsequence corresponds to the 6-nucleotide mutation determined by the rational selection algorithm. Mutant rrnB operons generated in pZA31-WT were subsequently subcloned by KpnI and PstI digestion into pZE31 (ColE1 origin, chloramphenicol resistance) resulting in pZE31-WT and derivatives pZE31-OR1 to pZE31-OR8. Similarly, the rrnB operon was cloned into the plasmid pTrc99A under the control of the strong, IPTG-inducible promoter Ptrc in two steps (32). First, the 23S and 5S rRNA segments were subcloned from pKK3535 into pTrc99A by XbaI and BspHI digestion and ligation resulting in pTrc23S. Second, the 16S rRNA portions of the rrnB operon was subcloned from pZA31-WT and pZA31-OR1 into pTrc23S by KpnI and XbaI digestion and ligation resulting in plasmids pTrcWT and pTrcOR1.
Orthogonal mRNA (o-mRNA) expression systems
To monitor the translation mediated by o-ribosomes and also to determine if the o-mRNA remained untranslated by host ribosomes, an optimized green fluorescent protein (GFP) variant, we term gfp (33) was used. For the o-mRNA reporter system, gfp was amplified using primers ATAGAATTCTAANNNNNNAAAAAAATGAGTAAAGGAGAAG and ATAAAGCTTTTATTATTTGTATAGTTC containing EcoRI and HindIII restriction sites (underlined), where the NNNNNN subsequence denotes the complementary mutation determined by the algorithm, followed by cloning into the EcoRI and HindIII sites of pZE12 (ColE1 origin, ampicillin resistance) under the control of the IPTG-inducible promoter PLlacO-1 (28).
Fluorescence and cellular growth measurements
To monitor fluorescent protein expression, cultures were grown overnight in noninducing LB liquid media. Cultures were then diluted 1:100 in fresh LB media supplemented with appropriate inducers. These cultures were then allowed to grow with vigorous aeration for 18 h before fluorescence and optical density measurements. All fluorescence measurements were made using a Tecan Safire2 plate reader.
To determine the impact on cell growth due to the expression of o-ribosomes, E. coli LC100 cells containing o-ribosome expression plasmids based on pZA31-WT, pZE31-WT and pTrcWT (see above) were grown overnight in LB liquid media. Cultures were then subcultured to an OD600 of 0.1 in fresh LB media followed by 1 h of noninduced growth. After 1 h of growth, aTc (200 ng/ml) or IPTG (1 mM) was added to allow for expression of the o-ribosome. Growth measurements were made every 1 h with an Amersham UltroSpec 10 spectrophotometer.
RESULTS
Computational procedure for designing o-ribosomes
We sought to develop a computational approach for engineering o-ribosomes as an alternative to random mutagenesis. Working under the assumption that base-pair interactions between the ASD and SD serve as the primary mode for ribosome binding and translational initiation, we computationally generated a library of mutations to the ASD and then selected those predicted to minimally interact with the native translation process yet still efficiently translate genes with the orthogonal SD sequences (Figure 2).
Figure 2.
Illustration of the selection algorithm. (a) Enumeration. All possible ASD–SD pairs are enumerated. (b) Similarity. Mutant ASD–SD pairs are selected with similar binding energies as the wild-type, canonical ASD–SD pair and that also lack RNA secondary structure within the ASD region. (c) Orthogonality. ASD–SD pairs are enriched for pairs that do not cross-react with the wild-type ASD–SD pair and vice versa. (d) Host cross-reactivity. The candidate o-ribosomes are ranked ordered based on the number of times they have stronger interactions with host genes than the native ribosomes.
The first two steps in the algorithm involve enumerating all possible mutant ASD–SD complementary sequence pairs and then comparing their binding energies to the wild-type, canonical ASD–SD pair using the RNAfold and RNAduplex programs from the Vienna RNA Package (34). To fix the size of the mutant library, we only considered randomizing the six bases from region 1535 to 1540 of the rrnB 16S rRNA (Figure 2a). This segment is known to bind the mRNA translation initiation region (TIR) (5,20,22,25). For each 16S rRNA mutant, a cognate (reverse complement) SD sequence was determined to create an o-ribosome–mRNA pair. In this way, it selected only o-ribosomes with their cognate mRNA pair unlike other experimental approaches that select both cognate and potentially noncognate ASD–SD pairs (7,8).
