Abstract
How folding of proteins is coupled to their synthesis remains poorly understood. Here, we apply single-molecule fluorescence imaging to full protein synthesis in vitro. Ribosomes were specifically immobilized onto glass surfaces and synthesis of green fluorescent protein (GFP) was achieved using modified commercial Protein Synthesis using Recombinant Elements that lacked ribosomes but contained purified factors and enzyme that are required for translation in Escherichia coli. Translation was monitored using a GFP mutant (F64L/S65T/F99S/M153T/V163A) that has a high fluorophore maturation rate and that contained the Secretion Monitor arrest sequence to prevent dissociation from the ribosome. Immobilized ribosomal subunits were labeled with Cy3 and GFP synthesis was measured by colocalization of GFP fluorescence with the ribosome position. The rate of appearance of colocalized ribosome GFP was equivalent to the rates of fluorescence appearance coupled with translation measured in bulk, and the ribosome–polypeptide complexes were stable for hours. The methods presented here are applicable to single-molecule investigation of translational initiation, elongation and cotranslational folding.
INTRODUCTION
Protein synthesis rates during translation by the ribosome control many key cellular processes. Protein folding in living cells is inherently coupled to protein synthesis and chain elongation. There is considerable evidence that some nascent chains fold into their native structures in a cotranslational manner before release from the ribosome (1). Furthermore, chaperones, such as GroEL, are estimated to assist folding of only ∼5% of newly synthesized protein in growing Escherichia coli (2). Translational elongation events such as pausing and frameshifting are linked to protein folding, export and expression.
Elongation rates have been measured using in vivo and in vitro translation assays in prokaryotic system. In vivo, protein synthesis occurs at a rate of ∼10–20 amino acids per second (3), and dilution in extracts leads to slower rates in vitro (4). Nascent chains are extruded from the ribosome through a long, narrow exit tunnel in the large subunit. This tunnel can accommodate ∼22–26 amino acids, and constricts folding of higher order structure for the nascent chain (5,6). A growing body of data suggests strong interplay of the nascent chain composition, the exit tunnel and elongation rates (5,6).
The dynamics of full protein synthesis and cotranslational folding of nascent polypeptide remain unclear. Current bulk methods do not measure protein synthesis in real time; instead discrete time points are obtained to measure protein concentration biochemically, or enzymatic reporter systems are used, such as luciferase. Ensemble averaging in these bulk experiments blurs the detailed dynamic behavior of individual molecules. Single-molecule analysis of translation removes ensemble analysis and allows detailed examination of dynamics. However, single-molecule studies on protein synthesis to date have focused on individual kinetic steps, or overall protein synthesis in cells (7); new methods are needed to couple ribosomal dynamics with protein synthesis and folding rates.
Here, we present a simple single-molecule system to monitor real-time translation and folding of proteins. Ribosomes are specifically immobilized on glass surfaces, and protein synthesis and folding is monitored in real time through synthesis and rapid maturation of a single green fluorescent protein (GFP) on the immobilized ribosome. However, GFP requires slow posttranslational modification of the folded protein to become fluorescent. Here, we use a fast-maturing mutant of GFP that provides improved time resolution for fluorescence. Nascent chains are stalled on immobilized ribosomes using the Secretion Monitor (SecM) arrest sequence, which lodges in the peptide exit tunnel and halts elongation. Thus, we can observe appearance of GFP fluorescence as a surrogate marker for coupled translation protein folding and chromophore maturation.
METHOD
Sample preparation and experiments
Isolated ribosomes with a 23 nt extension in helix 44 of the 16S rRNA (8) were hybridized with biotin and Cy3-modified oligonucleotide at 30°C for 30 min and purified by gel filtration (MicroSpin S300 HR, GE Healthcare UK Ltd, Buckinghamshire, UK). The glass surface was coated with 3 mg/ml biotinylated BSA for 2 min and derivatized with 0.33 mg/ml streptavidin for 2 min. After binding of ribosome–oligonucleotide complexes to the derivatized surface, template DNA and the Protein Synthesis using Recombinant Elements (PURE) system ▵ ribosome (see text) were added. At each step the surface was rinsed with the sample buffer (50 mM Tris-acetate (pH 7.5), 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA and 6 mM β-mercaptoethanol) to remove any unbound material. The fluorescence images were recorded by a sensitive CCD camera (C9100-12, Hamamatsu Photonics, Hamamatsu, Japan) combined with Olympus IX70 microscope. GFP fluorescence image was taken with 200-ms time exposure right after the stage was moved quickly to avoid photobleaching molecules due to the absence of O2-scavenging system. GFP and Cy3 fluorescence was detected upon 488 and 532 nm excitation. After GFP images were recorded with 200-ms time exposure for 20 s, Cy3 images were detected at same image position by changing the dichroic filter and laser source quickly for image colocalization. The colocalization position for both GFP and Cy3 images was calibrated by non-specifically immobilized fluorescent beads. The cover glass was treated by plasma cleaning to get rid of fluorescent impurities and non-specific attachment of ribosome and other components beforehand every time. Nearly 170 × 170 pixels of GFP and Cy3 images were picked up from center of 512 × 512 pixels of whole image area and subtracted images filtered by Gaussian blurring with five pixels radius from original images to obtain high signal-to-noise using Adobe Photoshop.
