Abstract
Bacteriophage ϕC31 encodes an integrase, which acts on the phage and host attachment sites, attP and attB, to form an integrated prophage flanked by attL and attR. In the absence of accessory factors, ϕC31 integrase cannot catalyse attL x attR recombination to excise the prophage. To understand the mechanism of directionality, mutant integrases were characterized that were active in excision. A hyperactive integrase, Int E449K, gained the ability to catalyse attL x attR, attL x attL and attR x attR recombination whilst retaining the ability to recombine attP x attB. A catalytically defective derivative of this mutant, Int S12A, E449K, could form stable complexes with attP/attB, attL/attR, attL/attL and attR/attR under conditions where Int S12A only complexed with attP/attB. Further analysis of the Int E449K-attL/attR synaptic events revealed a preference for one of the two predicted synapse structures with different orientations of the attL/attR sites. Several amino acid substitutions conferring hyperactivity, including E449K, were localized to one face of a predicted coiled-coil motif in the C-terminal domain. This work shows that a motif in the C-terminal domain of ϕC31 integrase controls the formation of the synaptic interface in both integration and excision, possibly through a direct role in protein–protein interactions.
INTRODUCTION
In order to establish a lysogenic life style, many bacteriophages encode an integrase to integrate the phage genome into the host chromosome (1,2). In association with an accessory factor, the recombination directionality factor (RDF) or Xis, the integrase also excises the phage genome on induction into the lytic pathway (2,3). During integration, the phage integrases act site-specifically by recombining the host and phage attachment sites, attB and attP, to form the hybrid products attL and attR. In excision attL and attR recombine to regenerate attB and attP. Phage integration and excision has to be a highly regulated process to ensure efficient switching between the lytic pathway and lysogeny (1). As some integrases are unidirectional in the absence of accessory factors, phage integrases can be used for site-specific genome manipulations and for the stable integration of transgenes (4). The integrase from the Streptomyces temperate phage ϕC31 is currently the most widely used phage integrase for genome manipulations (5–14).
The ϕC31 integrase is a member of a large family of site-specific recombinases, the serine recombinases (15,16). The best understood members of this family are the resolvase/invertases, in particular γδ and Tn3 resolvases and Hin invertase. The resolvase/invertases have an N-terminal catalytic domain (∼1–140 aa) and a small (∼141–183 aa), C-terminal DNA-binding domain. The ϕC31 integrase is a member of the large serine recombinases (17). These proteins have similar N-terminal catalytic domains to those of the resolvase/invertases, but they have much larger C-terminal domains of ∼300–500 aa. The large serine recombinases also include some transposases such as TnpX from the Clostridium perfringens transposon, Tn4451, and TndX from the C. difficile conjugative transposon CTn5397 (18–20). Tn4451 and CTn5397 confer resistances to chloramphenicol and tetracycline, respectively. TnpX and TndX have been shown to be capable of integrating and excising their respective transposons with no requirement for any additional transposon-encoded accessory factors (20,21).
The first step in a site-specific recombination pathway is recognition and binding of the substrates by the recombinase (22). Protein–protein interactions between the recombinase subunits then bring the two substrates together in a synaptic complex. Ghosh et al. (23) showed that dimers of mycobacteriophage Bxb1 integrase (a large serine recombinase) bind the substrates attB and attP, which are then probably brought together to form a synaptic tetramer. Post-synapsis, the resolvase/invertases and the large serine recombinases appear to use the same catalytic mechanism (16,22–27). DNA cleavage generates 2 bp staggered breaks in both substrates with the concomitant formation of transient covalent phospho-serine bonds between the recombinases and the recessed 5′ ends. Strand exchange is thought to occur by 180° rotation of two recombinase subunits bound to half sites relative to the other two subunits (28–30). Joining of the half sites to form recombinant products is inhibited when the two base overhangs at the staggered breaks cannot base pair due to mismatches (31,32). In the serine integrases, the non-palindromic nature of the dinucleotides at the crossover site are critical in determining the left–right polarity of the sites (24,26).
In the absence of any accessory factors ϕC31, Bxb1 and ϕRv1 integrases only catalyse the integration reaction attP x attB to form attL and attR (33–35). In binding assays, synaptic complexes can only be detected when integrase is provided with attP and attB and not when provided with attL and attR or any other pairwise combination of att sites (23,25). These observations have led to the idea that integrase has specific conformations when bound to attP and attB that enable the protein–protein interactions required for synapsis and subsequent recombination (23,25,27). A mutation analysis of the ϕC31 attB site showed that there are indeed sequences in attB that appear to be required for stable synapsis (36). An RDF (gp47) has been reported from Bxb1 that is essential for excisive recombination by Bxb1 integrase, (gpInt) (37). In this system, correct excision is dependent on gp47 to construct a synaptic complex containing gpInt and the DNA sites in a specific alignment that enables attL x attR recombination and prevents aberrant DNA rearrangements (38).
