Abstract
Four myogenic regulatory factors (MRFs); MyoD, Myf-5, MRF4 and Myogenin direct muscle tissue differentiation. Heterodimers of MRFs with E-proteins activate muscle-specific gene expression by binding to E-box motifs d(CANNTG) in their promoters or enhancers. We showed previously that in contrast to the favored binding of E-box by MyoD-E47 heterodimers, homodimeric MyoD associated preferentially with quadruplex structures of regulatory sequences of muscle-specific genes. To inquire whether other MRFs shared the DNA binding preferences of MyoD, the DNA affinities of hetero- and homo-dimeric MyoD, MRF4 and Myogenin were compared. Similarly to MyoD, heterodimers with E47 of MRF4 or Myogenin bound E-box more tightly than quadruplex DNA. However, unlike homodimeric MyoD or MRF4, Myogenin homodimers associated weakly and nonpreferentially with quadruplex DNA. By reciprocally switching basic regions between MyoD and Myogenin we demonstrated dominance of MyoD in determining the quadruplex DNA-binding affinity. Thus, Myogenin with an implanted MyoD basic region bound quadruplex DNA nearly as tightly as MyoD. However, a grafted Myogenin basic region did not diminish the high affinity of homodimeric MyoD for quadruplex DNA. We speculate that the dissimilar interaction of MyoD and Myogenin with tetrahelical domains in muscle gene promoters may differently regulate their myogenic activities.
INTRODUCTION
Skeletal muscle develops from pluripotent mesodermal stem cells by multiple consecutive steps. In an initial determination phase, external signals emanating from neighboring cells induce mesodermal cells to differentiate into dividing myoblasts that become committed to the myogeneic lineage. In subsequent differentiation stages, additional external signals induce the myoblasts to differentiate into nondividing myocytes that ultimately fuse to form syncitial myotubes (1,2). The development of muscle tissue is directed by four myogenic regulatory factors (MRFs); MyoD, Myf-5, MRF4 and Myogenin (Myf4) that comprise a sub-group within the basic helix–loop–helix (bHLH) superfamily of proteins (3). MRFs act as master transcription factors that induce the expression of a wide array of muscle-specific genes (4). Conversion of mesenchymal cells into dividing myoblasts during the initial determination step is directed by three primary MRFs; MyoD, Myf5 and MRF4 (5,6). Progression from myoblasts to nondividing myocytes and then to myotubes during the subsequent differentiation stage is mediated by Myogenin together with MRF4 (7–10). The different members of the MRF family appear to have distinct but overlapping roles in that each protein induces the expression of different but partially overlapping sets of muscle-specific proteins (11).
The HLH segment of the bHLH domain in MRFs mediates their dimerization, whereas the basic region serves as a DNA-binding site (12). Each MRF either forms homodimers or generates heterodimers by associating with Class I bHLH E-proteins; E12, E47 or E2-2/ITF2 (3,12,13). Studies of muscle cell differentiation in culture revealed that transcription of muscle-specific genes is activated by the binding of MyoD–E protein heterodimers to conserved E-box, d(CANNTG), motifs in the promoter or enhancer regions of the genes. Such heterodimers bind E-box significantly more tightly than MyoD homodimers (13–15). E-box elements are highly abundant in regulatory regions of a large number of both muscle-specific and nonmuscle genes. However, while MRF heterodimers selectively induce transcription of muscle-specific genes, they do not affect the expression of E-box regulated nonmuscle genes. Multiple factors appear to contribute to this specificity of gene activation. One factor is the presence in the bHLH region of MRFs, but not of other bHLH proteins, of three amino acids Ala, Thr and Lys (residues 114, 115 and 124 in the prototypical MyoD) (12,16–18). Other attributes are the identity of the two variable residues d(CANNTG) within E-box (19), the effects of cis-acting suppressor elements (20) and the interaction of MRFs with additional transcription factors.
As conjectured by Larsen and Weintraub (21), the activity of transcription factors may also be regulated by their differential interaction with noncanonical DNA structures. In line with this proposal, quadruplex structures in promoter or upstream sequences were implicated in the negative or positive regulation of the expression of genes such as those that encode insulin (22–25), c-MYC (26–28), c-kit (29,30), bcl-2 (31), VEGF (32) and PDF-A (33). Correspondingly, some transcription factors were found to interact specifically with tetraplex formations of gene regulatory regions (15,25). That homodimeric MyoD may preferentially recognize quadruplex structure in DNA was initially indicated by the finding that it formed weaker complexes with E-box than with tetrahelical structures of a guanine-rich mouse creatine kinase enhancer sequence or of Tetrahymena telomeric DNA (34). Promoter and enhancer regions of several muscle-specific genes were later found to contain a disproportional high frequency of clusters of contiguous guanine residues that readily formed hairpin and parallel-stranded G′4 unimolecular and G′2 bimolecular quadruplex structures (35). Subsequently, MyoD homodimers were shown to bind bimolecular tetraplex formations of the muscle-specific regulatory sequences significantly more tightly than double-stranded E-box (15). Since these sequences are present as single copies in the genome, they cannot associate in vivo to form G′2 structures. However, a bimolecular-like quadruplex was also generated by the association of hairpin structures of two neighboring guanine-rich sequences in the integrin promoter region (35). This more physiologically relevant tetraplex structure was also bound by MyoD more tightly than E-box (15). In contrast, MyoD–E47 heterodimers bound E-box more tightly than G′2 tetraplex DNA structures. Additional investigation revealed that the MyoD basic region served as the binding site for E-box as well as quadruplex DNA. However, whereas complex formation with E-box required integrity of the complete basic domain, MyoD whose basic region was largely deleted except for a single remaining cluster of three basic amino acids, maintained its capacity to bind quadruplex DNA (36).