RESULTS AND DISCUSSION
Use of a purified E. coli translation system
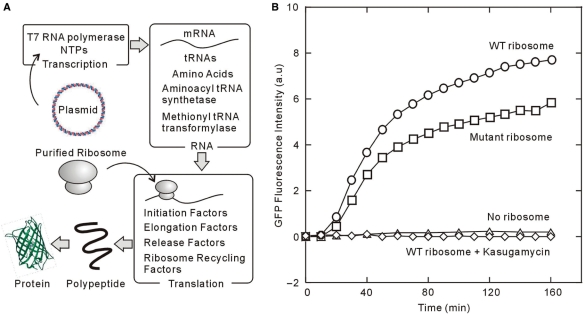
We have used a reconstituted, cell-free protein synthesis system to observe single-molecule translation. This PURE (Post Genome Institute, Tokyo, Japan) system is commercially available and is composed of individually prepared components required for gene expression in E. coli (9,10). As such, it is possible to exclude, add, or optimize the concentration of components to verify their effects on the translation system and production of higher order complexes (Figure 1A). The modified PURE system ‘▵ ribosome’ is lacking ribosomes, which allows our use of purified mutant ribosomes (Figure 1A).
Figure 1.
(A) The schematic illustration showing the PURE system ▵ ribosome. The PURE system ▵ ribosome is the cell-free protein synthesis system composed of every component except for the ribosome for gene expression. The target protein will be expressed, which is started by mixing the template DNA introduced into pET vector and our purified ribosome. (B) The time course of the fluorescent intensity of GFPS65T in the presence of wild-type ribosome (open circle), C68 mutant ribosome (open square) and wild-type ribosome in 10 mM kasugamycin (open diamond) or in the absence of ribosome (open triangle). The ribosome concentrations are 300 nM for every condition. The emission spectrum was checked every 10 min except for no ribosome condition and taken their peak value at 515 nm when excited at 488 nm.
To test the efficiency of the PURE system, we monitored appearance of GFP mutant Ser 65→Thr fluorescence as a function of time during a translation reaction. GFPS65T optimized the excitation wavelength at 488 nm and increases the quantum yield of fluorescence intensity and maturation rate approximately four-fold over that of wild-type GFP in vivo (11). Translation of mutant GFP was detected by the fluorescence of GFPS65T; the reaction was initiated by using the PURE system ▵ ribosome extract and adding ribosomes and template DNA. The synthesis and subsequent maturation of GFPS65T by wild-type E. coli ribosomes gradually increases in an hour (Figure 1B), which agrees well with prior observations (12). We also prepared C68 mutant ribosome for single-molecule imaging, which is designed to hybridize to an extension genetically engineered into the 16S rRNA (8,13) for immobilization on the surface. To test the effect of the C68 extension loop on synthesis activity, the translation of GFPS65T by C68 mutant ribosome was compared with that of wild-type ribosomes. Figure 1B indicates that the C68 mutant ribosome has 75% GFPS65T synthesis activity in comparison with the wild-type ribosome in the bulk solution; no translation was observed in the absence of the ribosome or in the presence of the ribosome including 10 mM kasugamycin, which inhibits translational initiation. These data agree with our prior genetic and biochemical data on the C68 mutant ribosome that demonstrated its functionality (8).
Slow oxidative maturation of the GFP chromophore hinders usage for real-time experiments. Newly translated GFP must first fold into a catalytically active conformation that facilitates the formation of the chromophore. Second, mature GFP must then be correctly folded to maintain its fluorescent properties, presumably to protect the chromophore from solvent effects. Wild-type GFP maturation rates are 120 min, whereas they are 27 min for GFPS65T (11). To accelerate the chromophore maturation rate of GFPS65T, we designed a construct of GFPuv3 (14), which has F64L/S65T into the cycle 3 (F99S/M153T/V163A) mutation (15). This GFPuv3 has maximum excitation and emission at 488 and 515 nm, respectively (14,15). As observed below, the maturation rate for this mutant is ∼15 min (Supplementary Figure 2).