In our free-energy calculations, we also considered the neighboring residues flanking the mutated six bp region of the 16S rRNA (e.g. 5′-AUCAnnnnnnUA-3′, where n denotes the mutated base pair). Of the 4096 possible mutant ASD–SD pairs, the algorithm selects those ASD–SD pairs with free energies of binding within 0.5 kcal/mol of the wild-type, canonical ASD–SD pairs (Figure 2b). Extended base pairing between 16S rRNA and the TIR has previously been shown to inhibit translation, presumably due to the formation of a tightly bound complex between the 30S ribosomal subunit and mRNA that does not allow the ribosome to proceed with elongation (35). Similarly, weak interactions between 16S rRNA and the TIR are believed to result in inefficient translational initiation (7). We, therefore, selected only those ASD–SD pairs with binding strengths similar to the wild-type ASD–SD interaction (−9.2 kcal/mol). We also discarded 16S rRNA mutants that formed any RNA secondary structures within the ASD region in order to ensure efficient mRNA binding. This region is unstructured in the wild-type ribosome (22,36) and we hypothesized that hairpins resulting from mutations to this region would inhibit the binding of the 30S ribosomal subunit to mRNA.
The third step in the algorithm involves computationally enriching for mutant ASD–SD pairs that are orthogonal to the wild-type, canonical ASD–SD pair (Figure 2c). The algorithm enforces this orthogonality constraint by removing those mutant-wildtype pairs whose mutual binding energies are less than −1 kcal/mol. In other words, the mutant ribosome should not be able to bind the canonical SD sequence nor should the wild-type ribosome be able to bind the corresponding mutant SD sequence, where failure to bind is defined by a −1 kcal/mol threshold. This threshold is well above the average energy of −4.2 kcal/mol associated with the binding of the wild-type ASD to the core SD motifs AGGA, GGAG and GAGG (37–40). Note that this latter binding energy is far weaker than the threshold used in the second step. Our rational is that many genes do not possess a full, canonical SD sequence (AGGAGG) but instead contain subsets of this sequence, the core motifs mentioned above. This measure (−4.2 kcal/mol) was also previously used by Ma and coworkers (38) as a threshold criterion for ribosome binding.
The result of the first three steps of the algorithm is a set of candidate o-ribosomes. In order to rank these candidates, we included a fourth step that accounts for cross-reactivity with host mRNA. While the third step also addresses cross-reactivity, it does so only for the canonical SD. However, many mRNA TIR regions lack a conserved SD. Therefore, we also considered all known TIR regions irrespective of whether they had a SD sequence or not. A 30-bp window, 20 bases upstream and 10 bases downstream of the predicted start codon, was used to measure cross-reactivity. This region is known to be the primary binding site for wild-type ribosomes (21,41); downstream regions of the host mRNA are often saturated with translating ribosomes and, therefore, unlikely sites for interference by the mutant ribosomes (though we note that internal translation initiation sites have been documented) (42–45). To determine the extent of cross-reactivity for a given o-ribosome, we counted the number of TIR regions that it was able to bind more strongly than the host, wild-type ribosome. We then rank ordered the o-ribosomes based on this number (Table S1).
Experimental validation of the computationally designed o-ribosomes
We next sought to experimentally validate the algorithm predictions. We chose four designs among the top eight mutations predicted by the algorithm (Table 1). We then tested translation by these o-ribosomes using a two-plasmid expression system, one for the o-ribosome and the other for the cognate mRNA reporter. To express the o-ribosome, we cloned the E. coli rrnB rRNA operon with the desired mutations under the control of the aTc-inducible promoter PLtetO-1 into a medium number copy plasmid. The PLtetO-1 promoter, a phage lambda PL derivative containing engineered TetR operator sites, has been previously observed to remain tightly repressed in noninducing conditions for low and medium copy number plasmids (28). Similarly, the corresponding o-mRNA reporter was produced by cloning the GFP with the cognate SD sequence under the control of the IPTG-inducible promoter PLlacO-1 on a high copy number plasmid. The PLlacO-1 promoter is analogous to the PLtetO-1 promoter except that is uses LacI operator sites in place of TetR operator sites (28). Use of the PLlacO-1 promoter on a high copy plasmid allowed for high levels of expression for the o-mRNA in inducing conditions and also provided a sensitive platform for detecting weak GFP translation. The o-ribosome and reporter plasmids were then transformed into the E. coli strain LC100.