In this work, we inquired whether or not the differential affinities of MyoD homo- and hetero-dimers for E-box and quadruplex DNA were shared by other MRFs. We report that similarly to MyoD, homodimeric MRF4 associated more tightly with quadruplex than with E-box DNA and that its heterodimers with E47 protein bound E-box more strongly than tetraplex DNA. Distinctly, whereas Myogenin-E47 heterodimers behaved like heterodimeric MyoD and MRF4 by preferentially binding E-box, unlike homodimeric MyoD and MRF4, Myogenin homodimers bound tetraplex DNA weakly and nonpreferentially. By reciprocally exchanging complete basic regions between homodimers of MyoD and Myogenin, we showed that MyoD dominantly dictated high affinity for quadruplex DNA. The distinct affinities of homodimeric MyoD and MRF4 on one hand and of Myogenin on the other for tetrahelical structures of regulatory sequences of muscle-specific genes may contribute to their differently modulated activities during myogenesis.
MATERIALS AND METHODS
Hairpin, double-stranded and monomolecular and bimolecular quadruplex DNA structures
Nucleotide sequences of synthetic DNA oligomers (products of Sigma/Genosys Rehovot, Israel) represented guanine-rich segments of promoter regions of the α7 integrin and sarcomeric mitochondrial creatine kinase (sMtCK) genes (35), included the core E-box DNA sequence or encoded the basic regions of MyoD or Myogenin (Table 1). Following purification of the single-stranded oligonucleotides by denaturing gel electrophoresis in 8.0 M urea, 12% polyacrylamide (acryl/bisacrylamide, 19:1) (37), their 5′ ends were labeled by 32P in bacteriophage T4 polynucleotide kinase (PNK)-catalyzed reaction (38). Hairpin, monomolecular and bimolecular tetraplex structures of the integrin and sMtCK DNA oligomers were formed as we described (35). E-box or basic region encoding DNA duplexes were prepared by annealing under described conditions (39) equimolar amounts of 5′ and 3′ complementary oligomers.
Table 1.
DNA oligomers used in this study
| Oligomer designation | Bases | Nucleotide sequence |
|---|---|---|
| Integrin DNA | 26 | 5′-d(CATGGGGGCGGGAAGGGGCGGGGTCT)-3′ |
| sMtCK DNA | 24 | 5′-d(CTGAGGAGGGGCTGGAGGGACCAC)-3′ |
| 5′ E-box | 26 | 5′-d(TCGATCCCCCAACACCTGCTGCCTGA)-3′ |
| 3′ E-box | 26 | 5′-d(TCAGGCAGCAGGTGTTGGGGGACGA)-3′ |
| 5′- Myogenin basic region | 89 | 5′-d(CAAGGCGTGTAAGAGGAAGTCTGTGTCTGTGGACCGGCGGAGGGCAG CCACACTGAGGGAGAAGCGCAGGCTCAGCAAAGT GAATGAGG)-3′ |
| 3′- Myogenin basic region | 91 | 5′-d(TCATTCACTTTGCTGAGCCTGCGCTTCTCCCTCAGTGTGGCTGCCCTCC GCCGGTCCACAGACACAGACTTCCTCTTACACGCCTTGCATG)-3′ |
| 5′- MyoD basic region | 89 | 5′-d(CAAGGTGTGCAAGCGCAAGACCACCAACGCTGATCGCCGCAAGGCCG CCACCATGCGCGAGCGCCGCCGCCTGAAGAAAGTGAATGAGG)-3′ |
| 3′- MyoD basic region | 91 | 5′-d(TCATTCACTTTCTTCAGGCGGCGGCGCTCGCGCATGGTGGCGGCCTTGC GGCGATCAGCGTTGGTGGTCTTGCGCTTGCACACCTTGCATG)-3 |
The integrin and sMtCK oligomers are tetraplex-forming guanine-rich sequences derived from promoter regions of the respective genes (35). Underlined are guanine clusters that participate in quadruplex formation. The core E-box sequence is underlined in the 5′ and 3′ E-box complementary oligomers. Sequences representing the Myogenin and MyoD basic regions are italicized in the respective basic region 5′ and 3′ complementary oligomers and the BsmI and SphI complementing ends are underlined.
Preparation, purification and expression of full-length and mutant recombinant MyoD, MRF4 and Myogenin
Plasmids harboring cDNA that encoded full-length unmodified member proteins of the MRF family were: pGEX-6P-MyoD (Mus musculus) (15); pGEX-MRF4 (Rattus norvegicus) (gift of Dr P. Muñoz-Cánoves, Center for Genomic Regulation, Barcelona, Spain); pEMSV- Myogenin (Rattus norvegicus) (contributed by Dr S.J. Tapscott, FHCRC, Seattle, WA, USA) was subcloned into a pGEX-6P vector. The pGEX4T1-E47 harboring cDNA that encoded full-length Mus musculus E47 protein was a gift of Dr A. Cano (CSIC-UAM, Madrid, Spain).
To create mutant Myogenin with basic amino acid clusters identical to those of MyoD and MRF4, an R84K mutation was introduced by PCR into the second triad using the 5′ and 3′ primers; 5′-d(GTCTGTGGACCGGCGGAAGGCAGCCCACACTGAGGG)-3′ and 5′-d(CCCTCAGTGTGGGCTGCCTTCCGCCGGTCCACAGAC)-3′, respectively, and full-length pGEX-6P-Myogenin cDNA template. Following verification of the mutation by sequencing, an additional K91R mutation was introduced by PCR into the third triad using a pGEX-6P-myogenin R84K cDNA template and the respective 5′ and 3′ primers; 5′-d(GCCACACTGAGGGAGAGGCGCAGGCTCAAGAAAG)-3′ and 5′(CTTTCTTGAG CCTGCGCCTCTCCCTCAGTGTGGC)-3′.