Stalling of ribosomes during translation using the SecM sequence
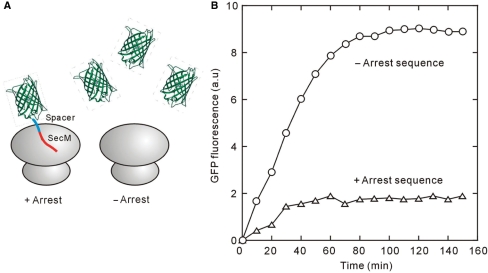
To allow GFP to fold on the ribosome, the protein must be extended by 23–26 amino acids at the C-terminus to fill the ribosome exit tunnel (5,6). The C-terminal segment of unextended GFP enzyme is held within the ribosome, preventing folding of the polypeptide into the native conformation. Therefore, we introduced a ‘spacer sequence’ downstream of the GFP gene, as described in Supplementary Figure 1. To trap folded GFP on ribosomes, we incorporated an ‘arrest sequence’ that stops translational elongation, derived from the E. coli SecM gene. The arrest sequence, FXXXXWIXXXXGIRAGP interacts with the ribosomal exit tunnel, stalling translational elongation (5,16) (Supplementary Figure 1). Translational assays using the PURE system showed a far lower yield of fluorescent protein when the arrest sequence is included, suggesting that mature GFPuv3 did not dissociate from the ribosome, thus preventing polysomal turnover on each mRNA (Figure 2A and B).
Figure 2.
(A) Schematic of the designed GFP arrest ribosome. To trap folded GFP on ribosomes, we incorporated an ‘arrest sequence’ that stops translational elongation, derived from the E. coli SecM gene. The arrest sequence interacts with the ribosomal exit tunnel, stalling translational elongation (see Supplementary Figure 1). (B) The time course of the fluorescent intensity of GFPuv3 in the presence or absence of the arrest sequence. Solvent condition is the same as that in Figure 1B.
Specific surface immobilization of ribosomes maintains activity
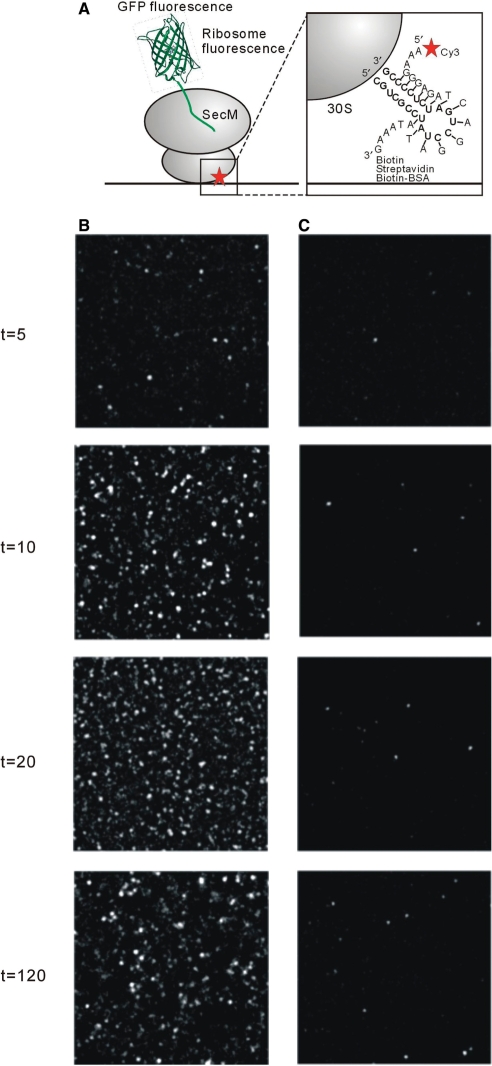
To monitor translation at a single-molecule level, we specifically immobilized ribosomes onto glass surfaces. C68 mutant ribosomes were hybridized with a biotinylated oligonucleotide that was complementary to an extension genetically engineered into the 16S rRNA (8,13) (Figure 3A). Ribosomes were then immobilized through a surface-bound streptavidin linkage with the biotinylated BSA; ribosomes without a biotinylated oligonucleotide do not bind to the surface, indicating that the BSA surface is specific. Translation was monitored by the appearance of GFPuv3 fluorescence, after initiation of the reaction by addition of DNA template and Δ ribosome PURE solution. When a high concentration of unlabeled mutant ribosomes (∼5 nM) was introduced on a glass surface, the number of fluorescent molecules gradually increased within ∼20 min (Figure 3B). These results reveal that the SecM arrest sequence mediates stable association of the folded GFP on the immobilized ribosomes. Deletion of the arrest sequence eliminates the large number of fluorescent molecules observed during translation (Figure 3C). This strongly suggests that the arrest sequence stabilizes GFP on the ribosome, and that in its absence, folded GFP is released into solution and does not adhere to the surface. To measure the stability of arrested GFP on the ribosome, the translation mix was washed away after GFP synthesis. Dissociation of GFP from the ribosome leads to loss of fluorescent molecules; this experiment revealed that stalled GFP is bound to ribosome for hours (Supplementary Figure 3), consistent with prior biochemical studies on the SecM system (17,18).