The role of the large C-terminal domain in the recombination mechanism by the phage serine integrases is poorly understood. It seems likely that the C-terminal domains are involved in DNA binding and in the control of directionality. The aim of this investigation was to understand the mechanism of directional control in ϕC31 integrase through the isolation and characterization of ‘hyperactive integrases’ that could recombine attL x attR sites. This work shows that a motif in the C-terminal domain of ϕC31 integrase controls the formation of a synaptic interface, possibly through a direct role in protein–protein interactions.
MATERIALS AND METHODS
Bacterial strains, plasmids and recombinant DNA techniques
Escherichia coli DH5α (F- ϕ80lacZΔM15,Δ.(lacZYA-argF), recA1, endA1, hsdR17, phoA, supE44, thi-1, gyrA96, relA1, λ) was used as a general host for plasmid preparation, SDM and for in vivo recombination assays (39). DH5α was grown routinely in 2YT (39). Plasmid DNAs (Table 1 and Supplementary Material) were introduced by transformation or electroporation (39).
Table 1.
Reporter and att site plasmids used in this study
| Plasmid | att sitesa | Reference |
|---|---|---|
| pRT504 | attB373-lacZ-attP464 | (25) |
| pRT508 | attB373-lacZ-attP464 | (25) |
| pPAR1000 | attL360-lacZ-attR475 | This work |
| pRT600 | attB51 | (26) |
| pRT702 | attP50 | (26) |
| pMSX18 | attL360-lacZα-attR475 | This work |
| pMSX22 | attP464-lacZα-attP50 | This work |
| pMSX24 | attL360-lacZα-attP50 | This work |
| pMSX26 | attB51-lacZα-attL360 | This work |
| pMSX28 | attB51-lacZα-attR475 | This work |
| pMSX30 | attP50-lacZα-attR475 | This work |
| pMSX32 | attB373-lacZα-attB51 | This work |
| pMSX34 | attL360-lacZα-attL62 | This work |
| pMSX36 | attR62-lacZα-attR475 | This work |
| pPARX18LAA | attL360_AA-lacZα-attR475 | This work |
| pPARX18RAA | attL360-lacZα-attR475_AA | This work |
| pMS92 | attL52_TA | This work |
| pMS93 | attR49_TA | This work |
| pMS94 | attL52 | This work |
| pMS95 | attR49 | This work |
aNumbers in superscript show the length of the att site in base pairs and the sequence of the central dinucleotides if different from wt.
In vivo recombination assays were performed by introducing ∼50 ng of an integrase expression plasmid into competent DH5α containing either the integration reporter plasmid, pRT504, or the excision reporter plasmid, pPAR1000 (Table 1), and plating the transformants on 2YT plates containing carbenicillin (50 μg/ml), kanamycin (20 μg/ml), IPTG (50 μg/ml) and Xgal (160 μg/ml). The recombination frequency was calculated as the total number of white colonies (including white colonies with blue sectors) divided by the total number of colonies.
Mutagenesis
A library of random mutants of the integrase expression plasmid, pHS62, was prepared in the E. coli mutator strain, XL1-Red (Stratagene, CA, USA). Over 200 transformants of XL1-Red (pHS62) were pooled and amplified. The extracted plasmids were screened for loss-of-function or gain-of-function phenotypes as described in the Results section.
SDM of the int gene was adapted from the method described in the QuikChange® SDM kit (Stratagene, CA, USA), with primers for SDM designed according to the manufacturer's recommendations. Primers are listed in the Supplementary Material. The DNA polymerase, PfuI (Promega, WI, USA), was used for PCR with pHS62 as the template and the PCR programme set to 98°C denaturation (1 min), 55°C annealing (30 s), 72°C extension (12 min) for 30 cycles. PCR reactions were digested with DpnI overnight and the products were introduced into DH5α and plated onto 2YT plates containing carbenicillin. Mutations in the int gene were verified by sequencing.
Expression and purification of integrase
E. coli BL21 DE3 (pLysS) was used as a host for overexpression of integrase. Integrase purifications were carried out as described previously (25). Protein concentrations were determined using the Quickstart Bradford protein reagent (BioRad, CA, USA).