To construct chimerical MyoD, which had its native basic region replaced by a Myogenin basic region (MyoD/Myogeninb), SphI and BsmI cleavage sites were generated at positions 99 and 126, respectively, upstream and downstream to the MyoD basic region (residues 102–121). A BsmI restriction site was formed by PCR as we described (36) using as template GST-fused full-length MyoD cDNA in pGEX-6P and the 5′ and 3′ primers; 5′-d(GCAAAGTGAATGAGGCATTCGAGACGCTCAAGC)-3′ and 5′-d(GCTTGAGCGTCTCGAATGCCTCATTCACTTTGC)-3′, respectively. Presence of the BsmI restriction site was verified by sequencing and the mutated cDNA was used as template to generate by PCR a SphI restriction site using the respective 5′ and 3′ primers; 5′-d(CTGCTTGCTGTGGGCATGCAAGGCGTGCAAG)-3′ and 5′-d(CTTGCACGCCTTGCATGCCCACAGCAAGCAG)-3′. The doubly mutated MyoD DNA was cut by SphI, treated with calf intestinal phosphatase (CIP), cleaved by BsmI, resolved by gel agarose electrophoresis and isolated. Myogenin basic region 5′ and 3′ oligomers (Table 1) that were annealed and phosphorylated at their 5′-termini by PNK were ligated into the restricted MyoD cDNA. The insert had at its ends SphI and BsmI complementary sites and 5′ and 3′ sequences that encoded MyoD residues 99–101 and 122–126, respectively whereas its core encoded the Myogenin basic region (residues 74–93). Reciprocal chimerical Myogenin that contained a MyoD basic region (Myogenin/MyoDb) was prepared as follows: GST-fused full-length Myogenin cDNA in pGEX-6P vector served as a template into which a BsmI restriction site was introduced by PCR using the 5′ and 3′ primers; 5′-d(GAAAGTGAATGAGGCATTCGAGGCTCTGAAGAGAAGC)-3′ and 5′-d(GCTTCTCTTCAGAGCCTCGAATGCCTCATTCACTTTC)-3′, respectively. To have the generated BsmI site as a single a BsmI restriction sequence in the Myogenin cDNA, PCR was employed to eliminate a native site at position 486 by using the respective 5′ and 3′ primers; 5′-d(CCCAGTGAATGTAACTCCCACAGCGCCTCC)-3′ and 5′-d(GGAGGCGCTGTGGGAGTTACATTCACTGGG)-3′ and the modified Myogenin cDNA template. A SphI restriction site was introduced into the mutated Myogenin cDNA template by PCR using the respective 5′ and 3′ primers; 5′-d(CAGTGCCTGCCCTGGGCATGCAAGGTGTGTAAGAG)-3′ and 5′-d(CTCTTACACACCTTGCATGCCCAGGGCA GGCACTG)-3′. Restriction and isolation of linear Myogenin DNA was conducted as described for MyoD DNA. MyoD basic region 5′ and 3′ oligomers (Table 1) were annealed, phosphorylated and ligated into the restricted Myogenin DNA as detailed above. The insert had at its ends SphI and BsmI complementary sites and 5′ and 3′ sequences that encoded Myogenin residues 71–73 and 94–98, respectively, whereas its core encoded the MyoD basic region (residues 102–121).
Recombinant unmodified or mutant proteins were expressed and purified as follows: pGEX-6P plasmids harboring the respective cDNA sequences were electroporated into competent Escherichia coli BL21 (DE3)pLysS cells that were grown in LB medium containing ampicillin and chloramphenicol to an A600 of ∼0.6. Synthesis of the GST-fused proteins was induced by exposure to 100 μM IPTG for 3 h. Purification of the recombinant proteins from the bacterial cell extracts to >95% homogeneity was attained by glutathione-agarose (Sigma) affinity column chromatography. The GST residue was cleaved from the MyoD or Myogenin fusion proteins by incubating 100 μg protein at 4°C for 4 h with 1.0 unit preScission protease (GE Healthcare, Bucks, UK). The DNA-binding capacity of Myogenin that was used in some experiments as an intact fusion protein was found to be indistinguishable from that of the preScission protease cleaved protein. MRF4 and E47 were used as uncleaved fusion proteins.
Electrophoresis mobility shift separation of protein–DNA complexes and determination of their dissociation constants
Heterodimers of MyoD, MRF4 or Myogenin with E47 were generated prior to their binding to the various DNA probes by incubating at 37°C for 10 min purified recombinant MRF protein with an equimolar amount of E47 in reaction mixtures that contained in a final volume of 10 μl: 45 mM KCl, 4.5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 20% glycerol in 20 mM Tris–HCl buffer, pH 8.0. Protein–DNA binding was conducted in reaction mixtures that contained in a final volume of 10 μl: specified amounts of homodimeric MRF proteins or their heterodimers with E47 and 5′-32P labeled DNA probe, 10 mM KCl, 1.0 mM EDTA, 0.5 mM DTT, 20% glycerol in 25 mM Tris–HCl buffer, pH 8.0. Mixtures for the binding of end-labeled bimolecular tetraplex DNA structures of muscle-specific regulatory sequences were also supplemented with 10 mM KCl. The mixtures were incubated at 30°C for 20 min and protein–DNA complexes were resolved from free DNA by electrophoresis at 4°C and under 200–250 V in nondenaturing 4 or 6% polyacrylamide gel (acryl/bisacrylamide, 19:1) in 10 mM KCl in 0.25× TBE buffer (0.5 mM EDTA in 22.5 mM Tris–borate buffer, pH 8.3). Electrophoresis of the DNA was conducted until a bromophenol blue marker dye migrated 7.5 cm into the gel. The gels were dried on DE81 filter paper and the relative proportions of bands of free and protein-bound DNA were quantified by phosphor imaging analysis.
Dissociation constants, Kd, values of complexes of homodimeric MRF proteins or their heterodimers with E47 with E-box DNA or with bimolecular tetraplex structures of guanine-rich muscle-specific DNA sequences were determined as follows: increasing amounts of 32P-labeled DNA were incubated with a constant amount of protein in the above described protein–DNA binding reaction mixtures. Following electrophoretic resolution of the protein–DNA complexes from free DNA by mobility shift, their relative amounts were determined by phosphor imaging quantification of the respective radioactive bands. Kd values were deduced from the negative reciprocal of the slope of a Scatchard plot of the results as described elsewhere (40).