Table 1.
Orthogonal ASD-SD pairs used in this work
| Name | ASD sequence | SD sequence | ΔGbinding (kcal/mol) | Reference |
|---|---|---|---|---|
| OR1 | AUCACGAGACUA | GUCUCG | −9.1 | This work |
| OR2 | AUCAACGAGGUA | CCUCGU | −8.7 | This work |
| OR3 | AUCAACGAGCUA | GCUCGU | −9.4 | This work |
| OR4 | AUCACGGAACUA | GUUCCG | −8.8 | This work |
| OR5 | AUCAGGGGGGUA | CCCCCC | −13.2 | This work |
| OR6 | AUCAGGGCGCUA | GCGCCC | −13.1 | This work |
| OR7 | AUCAUGGCUGUA | CAGCCA | −9.3 | This work |
| OR8 | AUCAUGGCGUUA | ACGCCA | −8.7 | This work |
| OR-HUI | AUCACACACUUA | AGUGUG | −6.7 | (5) |
Binding energies were determined by the RNAduplex software component of the Vienna RNA Package (34).
Cells harboring o-ribosome and o-mRNA cognate plasmid pairs were grown for 18 h in LB media at 37°C in order to determine whether the o-ribosome candidate designs were able to efficiently translate their cognate o-mRNA. When grown under inducing conditions (IPTG at 1 mM and aTc at 200 ng/ml), all four candidate designs resulted in high levels of GFP expression as determined by fluorescence measurements (Figure 3a). However, when grown in the absence of aTc but presence of IPTG, such that only the o-mRNA was expressed, negligible fluorescence relative to the empty plasmid control was detected. These results indicate that the o-mRNA is not translated in the absence of o-ribosome expression. Note that much greater fluorescence was observed using the o-ribosome relative to the wild-type control, most notably for o-ribosomes OR1 and OR4. In the case of the wild-type control, the native rrnB operon was expressed from the aTc-inducible promoter and GFP with the canonical SD (AGGAGG) was expressed from the IPTG inducible promoter. We suspect that the increased expression by the o-ribosomes relative to the control is due to the lack of competing substrates arising, at least in part, from the orthogonality constraint imposed in the selection algorithm. As partial evidence in support of this conclusion, expressing the wild-type rrnB operon from the aTc-inducible promoter increased translation of gfp with the canonical SD, though not to the same levels as the o-ribosome/o-mRNA pairs. Collectively, these results demonstrate the candidate designs work as predicted. Finally, as a point of reference, we also tested the o-ribosome previously characterized by Hui and de Boer (5) using our two plasmid expression system (Figure 3b). This o-ribosome (OR-HUI) was able to translate its cognate o-mRNA at levels roughly equal to wild type. Note that this o-ribosome was not selected by our algorithm.
Figure 3.
Verification of o-ribosomes selected by algorithm in E. coli. Cells were grown in LB with specified inducers for 18 h prior to fluorescence measurements. The control is LC100 cells containing empty plasmids. WT denotes cells expressing wild-type rrnB rRNA and GFP with the canonical SD. (a) Optimally selected o-ribosome–mRNA pairs. (b) Previously reported o-ribosome of Hui and de Boer (5) and o-ribosomes designed to test different steps in the selection algorithm.
In order to test the significance of the different steps of the algorithm, we also designed two additional types of o-ribosomes. The first type was used to test the similarity constraint (step 2). To test this constraint, we designed two o-ribosomes OR5 and OR6 (Table 1) that bound their cognate ASD at energy levels significantly greater than the cognate ASD–SD pair (an average of −13 kcal/mol versus −9 kcal/mol for the algorithm-selected o-ribosomes) and thus violated the second selection step in algorithm. Consistent with our hypothesis, these o-ribosomes were only able to weakly translate GFP with their cognate ASD (Figure 3b). Presumably, this inhibition is due to the inability of the ribosome to release from the TIR during elongation as too tight of a complex forms. Similar results have been observed when there is extended base-pairing between the TIR and wild-type 16S rRNA in E. coli (35). Collectively, these results suggest that the binding energy for an o-ribosome ASD–SD pair cannot be too strong, thereby justifying the similarity step in the algorithm. Note, we did not directly consider the reciprocal case where binding was too weak as equivalent experiments were performed with our controls and orthogonality experiments (see below). Suffice to say, when the binding energy is too weak between an ASD–SD pair, translation is severely inhibited.