DNA synapsis assays
DNA synapsis assays were performed as described previously (25,27), except that annealed oligonucleotides (Supplementary Material) were used to generate att site probes of identical lengths. Oligonucleotides were annealed (95°C, 10 min, followed by gradual cooling overnight to room temperature), purified by polyacrylamide gel electrophoresis (PAGE) using 5% polyacrylamide and 1 × TBE (Tris–borate–EDTA) buffer. After electrophoresis gels were stained with ethidium bromide and the bands containing dsDNA were excised and eluted into TE (10 mM Tris pH 8, 1 mM EDTA) using the using the ‘crush and soak’ method (39).
The annealed DNA fragments (∼1 pmol) were labelled with α-32P-dCTP by polymerization of the recessed 3′ ends by DNA polymerase I, Klenow Fragment (New England Biolabs) (39). Excess α-32P-dCTP was removed using MicroCLEAN (Microzone Ltd, West Sussex, UK) according to the supplier's instructions.
DNA binding was performed using 125 nM integrase and 1.5 nM of radio-labelled att site DNA in binding buffer (25). To assay synapsis, 1.5 nM of radio-labelled att site and between 14 and 32 nM of unlabelled PCR fragment encoding a second att site were incubated with 125 nM integrase in binding buffer. Samples were incubated for 2 h at 30°C and the products were separated by PAGE (5% polyacrylamide, 1 × TBE, 4°C, 5 W, 2 h). Gels were dried and exposed to Fuji phosphorimager screens. Images from the phosphorimager (Fuji FLA-3000) were analysed using AIDA software (Raytest).
In vitro recombination assays
Recombination in vitro was assayed either with unlabelled, supercoiled plasmids or with linear DNA fragments, one of which was radio-labelled. In a standard reaction using plasmid substrates containing att sites, ∼100 ng of plasmid substrates (final concentration ∼1.5 nM) were incubated (30°C, 2 h) in R buffer (10 mM Tris pH 7.5, 1 mM EDTA pH 8, 100 mM, NaCl, 5 mM DTT, 5 mM spermidine, 4.5% glycerol and 0.5 mg/ml bovine serum albumin) with integrase (733 nM) then heat denatured (80°C, 10 min) (25). Direct detection of recombinant molecules was by restriction analysis and agarose gel electrophoresis (25). A quantitative assay for recombination was also used which measured the deletion of lacZ by recombination between flanking att sites. In these assays, the plasmid substrate concentration was lowered to ∼0.15 nM but the recombination conditions were otherwise the same as in the standard reaction. Recombinants were detected by introducing the whole reaction mixture into E. coli DH5α by transformation and selecting transformants on media containing carbenicillin, IPTG and XGal. Recombinants (white colonies) were scored amongst a background of non-recombinant (blue colonies).
Recombination of linear fragments was performed using an attP (98 bp) fragment prepared by digestion of pRT702 (HindIII/BglII) or attL (90 bp) by digestion of pRT600702 (BglII/SphI) (26). Fragments were labelled as described for the synapse assays. Labelled probe (∼1.3 nM) and a PCR-generated fragment (∼10.6 nM) encoding a 235 bp partner att site and integrase (733 nM) were incubated in R buffer over a time course at 30°C. The reactions were stopped by heat denaturation (80°C, 10 min) and subtilisin (Sigma, Sigma-Aldrich Co. St Louis, MO, USA; 0.016 U, 37°C, 20 min). Subtilisin was inactivated (80°C, 10), the products were separated by PAGE (5% polyacrylamide, 1 × TBE) and analysed as described for the DNA-binding assays.
RESULTS
Integrase mutants that can recombine attL x attR
Plasmids encoding recombination defective or hyperactive integrases were identified using reporter strains of E. coli. The pRT504 encodes attP and attB flanking lacZ (25) and pPAR1000 encodes attL and attR flanking lacZα (Table 1). Transformation of E. coli DH5α (pRT504) or DH5α (pPAR1000) with a plasmid, pHS62 (33), that expresses the wild-type (wt) integrase (Int) yielded 100% white and 100% blue colonies, respectively, when grown on plates containing Xgal, IPTG and antibiotics. Thus, wt Int was fully active on the attP x attB reporter plasmid yielding recombinants that had lost the lacZ gene but was inactive on the attL x attR reporter plasmid. The pHS62 was introduced into the mutagenic E. coli strain XL1-Red and plasmid DNA was prepared from the pooled colonies. When the XL1-Red derived pHS62 plasmids were introduced into DH5α (pRT504), ∼3% of the transformants plated on agar containing Xgal, IPTG and antibiotics were dark blue, light blue or sectored blue suggestive of a defective integrase. When the pHS62-derived library was introduced into DH5α (pPAR1000), <1% appeared white on the same media and these were candidate gain-of-function mutants. The pHS62 derivatives conferring either defective or gain-of-function recombination phenotypes were purified and the int genes were sequenced to identify the mutations.