RESULTS
Homodimeric Myogenin and MRF4 selectively bind and stabilize bimolecular quadruplex forms of muscle-specific promoter sequences
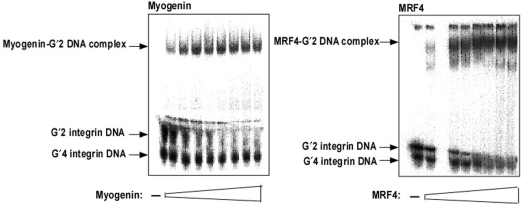
Tetrahelical forms of the α7 integrin promoter DNA consist of a mixture of unimolecular, G′4, and bimolecular, G′2, tetraplexes and the G′2 structure of sMtCK promoter DNA is in mixture with single strands of this sequence (35). We showed recently that homodimers of MyoD bind preferentially only the bimolecular tetraplex forms of integrin and sMtCK DNA (15). To examine the binding preferences of homodimeric recombinant Myogenin or MRF4, increasing amounts of the proteins were incubated under binding conditions with a mixture of G′2 and G′4 integrin DNA. Results indicated that both Myogenin and MRF4 associated only with the bimolecular quadruplex structures of integrin DNA (Figure 1A and B, respectively) without forming appreciable complexes with the unimolecular quadruplex structure of this sequence. Similarly, homodimeric Myogenin and MRF4 were also found to selectively bind a G′2 tetraplex structure of sMtCK DNA without associating with the single-stranded form of this oligomer (data not shown). Further, both Myogenin and MRF4 failed to detectably bind double-stranded structures of the guanine-rich integrin and sMtCK sequences (data not shown). Thus, homodimers of all the three examined MRFs; MyoD, Myogenin and MRF4 share similar binding preference for bimolecular quadruplex forms of muscle-specific promoter sequences.
Figure 1.
Homodimeric Myogenin and MRF4 bind preferentially bimolecular quadruplex structures of integrin DNA. Increasing amounts of homodimers of recombinant Myogenin or MRF4 were incubated under binding conditions with, respectively, 0.18 or 1.0 pmol of 5′-32P quadruplex integrin DNA. Protein-bound G′2 DNA was resolved from free DNA by nondenaturing 4% polyacrylamide electrophoresis. (A) Binding of Myogenin to G′2 integrin DNA. Shown are the retarded protein–G′2 integrin DNA complex and the G′2 and G′4 forms of free DNA. (B) Binding of MRF4 to G′2 integrin DNA.
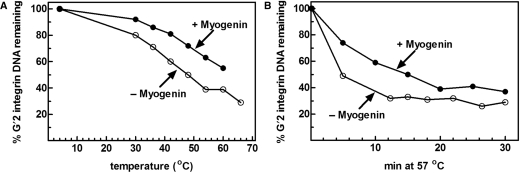
To examine whether the DNA maintained its quadruplex conformation when in complex with protein, G′2 integrin DNA was bound to homodimers of recombinant Myogenin and aliquots of the binding mixtures were either incubated for 10 min each at increasing temperatures or heated at 57°C for different periods of time. To follow the heat denaturation of free G′2 DNA, control mixtures that did not contain Myogenin were similarly heat-treated. DNA denaturation was terminated by placing the mixtures on ice and stripping the bound protein by 0.5% sodium dodecyl sulfate (SDS). Remaining G′2 integrin DNA was resolved from its G′4 form by nondenaturing gel electrophoresis and their amounts were quantified by phosphor imaging analysis. As seen in Figure 2A and B, not only did the Myogenin-bound G′2 DNA maintain its quadruplex structure, but also became more heat resistant relative to free DNA. Thus, similarly to MyoD, (15) Myogenin stabilized the bound G′2 DNA.
Figure 2.
Homodimeric Myogenin raises the heat stability of bound G′2 integrin DNA. (A) Heat denaturation of Myogenin-bound and free G′2 integrin DNA. Reaction mixtures that contained each 0.18 pmol of 5′-32P labeled G′2 integrin DNA were incubated under binding conditions with or without saturating amounts of recombinant homodimeric Myogenin. Following completion of the binding reaction, the mixtures were heated at the indicated temperatures for 10 min. Heat denaturation was terminated by rapid cooling of the mixtures to 4°C and the addition of 0.5% SDS to strip the protein off the DNA. Residual G′2 integrin DNA was resolved from G′4 DNA by nondenaturing 10% polyacrylamide gel electrophoresis and their relative amounts were quantified by phosphor imaging analysis. Shown is a plot of the remaining amount of G′2 DNA in mixtures that contained or were devoid of Myogenin as a function of the increasing temperature. (B) Kinetics of heat denaturation of Myogenin-bound or free G′2 integrin DNA. Binding conditions, heat denaturation and electrophoretic resolution of remaining G′2 DNA were as described in (A) except that all the mixtures were heated at 57°C for the indicated increasing periods of time. Shown is a plot of the residual amount of G′2 DNA in mixtures that contained or were devoid of Myogenin as a function of increasing time at 57°C.