The second type of o-ribosome was used to test the cross-reactivity constraint, the last step in the selection algorithm. To test this constraint, we built two o-ribosomes, OR7 and OR8 (Table 1) that were the two worst ranked among 216 candidate designs listed in Table S1. Despite ranking at the bottom of the list, however, these two ribosomes were able to efficiently translate their cognate o-mRNA despite greater predicted cross-reactivity with host mRNA (Figure 3b). As these results demonstrate, this step in the algorithm is not necessary. The reason that we included it in the algorithm was in order to provide some measure for selecting among the various candidate designs. While all of the top-ranked o-ribosome were able to efficiently translate their cognate o-mRNA, the same was also true for the low ranking ones. We do note that all of the candidate designs are predicted to have a high degree of cross-reactivity. The top ranking o-ribosome in E. coli, for example, is predicted to out compete the wild-type ribosome at 1042 TIR's. As a comparison, the bottom ranking o-ribosome is predicted to out compete wild type at 2129 TIR's.
As a final step in characterizing our o-ribosomes in E. coli, we measured the degree of GFP translation at varying levels of o-ribosome and mRNA expression for OR1 and OR2. In order to produce sub-saturating amounts of o-ribosome or o-mRNA, one inducer (aTc or IPTG) was varied with the other fixed at a saturating concentration (Figure 4). The results from these experiments indicate that GFP translation is dose dependent, both with respect to o-ribosome and o-mRNA expression. Furthermore, these results indicate that o-ribosome translation can be tuned at the level of o-ribosome expression.
Figure 4.
Dose-dependent response of o-ribosome and o-mRNA expression. Cells harboring o-ribosome–mRNA pairs or empty plasmid controls were grown in (a) mRNA inducing conditions (IPTG at 1 mM) with varied concentrations of o-ribosome inducer (aTc). (b) o-ribosome inducing conditions (aTc at 200 ng/ml) with varied concentrations of o-mRNA inducer (IPTG). All cultures were grown in LB for 18 h prior to fluorescence measurements.
Mutual orthogonality of o-ribosomes: computational predictions versus experimental results
We next tested the mutual orthogonality of five proposed o-ribosome designs and compared the results to predictions based on the underlying free-energy calculations, the engine of our algorithm. In these experiments, we measured the level of GFP expression for each o-ribosome paired against the cognate o-mRNA for other o-ribosomes. In addition to the four optimal o-ribosomes (OR1–OR4), we also included one other previously discussed o-ribosome (OR7) to expand the analysis. As demonstrated in Figure 5a, the five o-ribosomes were almost perfectly orthogonal with respect to one other in terms of relative fluorescence measurements.
Figure 5.
Mutual orthogonality of o-ribosomes. OR and OM refer to cognate o-ribosome and o-mRNA, respectively. (a) Observed relative fluorescence. (b) Predicted binding energies of cognate and noncognate pairs. (c) Rescaled (logarithmic) relative fluorescence.
In order to compare these results with the predictions based on the free-energy calculations, we replotted the same data on a logarithmic scale (Figure 5c). As translational efficiency is believed to be correlated with the strength of the ribosome/mRNA interaction (assuming, of course, that it is not too strong), the logarithm of the relative fluorescence should scale proportionally with the free energy of binding. Comparing the scaled experimental data with the predicted free energies (Figure 5b versus Figure 5c), we observed some degree of correlation though the five o-ribosomes tested were more orthogonal than predicted by simple thermodynamic calculations. These results suggest that while the thermodynamic criteria provides predictable results at high free-energy values of binding, at lower free energies other factors likely become increasingly important with regards to determining the ribosome–mRNA interaction.