Re-transformation assays were used semi-quantitatively to characterize the activities of the mutant integrases. The defective mutants varied between 0% and 90% activity for attB x attP recombination (data not shown). In addition, two mutants, Int L460P and Int Y475H, demonstrated a low level of attL x attR recombination. Int E449G was isolated repeatedly from the gain-of-function screen and was 100% active in both integrative (attP x attB) and excisive (attL x attR) recombination using these in vivo assays.
In vitro recombination with hyperactive integrases
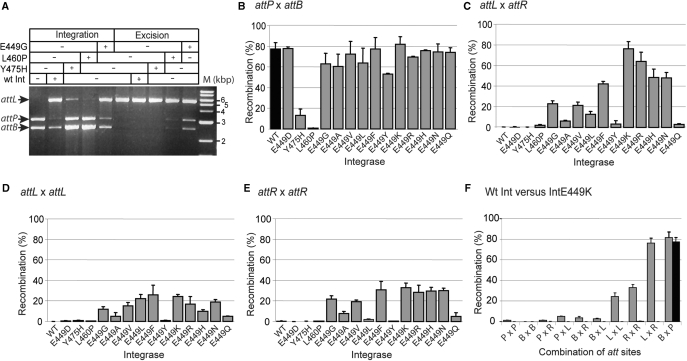
Integrase proteins were purified from E. coli strains expressing Int E449G, Int L460P and Int Y475H. These proteins were used in integration (attB x attP) and excision (attL x attR) assays using plasmid substrates in vitro (Figure 1A). Int E449G and Int L460P were active in both integration and excision, while Int Y475H, in this assay, appeared to be active only in integration.
Figure 1.
In vitro recombination with the hyperactive integrases. (A) Detection of recombination products by restriction analysis and agarose gel electrophoresis. Standard recombination conditions were applied and the reactions were digested with HindIII to detect integration and excision products. The product of integration, pRT602700, gave two fragments, 5435 bp (attL) and 91 bp (attR; data not shown), resulting from the fusion of the substrates, 3034 bp (pRT700, attP) and 2513 bp (pRT602, attB). For the excision reaction pRT602700 was used as a substrate. After HindIII digestion, the presence of the 3034 bp (attP) and 2513 bp (attB) indicated excision had occurred. The extreme right-hand lane contains molecular weight markers. (B–F) Quantitative in vitro recombination assays of Int mutants (grey bars) compared to wt Int (black bars). Recombinants were detected indirectly by excision of lacZ or lacZα flanked by two att sites using the following substrates: (B); attB x attP (pRT508), (C) attL x attR (pMSX18), (D), attL x attL (pMSX34) and (E), attR x attR, (pMSX36). Substrates were incubated with integrase and then introduced into DH5α by transformation. The percentage of white colonies obtained for each integrase is shown. (F) Activities of Int E449K and wt Int on all possible combinations of att sites using the following substrates; P × P (attP x attP; pMSX22), B × B (attB x attB; pMSX32), P × R (attP x attR; pMSX30), P × L (attP x attL; pMSX24), B × R (attB x attR; pMSX28), B × L (attB x attL; pMSX26), L × L (attL x attL; pMSX34), R × R (attR x attR; pMSX36), L × R (attL x attR; pMSX18) and B × P (attB x attP; pRT504). The data are the average and standard error for two replicates.