Figure 3.
(A) Schematic of interactions within the ribosome-synthesized protein complex. A biotin and Cy3-modified oligonucleotide at each ends was designed to hybridize to an rRNA loop extension on the small ribosomal subunit (see magnified view at right). ‘Biotin Streptavidin Biotin-BSA’ indicates the details of mRNA attachment to the surface. (B) and (C) The GFPuv3 fluorescent image of time course during the synthesis reaction by saturated ribosome molecules immobilization on the surface in the presence (B) or absence (C) of the arrest sequence. The image was taken with 200-ms time exposure right after the stage was moved quickly to avoid photobleaching molecules.
Single-molecule analysis of GFP synthesis
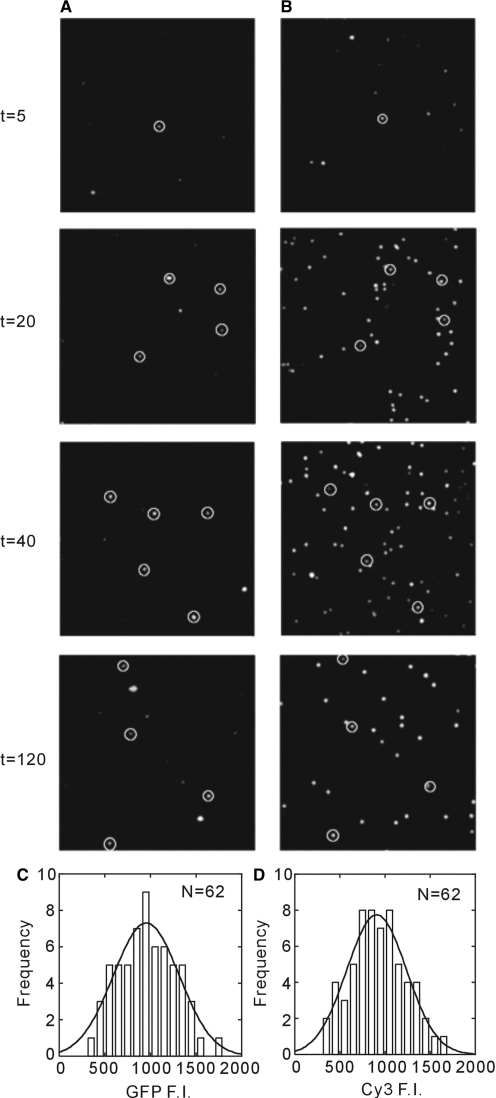
To test the specificity of GFPuv3 ribosome colocalization, translation was performed at greatly lower concentration of surface-immobilized ribosomes to ∼50 pM. In this experiment, ribosomes were immobilized using a biotinylated, Cy3-labeled oligonucleotide, and the position of ribosomes was established by Cy3 fluorescence. Colocalization of Cy3 and GFPuv3 fluorescence (Figure 4A) allowed correlation of protein synthesis and ribosome position. The proportion of colocalized molecules over total number of Cy3 fluorescence was increased gradually with time. This result represents the formation of stable ribosome-polypeptide complexes. Only a single molecule of GFPuv3 was observed colocalized with the ribosome as shown by the single-Gaussian fluorescent intensity histogram for colocalized molecules (Figure 4C and D). Our results provide direct evidence for the formation of single stable ribosome–polypeptide complexes.
Figure 4.
The colocalization image for GFPuv3 and Cy3 ribosome fluorescent. The GFPuv3 (A) and Cy3 (B) fluorescent image of time course during the synthesis reaction by low ribosome molecules immobilization on the surface. The white circle on the fluorescent molecule represents the colocalization for both GFPuv3 and Cy3 fluorescence. (C) and (D) Fluorescence intensity histogram for GFPuv3 (C) and Cy3 (D). Both histograms have been fit by single Gaussian function indicating that the colocalized molecule shows a single molecule.