The role of copy number and promoter strength on o-ribosome toxicity
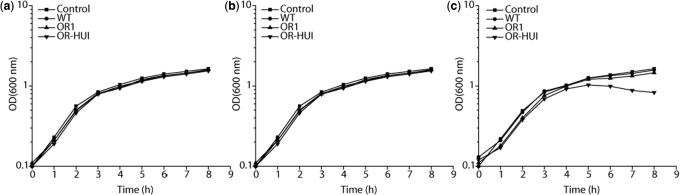
A number of studies have previously noted that the expression of o-ribosomes may be toxic to the cell (7,8,27). To determine if and under what conditions our o-ribosomes may be toxic, we measured the effect of expressing them on cell growth. As a control, we also included the Hui and de Boer o-ribosome, as it is known to retard cell growth under certain conditions. As illustrated in Figure 6a, we did not observe any growth defect when expressing OR1 (similar results were observed for the other o-ribosomes). We also did not observe any growth defect when expressing the Hui and de Boer ribosome, despite previous observations otherwise. We do note that no defect was observed with this o-ribosome in the original study by Hui and de Boer (5,6). Therefore, we hypothesized that the previously observed growth defects may be due to the choice of the expression system. Because our o-ribosome expression system was constructed on a medium copy number plasmid, it may not express the mutant rRNA as strongly as other expression systems, thereby minimizing any toxic effect. To test this hypothesis, we cloned the o-ribosomes into an otherwise identical expression vector utilizing a high copy number origin of replication. Once again, however, we did not observe any defect in growth (Figure 6b). These results indicate that the plasmid copy number does not have an effect. Next, we tested the promoter. Other studies observing growth defects have used the strong, hybrid Ptrc promoter (18) or PlacUV5 promoter (7). Likewise, the original Hui and de Boer studies, where no defect was observed, used the phage lambda PL promoter, the same promoter used in this study. Therefore, to test whether the promoter may have an effect, we cloned the o-ribosomes into a medium copy number plasmid under the control of the IPTG-inducible promoter Ptrc. In this case, we observed no defect in growth with OR1 but did observe a defect with the Hui and de Boer o-ribosome, similar to what was observed previously (Figure 6c).
Figure 6.
Growth of E. coli LC100 harboring different o-ribosome expression systems. Cells were subcultured to an OD600 of 0.1 in fresh LB media and grown for 1 h prior to induction with aTc (200 ng/ml) or IPTG (1 mM). Control and WT refer to cells harboring empty expression vector or the wild-type rrnB operon, respectively. (a) PLtetO-1 promoter (pZA31) with medium-copy p15A origin. (b) PLtetO-1 (pZE31) with high-copy ColE1 origin. (c) Ptrc promoter(pTrc99A) with medium- to low-copy pMB1 origin.
Collectively, the results indicate that, under the conditions used in our experiments, expression of the o-ribosomes has no effect on cell growth. However, when expressed from a strong promoter, some o-ribosomes may be toxic to the cell. Note that we did not directly address toxicity in our algorithm, so the lack of toxicity associated with our o-ribosomes is somewhat serendipitous. Finally, these results along with those for GFP expression (Figure 4) demonstrate that the expression system used in this study is highly effective for examining o-ribosome–mRNA interactions without the loss of cellular viability.
DISCUSSION
In this work, we have developed and experimentally validated a computational method for engineering o-ribosomes. The method involves enumerating all possible ASD–SD sequences and then selecting those that are orthogonal to the wild-type ASD–SD pair while also having similar binding strengths. The first constraint is used to ensure that the o-ribosomes do not bind too tightly to their target mRNA, something that can potentially inhibit translational efficiency, and the second to enforce orthogonality. As a number of ASD–SD pairs satisfy these two constraints, the algorithm then rank orders them based on the degree to which they interact with predicted TIR sequences within the genome of interest.
The two constraints based on similarity and orthogonality were motivated by a simple hypothesis regarding o-ribosome functionality. In the case of the similarity constraint, we were able to demonstrate that o-ribosomes predicted to bind too tightly to their ASD sequence inefficiently translate their cognate mRNA. Likewise, the orthogonality constraint ensures that only the o-ribosomes are able to translate their cognate mRNA. We were able to establish this latter condition for all our designs. These two constraints, however, do not yield a single solution. Rather, multiple ASD–SD pairs satisfy these constraints. Therefore, we introduced one additional step to distinguish between these potential designs based on cross-reactivity. We initially hypothesized that o-ribosomes with less cross-reactivity would be less toxic to the cell. However, as our results demonstrate, the cross-reactivity condition has no effect on performance, as the top and bottom ranking o-ribosomes are able to translate their cognate mRNA with equal efficiency. Furthermore, none of the o-ribosomes designed using our algorithm exhibited any toxicity, irrespective of ranking and the expression system. We, therefore, conclude that this step in the algorithm is unnecessary and that our rank ordering of the o-ribosomes is not relevant for design.