Homodimers of MyoD and MRF4 but not of Myogenin bind quadruplex DNA more tightly than E-box DNA
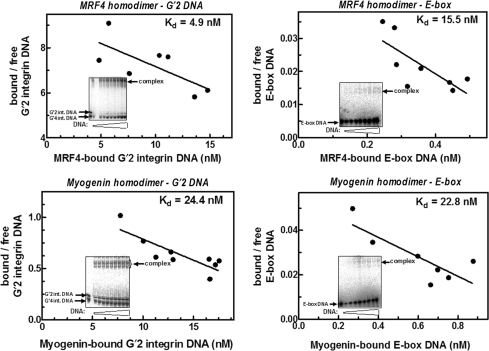
We previously reported that MyoD homodimers formed tighter complexes with G′2 quadruplex structures of guanine-rich regulatory sequences of muscle-specific genes than with E-box DNA (15). To inquire whether or not homodimers of other members of the MRF family also bind tetraplex DNA preferentially, we determined the dissociation constants, Kd, of complexes of homodimeric recombinant MRF4 and Myogenin proteins with G′2 quadruplex integrin DNA and with E-box. Typical results of such analysis (Figure 3) indicated that similarly to homodimeric MyoD, MRF4 homodimers bound G′2 integrin DNA more tightly than E-box as reflected by the lower Kd of its complex with the tetrahelical DNA. Distinctly, however, Myogenin homodimers did not display preference for the quadruplex DNA, forming similarly relatively weak complexes with tetraplex or E-box DNA (Figure 3). These representative results were substantiated in multiple independent determinations of the Kd values of complexes of homodimers of MyoD, MRF4 or Myogenin with quadruplex or E-box DNA (Table 2). As is evident from their calculated relative affinities, (Kd E-box/Kd G′2 DNA), homodimers of MyoD and MRF4, respectively, bound G′2 integrin DNA 21- and 4-fold more tightly than E-box DNA. By clear contrast, however, complexes of Myogenin with this DNA tetrahelix or with E-box had practically indistinguishable relatively high dissociation constants (Table 2). In a similar set of multiple measurements, we found that the Kd value of complexes of homodimeric MyoD with G′2 quadruplex sMtCK DNA was 27-fold lower than the dissociation constant of MyoD–E-box complexes. In contrast, Myogenin homodimers had a 1.8-fold higher relative affinity for E-box than for G′2 sMtCK DNA (data not shown). Thus, contrary to homodimeric MyoD or MRF4, Myogenin homodimers bound weakly and nonpreferentially quadruplex structures of regulatory sequences of the two examined muscle-specific genes.
Figure 3.
Homodimers of MRF4, but not of Myogenin, bind bimolecular quadruplex integrin DNA more tightly than E-box. Constant amounts of homodimers of recombinant MRF4 or Myogenin were incubated under DNA-binding conditions with increasing amounts of either 5′-32P labeled E-box or G'2 integrin DNA. Formed protein–DNA complexes were resolved from free DNA by nondenaturing polyacrylamide gel electrophoresis and their relative proportions were determined by phosphor imaging analysis (see Materials and methods section). Shown are representative electrophoregrams (insets) and Scatchard plots of the measured ratios of bound to free DNA as a function of the concentration of protein-bound DNA. The indicated Kd values in different plots were derived from the negative reciprocal of their slopes.
Table 2.
Dissociation constants of complexes of homodimeric and heterodimeric MRFs with E-box and G′2 quadruplex integrin DNA
| Protein | Kd (nM) [N] |
||
|---|---|---|---|
| E-box DNA | G′2 integrin DNA | G′2 integrin DNA/E-box relative affinity (Kd E-box/Kd G′2 integrin) | |
| MyoD homodimer | 91.8 ± 1.6 [4] | 4.4 ± 2.5 [10] | 20.9 |
| MyoD/E47 heterodimer | 1.7 ± 0.6 [4] | 11.2 ± 4.3 [5] | 0.15 |
| MRF4 homodimer | 21.3 ± 11.0 [4] | 5.3 ± 1.3 [3] | 4.0 |
| MRF4/E47 heterodimer | 0.2 ± 0.1 [5] | 19.5 ± 2.7 [4] | 0.01 |
| Myogenin homodimer | 25.4 ± 11.0 [5] | 30.0 ± 8.0 [4] | 0.85 |
| Myogenin/E47 heterodimer | 0.2 ± 0.1 [4] | 8.5 ± 2.0 [4] | 0.02 |
Dissociation constants, Kd, of the different protein–DNA complexes were determined as described under Materials and methods section and in the legends to Figures 1 and 2. Listed are average Kd values ± SD with [N] marking the number of independent measurements of each value.
Heterodimers of MyoD, MRF4 or myogenin with E47 bind E-box more tightly than quadruplex DNA
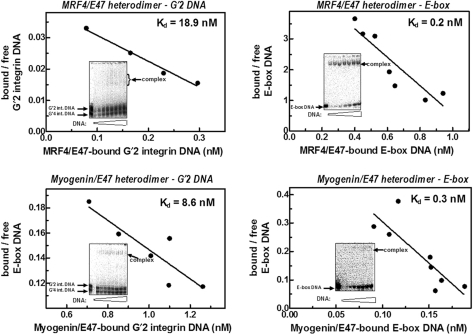
We showed in the past that in contrast to homodimeric MyoD, its heterodimers with E47 displayed greater affinity for E-box than for tetraplex DNA (15). Figure 4 presents typical results of determinations of the Kd values of complexes of MRF4-E47 or Myogenin-E47 heterodimers with G′2 integrin DNA or with E-box. It is evident that both types of heterodimers formed significantly tighter complexes with E-box than with quadruplex integrin DNA. That the heterodimeric forms of all the three examined MRFs prefer E-box over quadruplex DNA was confirmed by multiple determinations of the dissociation constants of the respective protein–DNA complexes. As shown in Table 2, the relative affinities of heterodimeric MyoD, MRF4 or Myogenin for E-box were, respectively, greater by 6.6-, 97- or 42-fold than for G′2 integrin DNA. In a parallel set of experiments, we found that MyoD-E47 and Myogenin-E47 heterodimers, respectively, bound E-box 15- and 128-fold more tightly than G′2 quadruplex sMtCK DNA (data not shown). Thus, heterodimers of all three MRFs displayed significant binding preference for E-box over tetraplex structures of promoter sequences of the two examined muscle-specific genes.
Figure 4.
Heterodimers of MRF4 and Myogenin with E47 bind E-box more tightly than G′2 quadruplex integrin DNA. Constant amounts of MRF4-E47 or Myogenin-E47 heterodimers that were prepared as described under Materials and methods section, were incubated under DNA-binding conditions with increasing amounts of either 5′-32P labeled E-box or G′2 integrin DNA. Electrophoretic resolution and phosphorimage analysis quantification of protein-bound and free DNA were conducted as detailed in the legend to Figure 5. Shown are representative electrophoregrams (insets) and Scatchard plots of the ratios of bound to free DNA as a function of the concentration of protein-bound DNA. The Kd values that were calculated for each plot were derived from the negative reciprocal of their respective slopes.