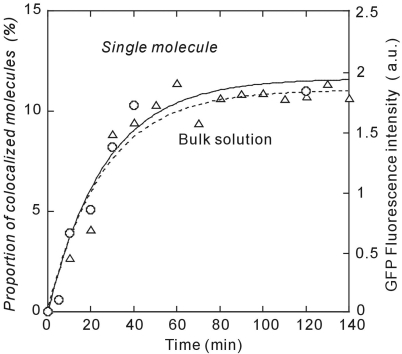
The time course of GFP synthesis as measured by colocalization agrees with that determined in bulk solution (Figure 5). The arrest sequence significantly increased the stability of ribosome-polypeptide complexes irrespective of the ribosome concentrations. The GFPuv3 synthesis including maturation time was achieved at plateau within 20 min, which is consistent with the time observed in bulk assay (Figure 2B). Despite the mutations in GFP, this rate is still dominated by fluorophore maturation. We measured this directly by performing surface-based translation in the presence of an O2-scavenging system, which allows protein folding on the ribosome but prevents maturation (Supplementary Figure 2). Upon washing with a saturated O2 solution, fluorescent molecules appear with a time constant of 15 min.
Figure 5.
The time course of the proportion of colocalized fluorescent molecules for low ribosome molecules immobilized (open circle) and the fluorescent intensity of GFPuv3 measured in bulk (open triangle). The smooth curves are exponential curve fits with time constants of 19.2 min (solid line) for single-molecule colocalization experiment and 26.2 min for bulk experiment (dotted line).
The method presented here allows single-molecule analysis of full protein synthesis and cotranslational folding. Prior studies have demonstrated that immobilized translation systems are functional for individual steps of protein synthesis, but our results show that full activity including folding activity is maintained. This method will complement existing single-molecule approaches that probe the dynamics of individual steps in protein synthesis, and allow comparison of these fundamental rate constants with the overall process of translation. The current time resolution of the method presented here is limited by the maturation time of the fluorescent protein chromophore. Faster maturing fluorescent proteins can be used in future applications. Alternatively, fluorescence in the folded protein can be monitored directly by incorporation of fluorescent amino acid analogs during translation. Extension of this method should reveal the dynamic interplay of translation and protein folding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to T. Iwagawa for the vector construction and colleagues in the Funatsu group. This work was supported in part by Takeda Science Foundation, grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and funded by grants to J.D.P. from the NIH and the Packard Foundation. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hsu ST, Fucini P, Cabrita LD, Launay H, Dobson CM, Christodoulou J. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc. Natl Acad. Sci. USA. 2007;104:16516–16521. doi: 10.1073/pnas.0704664104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorimer GH. A quantitative assessment of the role of chaperonin proteins in protein folding in vivo. FASEB J. 1996;10:5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- 3.Kennell D, Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol. 1977;114:1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- 4.Wagner GH, Jelenc PC, Ehrenberg M, Kurland CG. Rate of elongation of polyphenylalanine in vitro. Eur. J. Biochem. 1982;122:193–197. doi: 10.1111/j.1432-1033.1982.tb05866.x. [DOI] [PubMed] [Google Scholar]
- 5.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 6.Schaffitzel C, Ban N. Generation of ribosome nascent chain complexes for structural and functional studies. J. Struct. Biol. 2007;158:463–471. doi: 10.1016/j.jsb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu. Rev. Biochem. 2008 doi: 10.1146/annurev.biochem.77.070606.101431. in press. [DOI] [PubMed] [Google Scholar]
- 8.Dorywalska M, Blanchard SC, Gonzalez RL, Jr, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 11.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 12.Kolb VA, Makeyev EV, Ward WW, Spirin AS. Synthesis and maturation of green fluorescent protein in a cell-free translation. Biotechnol. Lett. 1996;18:1447–1452. [Google Scholar]
- 13.Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 14.Ito Y, Suzuki M, Husimi Y. A novel mutant of green fluorescent protein with enhanced sensitivity for microanalysis at 488 nm excitation. Biochem. Biophys. Res. Commun. 1999;264:556–560. doi: 10.1006/bbrc.1999.1541. [DOI] [PubMed] [Google Scholar]
- 15.Crameri A, Whitehorn EA, Tate E, Stemmer WPC. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 16.Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 17.Matsuura T, Yanagida H, Ushioda J, Urabe I, Yomo T. Nascent chain, mRNA, and ribosome complexes generated by a pure translation system. Biochem. Biophys. Res. Commun. 2007;352:372–377. doi: 10.1016/j.bbrc.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem. Biophys. Res. Commun. 2007;352:270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.