We note that, in developing the method, we have assumed that the ASD–SD interaction provides the key signal for ribosome binding and translation initiation. While sufficient for inducing initiation, the SD sequence is not necessary as many genes lack one. In fact, the only signal both necessary and sufficient for initiation is the start codon (46,47). Clearly, other sequence-dependent factors such as alternate recognition sites and secondary structure play a role in initiating translation both in conjunction and independently of the SD sequence (48). Despite these alternative mechanisms, our results illustrate the importance of the ASD–SD interaction during translational initiation and also demonstrate that certain properties of the wild-type ribosome must be preserved when engineering o-ribosomes. However, they also beg the question regarding the mechanism by which genes lacking SD sequences are translated.
The primary advantage of our strategy is that selection is performed on the computer, potentially reducing the time and resources necessary to vet different candidate designs. In addition, the algorithm can potentially be used to design o-ribosomes in diverse species of bacteria, the only requirement being a sequenced genome. Finally, we note that the choice of the expression system can have profound results. While none of our o-ribosomes were toxic to the cell, we demonstrated that the previously characterized o-ribosome of Hui and de Boer is toxic using some expression systems but not with others. Why some o-ribosomes are toxic but other are not is still not known.
In summary, o-ribosomes are a powerful tool for regulating the translation of mRNA not recognized by host ribosomes in a dose-dependent manner. In addition to being able to tune translation, o-ribosomes can also be used to probe ribosome structure (6,12–15), enhance the efficiency of synthetic amino acid incorporation (19), establish the translational coupling of adjacent genes (10,11) and explore the mechanism of ribosome binding and translational initiation (9,49). In conclusion, we were able to use a computational approach based on a few simple hypotheses for designing o-ribosomes. The computational methodology developed here simplifies some steps in the design process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Harry Noller for pKK3535, and Barry Canton for initial materials and helpful advice. This work was supported by the National Science Foundation CAREER Award CBET-0644744. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation CAREER Award CBET-0644744.
Conflict of interest statement. None declared.
REFERENCES
- 1.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 2.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J. Am. Chem Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 4.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 5.Hui A, de Boer HA. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl Acad. Sci. USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui A, Jhurani P, de Boer HA. Directing ribosomes to a single mRNA species: a method to study ribosomal RNA mutations and their effects on translation of a single messenger in Escherichia coli. Meth. Enzymol. 1987;153:432–452. doi: 10.1016/0076-6879(87)53070-1. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Holland-Staley CA, Cunningham PR. Genetic analysis of the Shine-Dalgarno interaction: selection of alternative functional mRNA-rRNA combinations. RNA. 1996;2:1270–1285. [PMC free article] [PubMed] [Google Scholar]
- 8.Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat. Chem. Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 9.Skorski P, Leroy P, Fayet O, Dreyfus M, Hermann-Le Denmat S. The highly efficient translation initiation region from the Escherichia coli rpsA gene lacks a shine-dalgarno element. J. Bacteriol. 2006;188:6277–6285. doi: 10.1128/JB.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govantes F, Andujar E, Santero E. Mechanism of translational coupling in the nifLA operon of Klebsiella pneumoniae. EMBO J. 1998;17:2368–2377. doi: 10.1093/emboj/17.8.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rex G, Surin B, Besse G, Schneppe B, McCarthy JE. The mechanism of translational coupling in Escherichia coli. Higher order structure in the atpHA mRNA acts as a conformational switch regulating the access of de novo initiating ribosomes. J. Biol. Chem. 1994;269:18118–18127. [PubMed] [Google Scholar]
- 12.Hui AS, Eaton DH, de Boer HA. Mutagenesis at the mRNA decoding site in the 16S ribosomal RNA using the specialized ribosome system in Escherichia coli. EMBO J. 1988;7:4383–4388. doi: 10.1002/j.1460-2075.1988.tb03337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poot RA, van den Worm SH, Pleij CW, van Duin J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998;26:549–553. doi: 10.1093/nar/26.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rackham O, Wang K, Chin JW. Functional epitopes at the ribosome subunit interface. Nat. Chem. Biol. 2006;2:254–258. doi: 10.1038/nchembio783. [DOI] [PubMed] [Google Scholar]