Two divergent amino acids in the core of the Myogenin basic region are not responsible for its low relative affinity for G′2 DNA
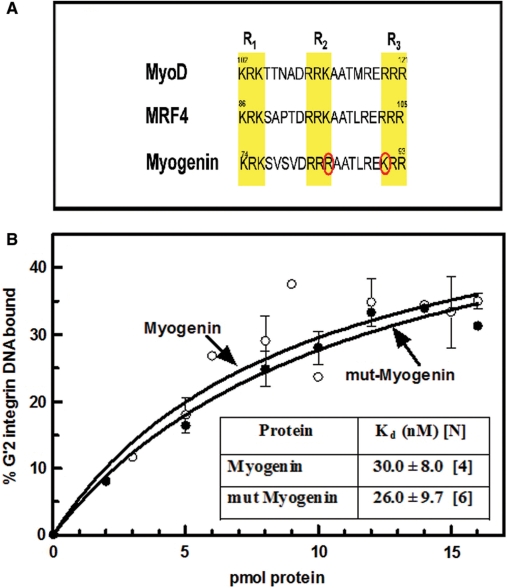
The binding of quadruplex DNA structures by MyoD was recently shown to be mediated by either one of three clusters of three conserved basic amino acids each in its basic region (36). Whereas the amino acid sequence of the three basic triads were identical in MyoD and MRF4, Myogenin had K84R and R91K conservative replacements in the second and third basic clusters, R2 and R3, respectively (Figure 5A). The similar preferential binding of quadruplex DNA by MyoD and MRF4 homodimers could potentially be due to their identical basic clusters, whereas the changed residues in the R2 and R3 triads of Myogenin could be the source of its relatively weak and nonpreferential association with tetrahelical DNA. To examine this possibility we constructed a Myogenin double mutant, mut-Myogenin whose three basic clusters were identical to those of MyoD and MRF4, and compared its G′2 integrin DNA-binding capacity to that of the unmodified protein. Results showed that the quadruplex DNA-binding curves of both proteins were essentially indistinguishable and that the Kd values of complexes of the two proteins with G′2 integrin DNA were also statistically interchangeable (Figure 5B). Thus, it appears that the distinctive two amino acids in the core basic triads were unlikely to cause of the relatively low affinity of homodimeric Myogenin for quadruplex DNA.
Figure 5.
Mutated Myogenin with clusters of basic amino acids identical to those of MyoD and MRF4 maintains low relative affinity for G′2 integrin DNA. To render the basic amino acids clusters in the Myogenin basic region identical to those of MyoD and MRF4, two mutations R84K and K91R were introduced into full-length Myogenin cDNA in pGEX-6P vector as detailed under Materials and methods section. G′2 integrin DNA binding by unmodified and mutated Myogenin (mut-Myogenin) recombinant proteins and Kd values of the respective complexes were measured (see Material and methods section). (A) Basic regions of MyoD, MRF4 and Myogenin. The three conserved clusters R1, R2 and R3 of three basic amino acids each are highlighted. The two amino acids, R84 and K91 that distinguish Myogenin from MyoD and MRF4 are circled in red. (B) Binding of quadruplex integrin DNA by Myogenin and mut-Myogenin. The indicated increasing amounts of the respective proteins were incubated under DNA-binding conditions with 0.18 pmol of 5′-32P labeled G′2 integrin DNA and the formed protein–DNA complexes were resolved by electrophoresis and quantified as detailed under Materials and methods section. Inset: average Kd values ± SD of complexes of G′2 integrin DNA with unmodified or mutated Myogenin that were determined in [N] independent measurements.
MyoD dominantly dictates high affinity for quadruplex DNA
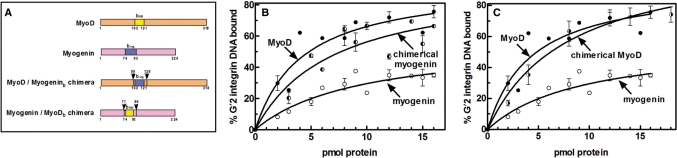
The basic regions of the three MRFs include between their R1 and R2 basic triads distinctly different intervening tracts of four residues. Also, leucine located in the stretch that separates the R2 and R3 clusters of MRF4 and Myogenin (residues 100 and 88, respectively) is substituted by methionine (residue 116) in MyoD (Figure 5A). We inquired whether these differences and/or the conformation of the basic region as a whole may dictate the dissimilar tetraplex DNA-binding affinities of homodimeric MyoD and Myogenin. To this end, we constructed chimerical MyoD and Myogenin proteins with reciprocally switched basic regions and compared their G′2 integrin DNA-binding capabilities to those of the unmodified proteins. As indicated by schemes of the unmodified and reconstructed proteins (Figure 6A), the chimerical MyoD or Myogenin maintained in full their respective original sequences except for the reciprocally exchanged basic regions. In line with their distinctly different affinities for G′2 integrin DNA (Table 2), unmodified Myogenin was significantly less efficient than native MyoD in binding this tetrahelix (Figure 6B). However, Myogenin whose basic region was replaced by that of MyoD bound quadruplex DNA nearly as efficiently as native MyoD (Figure 6B). In accord with this observation, measurements showed that the Kd value of complexes of Myogenin with G′2 integrin DNA was decreased by >4-fold when its native basic region was replaced by that of MyoD (Table 3). Surprisingly, however, reciprocal transplantation of the weakly binding basic region of Myogenin into MyoD did not significantly change the high-binding efficiency of the modified protein (Figure 6C) and the dissociation constant of its complex with the G'2 DNA remained statistically indistinguishable from that of native MyoD (Table 3). Thus, MyoD dictated high-binding affinity for quadruplex DNA not only through its basic region but also via the structure of the whole protein, which forced tight tetraplex DNA-binding capacity upon the transplanted Myogenin basic region (see Discussion section).
Figure 6.
The MyoD basic region and its peripheral domains dominantly dictate high affinity for G′2 integrin DNA. MyoD or Myogenin cDNA in pGEX-6P vectors were modified and restricted to remove their basic regions, which were then replaced by synthetic DNA duplexes that reciprocally encoded the respective Myogenin or MyoD basic regions (see Materials and methods section). Unmodified or proteins with exchanged basic regions were expressed and their capacities to bind 5′-32P labeled G′2 integrin DNA were determined. (A) Schemes of unmodified and basic region-switched chimeras of MyoD and Myogenin. The tract transplanted into MyoD at positions 102–121 corresponded to the amino acids sequence of the Myogenin basic region (bmg), whereas the flanking inserted tracts (residues 99–101 and 122–126) maintained the respective MyoD native sequences. The amino acid run transplanted into Myogenin (residues 74–93) represented the MyoD basic region (bMD), whereas the flanking inserted tracts (residues 71–73 and 94–98) retained the respective Myogenin native sequences. (B) Binding of G′2 quadruplex integrin DNA by MyoD, Myogenin and Myogenin/MyoDb chimerical protein. Increasing amounts of the respective proteins were incubated under DNA-binding conditions with 0.18 pmol of 5′-32P labeled G′2 integrin DNA and the protein–DNA complexes that were formed were resolved by electrophoresis and quantified as detailed under Materials and methods section. The presented results are averages ± SD of 3–5 independent measurements. (C) Binding of G′2 quadruplex integrin DNA by MyoD, Myogenin and MyoD/Myogeninb chimerical protein. Binding of G′2 integrin DNA by the MyoD/Myogeninb protein was assayed as described in (B) above. The results represent averages of four independent determinations ± SD. The binding curves for unmodified MyoD and Myogenin are the same as in (B).
Table 3.
Dissociation constants of complexes of G′2 quadruplex integrin DNA with unmodified and chimerical MyoD and Myogenin homodimers
| Protein homodimer | Kd (nM) of protein-G′2 integrin DNA complex [N] |
|---|---|
| Myogenin | 30.0 ± 8.0 [4] |
| Myogenin/MyoDb chimera | 7.1 ± 1.9 [4] |
| MyoD | 4.4 ± 2.5 [10] |
| MyoD/Myogeninb chimera | 6.0 ± 3.1 [5] |
Dissociation constants, Kd, of protein–DNA complexes were determined as described under Materials and methods section and in the legends to Figures 1 and 2. Shown are average Kd values ± SD with [N] denoting the number of independent measurements of each value.
DISCUSSION
Myogenesis is initiated and directed by MRF heterodimers with Class I bHLH E-proteins that act as master transcription factors, which activate muscle gene expression. The heterodimeric MRFs activate expression of muscle tissue genes by binding to E-box elements in their promoter or enhancer regions. Extensive studies on MyoD, the prototypical MRF, showed that its heterodimers with E protein induce the expression of a subset of muscle-specific genes by binding to E-box elements in their promoters. The E-box bound MyoD then summons ancillary proteins that affect chromatin remodeling and enable transcription (11). Less defined are the modes of gene activation by other members of the MRF family and the extent of their redundant or specific involvement in myogenesis. A recurrent question is how any given MRF activates only selected genes among all that contain an E-box in their promoters. It is likely that the selectivity of different MRFs is governed at several regulatory levels such as sequence variations within and at the flanks of E-boxes and the identity of different auxiliary proteins that associate with the MRF-E protein heterodimers. Heterodimeric MRFs bind E-box considerably more tightly than MRF homodimers and are thus more potent activators of gene expression (13–15). This raises the possibility that the transcriptional activity of MRFs may also be modulated by the equilibrium between their homo- and hetero-dimeric forms. One way to affect this equilibrium is to confine homo- and hetero-dimeric proteins to different DNA targets. In this context, the observed preferential binding of homodimeric MyoD to quadruplex structures of muscle-specific gene promoter sequences (15) may serve to attract it to the regulatory region of a target gene without activating its expression. By forming heterodimers with E-proteins, MyoD may then lose its tight association with the promoter quadruplex, bind with greater avidity to neighboring E-boxes and activate gene expression.
In the present study, we inquired whether the different binding affinities of MyoD homo- and hetero-dimers for quadruplex and E-box DNA are shared by other members of the MRF family. We first showed that similarly to MyoD (15), homodimers of Myogenin and MRF4 associated preferentially with bimolecular quadruplex forms of promoter sequences of the muscle-specific genes α7 integrin and sMtCK (Figure 1A and B) and that Myogenin increased the heat stability of the bound G'2 integrin DNA (Figure 2). Further, our results confirmed that in line with their role as muscle gene activators and similarly to heterodimeric MyoD, MRF4-E47 and Mygenin-E47 heterodimers also bound E-box considerably more tightly than quadruplex structure of a promoter sequence of the integrin gene (Figure 4 and Table 2). Also, in accord with the previously described preferential association of homodimeric MyoD with tetraplex DNA, MRF4 homodimers also bound G′2 integrin DNA more tightly than E-box (Figure 3 and Table 2). By clear contrast, however, Myogenin homodimers did not bind preferentially to quadruplex DNA and displayed similar relatively weak affinities for both integrin or sMtCK tetraplexes and E-box DNA (Figure 3, Table 2, and accompanying text). Thus, by failing to form tight complexes with quadruplex structures of promoter sequences of the two examined muscle-specific genes, homodimeric Myogenin was distinguished from homodimers of both MyoD and MRF4. Whereas MyoD and MRF4 are involved in the conversion of mesenchimal cells into dividing myoblasts during the initial determination step (5,6), Myogenin jointly with MRF4 mediates the subsequent phase of differentiation of myoblasts into nondividing myocytes and then to myotubes (7–10). It is tempting to speculate, therefore, that whereas quadruplex DNA structures may modulate the activity of MRFs during the earlier determination stage, they are not required for the regulation of MRFs action in the differentiation step.