- 15.Yassin A, Fredrick K, Mankin AS. Deleterious mutations in small subunit ribosomal RNA identify functional sites and potential targets for antibiotics. Proc. Natl Acad. Sci. USA. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin JW. Programming and engineering biological networks. Curr. Opin. Struct. Biol. 2006;16:551–556. doi: 10.1016/j.sbi.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Chin JW. Modular approaches to expanding the functions of living matter. Nat. Chem. Biol. 2006;2:304–311. doi: 10.1038/nchembio789. [DOI] [PubMed] [Google Scholar]
- 18.Rackham O, Chin JW. Cellular logic with orthogonal ribosomes. J. Am. Chem. Soc. 2005;127:17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- 19.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 20.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- 22.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 23.Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl Acad. Sci. USA. 1975;72:4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calogero RA, Pon CL, Canonaco MA, Gualerzi CO. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1988;85:6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob WF, Santer M, Dahlberg AE. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl Acad. Sci. USA. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brink MF, Verbeet MP, de Boer HA. Specialized ribosomes: highly specific translation in vivo of a single targetted mRNA species. Gene. 1995;156:215–222. doi: 10.1016/0378-1119(95)00030-a. [DOI] [PubMed] [Google Scholar]
- 27.Wood TK, Peretti SW. Construction of a specialized-ribosome vector for cloned-gene expression in E. coli. Biotechnol. Bioeng. 1991;38:891–906. doi: 10.1002/bit.260380811. [DOI] [PubMed] [Google Scholar]
- 28.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 30.Brosius J, Ullrich A, Raker MA, Gray A, Dull TJ, Gutell RR, Noller HF. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 31.Stemmer WP, Morris SK. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high-efficiency, site-directed mutagenesis. Biotechniques. 1992;13:214–220. [PubMed] [Google Scholar]
- 32.Brosius J, Erfle M, Storella J. Spacing of the -10 and -35 regions in the tac promoter. Effect on its in vivo activity. J. Biol. Chem. 1985;260:3539–3541. [PubMed] [Google Scholar]
- 33.Miller WG, Lindow SE. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 34.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komarova AV, Tchufistova LS, Supina EV, Boni IV. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noller HF, Woese CR. Secondary structure of 16S ribosomal RNA. Science. 1981;212:403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- 37.Lithwick G, Margalit H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 2003;13:2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Campbell A, Karlin S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 2002;184:5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osada Y, Saito R, Tomita M. Analysis of base-pairing potentials between 16S rRNA and 5′ UTR for translation initiation in various prokaryotes. Bioinformatics. 1999;15:578–581. doi: 10.1093/bioinformatics/15.7.578. [DOI] [PubMed] [Google Scholar]
- 40.Schurr T, Nadir E, Margalit H. Identification and characterization of E.coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 1993;21:4019–4023. doi: 10.1093/nar/21.17.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starmer J, Stomp A, Vouk M, Bitzer D. Predicting Shine-Dalgarno sequence locations exposes genome annotation errors. PLoS Comput. Biol. 2006;2:e57. doi: 10.1371/journal.pcbi.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iost I, Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquet M, Kepes A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied studied by measurement of its beta-galactosidase synthesizing capacity in vivo. J. Mol. Biol. 1971;60:453–472. doi: 10.1016/0022-2836(71)90181-1. [DOI] [PubMed] [Google Scholar]
- 44.Miller OL, Jr, Hamkalo BA, Thomas CA., Jr Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- 45.Smith RA, Parkinson JS. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl Acad. Sci. USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moll I, Grill S, Gualerzi CO, Blasi U. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 47.Nakamoto T. A unified view of the initiation of protein synthesis. Biochem. Biophys. Res. Commun. 2006;341:675–678. doi: 10.1016/j.bbrc.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 48.de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003;331:737–743. doi: 10.1016/s0022-2836(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 49.de Boer HA, Hui AS. Sequences within ribosome binding site affecting messenger RNA translatability and method to direct ribosomes to single messenger RNA species. Meth. Enzymol. 1990;185:103–114. doi: 10.1016/0076-6879(90)85011-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.