In a second part of this report, we determined the protein structure basis for the different affinities of homodimeric MyoD and Myogenin for quadruplex DNA. All MRFs bind DNA at their basic region and as we recently reported, full intactness of this region is a prerequisite for the binding of E-box (36). In contrast, MyoD mutants whose basic regions were largely deleted except for a single triad of basic amino acids maintained their ability to associate with quadruplex DNA (36). The basic region of Myogenin differs from that of MyoD by two conservative substitutions in the R2 and R3 triads, by an intervening tract of four residues between the R1 and R2 triads and a single L→M substitution in the stretch between R2 and R3 (Figure 5A). Comparing the quadruplex binding capacities of unmodified and mutant Myogenin homodimers, we first showed that the two different amino acids in its R2 and R3 basic triads were not the source of the weak association of its with tetraplex DNA (Figure 5B). However, replacing the Myogenin complete basic region with that of MyoD increased the affinity of its homodimers for quadruplex DNA to nearly that of homodimeric MyoD (Figure 6B, Table 3). Thus, the MyoD basic region was capable of maintaining its high affinity for quadruplex DNA in the foreign environment of the Myogenin homodimer. Yet, reciprocal substitution of the MyoD basic region by the weakly binding Myogenin homolog domain did not diminish significantly the high affinity of MyoD for quadruplex DNA (Figure 6C, Table 3). This outcome indicated that MyoD regions outside the basic region affected enhancement of tetraplex binding by the normally weak Myogenin basic region. Previous comparison of the DNA-binding efficacies of intact MyoD and its isolated bHLH domain had suggested that domains that surround the basic region modulate its affinity for DNA ligands. We reported that an isolated bHLH domain bound E-box at ∼10-fold greater efficiency than full-length MyoD, whereas its capacity to bind G′2 integrin DNA was decreased by close to 2-fold (15). Thus, peripheral domains in homodimers of intact MyoD quenched the affinity of its basic region for E-box, while increasing its avidity for quadruplex DNA. In a similar vein, results presented in Figure 6C and in Table 3 indicated that a MyoD periphery was capable of augmenting the quadruplex DNA affinity of a transplanted Myogenin basic region. Interestingly, however, the effect of domains outside the basic region on the binding of quadruplex DNA was restricted to MyoD as the Myogenin periphery did not extinguish the high affinity of an implanted MyoD basic region for G′2 DNA (Figure 6B and Table 3). It appeared, therefore, that both the basic region and peripheral domains of MyoD dominantly contribute to high avidity for tetraplex DNA.
ACKNOWLEDGEMENTS
This work was supported by Israel Science Foundation, United States-Israel Binational Science Foundation and the Fund for Promotion of Research at the Technion (to M.F.). Funding to pay the Open Access publication charges for this article was provided by the United States-Israel Binational Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tajbakhsh S. Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Curr. Opin. Genet. Dev. 2003;13:413–422. doi: 10.1016/s0959-437x(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 2.Schnorrer F, Dickson BJ. Muscle building; mechanisms of myotube guidance and attachment site selection. Dev. Cell. 2004;7:9–20. doi: 10.1016/j.devcel.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 4.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Munsterberg AE, Kitajewski J, Bumcrot DA, McMahon AP, Lassar AB. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9:2911–2922. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- 6.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 7.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 8.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 9.Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, Olson EN. Myogenin's functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev. Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 11.Chanoine C, Della Gaspera B, Charbonnier F. Myogenic regulatory factors: redundant or specific functions? Lessons from Xenopus. Dev. Dyn. 2004;231:662–670. doi: 10.1002/dvdy.20174. [DOI] [PubMed] [Google Scholar]
- 12.Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 13.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 14.Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 15.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J. Biol. Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 16.Brennan TJ, Chakraborty T, Olson EN. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl Acad. Sci. USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis RL, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 18.Black BL, Molkentin JD, Olson EN. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell. Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 20.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 21.Larsen A, Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982;29:609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- 22.Hammond-Kosack MC, Docherty K. A consensus repeat sequence from the human insulin gene linked polymorphic region adopts multiple quadriplex DNA structures in vitro. FEBS Lett. 1992;301:79–82. doi: 10.1016/0014-5793(92)80214-2. [DOI] [PubMed] [Google Scholar]
- 23.Hammond-Kosack MC, Kilpatrick MW, Docherty K. Analysis of DNA structure in the human insulin gene-linked polymorphic region in vivo. J. Mol. Endocrinol. 1992;9:221–225. doi: 10.1677/jme.0.0090221. [DOI] [PubMed] [Google Scholar]
- 24.Hammond-Kosack MC, Kilpatrick MW, Docherty K. The human insulin gene-linked polymorphic region adopts a G-quartet structure in chromatin assembled in vitro. J. Mol. Endocrinol. 1993;10:121–126. doi: 10.1677/jme.0.0100121. [DOI] [PubMed] [Google Scholar]
- 25.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 28.Kumar P, Verma A, Maiti S, Gargallo R, Chowdhury S. Tetraplex DNA transitions within the human c-myc promoter detected by multivariate curve resolution of fluorescence resonance energy transfer. Biochemistry. 2005;44:16426–16434. doi: 10.1021/bi051452x. [DOI] [PubMed] [Google Scholar]
- 29.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh K, Gualberto A. MyoD binds to the guanine tetrad nucleic acid structure. J. Biol. Chem. 1992;267:13714–13718. [PubMed] [Google Scholar]
- 35.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2007;35:7087–7095. doi: 10.1093/nar/gkm746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Labeling oligonucleotide probes using 32P. Short Protocol in Mol. Biol. 2002;2:14-51–14-52. [Google Scholar]
- 39.Weisman-Shomer P, Naot Y, Fry M. Tetrahelical forms of the fragile X syndrome expanded sequence d(CGG)n are destabilized by two heterogeneous nuclear ribonucleoprotein-related telomeric DNA-binding proteins. J. Biol. Chem. 2000;275:2231–2238. doi: 10.1074/jbc.275.3.2231. [DOI] [PubMed] [Google Scholar]
- 40.Sarig G, Weisman-Shomer P, Fry M. Telomeric and tetraplex DNA binding properties of qTBP42: a homologue of the CArG box binding protein CBF-A. Biochem. Biophys. Res. Commun. 1997;237:617–623. doi: 10.1006/bbrc.1997.7198. [DOI] [PubMed] [Google Scholar]