The properties of the mutant integrases were investigated further using a more sensitive and quantitative recombination assay. Plasmid substrates were constructed, which had every combination of two att sites in head-to-tail orientation flanking lacZ or lacZα (Table 1). These substrates were used in in vitro recombination reactions and, after inactivation of the integrase, the reaction products were introduced into E. coli DH5α by transformation. The presence of white colonies on plates containing Xgal, IPTG and carbenicillin indicated loss of lacZ or lacZα by recombination. The frequency of white colonies derived from a recombination reaction with attP x attB as substrates (pRT508) in the absence and presence of wt integrase was routinely about 5 × 10−4 and ∼0.8, respectively. To verify that a white colony contained the expected recombination product from pRT508, plasmid DNA was extracted and subjected to restriction analysis. All the colonies analysed in this way contained the expected deletion derivative of pRT508 (data not shown). The frequencies of white colonies obtained after incubation of wt Int with pairwise combinations of att sites other than attP x attB were not significantly higher than the background level (Figure 1F). Plasmids were extracted from the few white colonies that were obtained and no recombination products with the expected restriction fragments were observed with any the att site combinations except for attP x attP. The hyperactive integrase, Int E449G, was nearly as active as wt Int in attB x attP recombination and was able to recombine attL x attR, attR x attR and attL x attL (Figure 1B–E). Int E449G activity on the remaining pairwise combinations of sites (attP x attP, attB x attB, attP x attL, attP x attR, attB x attR, attB x attL) gave rise to rare white colonies, all of which contained genuine recombinant products except for those derived from the attB x attB plasmid. Int L460P and Int Y475H showed much lower, but detectable activities in attL x attR recombination (Figure 1C). Taken together these data indicated that a region of integrase from E449 to Y475 is required for integrase activity and is involved in controlling the directionality of recombination.
The residue at position 449 determines the extent of attL x attR recombination
Int E449G was the most hyperactive integrase mutant isolated from the XL1-Red mutagenesis experiment. E449 was targeted for site-directed mutagenesis. The mutant proteins were purified and their activities measured by in vitro recombination (Figure 1B–E). Except for E449D, all the E449 substitutions tested were active in excision to varying degrees (Figure 1C). This suggests that the negative charge prevented attL x attR recombination. Int E449Q and Int E449Y were also relatively poor at attL x attR recombination suggesting that a polar uncharged side chain can also inhibit excision. Generally, the substitutions at E449 were not greatly affected in attP x attB recombination (Figure 1B). The most affected substitution was Int E449Y but as Int E449F was as active as wt Int, steric hindrance alone cannot account for this reduced activity.
Int mutants containing substitutions at E449 that were competent for excision were also able to perform attL x attL and attR x attR recombination (Figure 1D and E). Little or no recombination was observed between other pairwise combinations of att sites (attP x attP, attB x attB, attP x attL, attP x attR, attB x attR, attB x attL; (Figure 1F and data not shown). The Int mutant most active in attL x attR recombination and most similar to wt integrase for attP x attB recombination was E449K (Figure 1B and C).
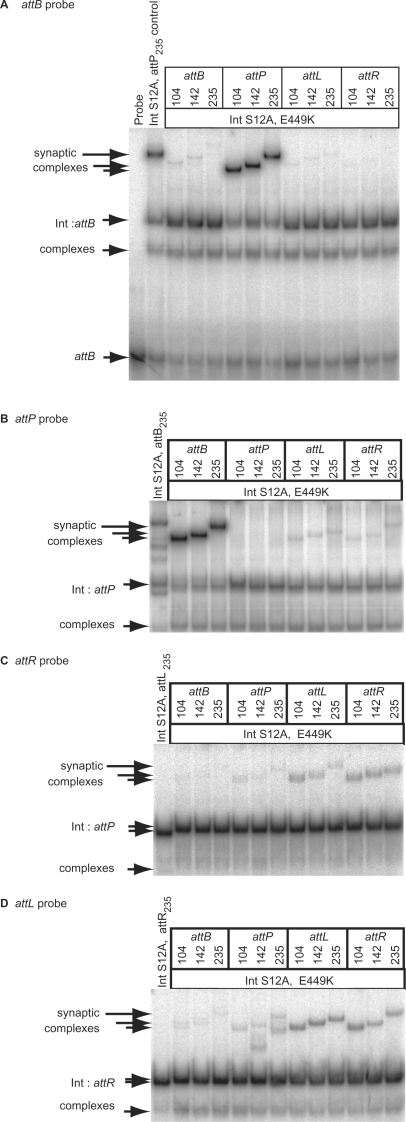
The hyperactive integrase, Int E449K, synapses attL and attR and other pairs of att sites
Previous studies have shown that wt Int does not synapse sites other than attP with attB (25,27). Synapse assays make use of an active site mutant of integrase, Int S12A, in which the serine nucleophile is altered and this prevents DNA cleavage (25). An integrase double mutant containing both S12A and E449K was purified. The intensities of the synaptic complexes obtained with Int S12A, E449K were highest with attP/attB, attL/attR, attL/attL and attR/attR as substrates (Figure 2). This was consistent with the ability of Int E449K to recombine attP x attB, attL x attR, attL x attL and attR x attR (Figure 1). However, synaptic complexes were also observed with Int S12A, E449K with pairs of att sites which, in recombination assays using Int E449K, did not yield products, e.g. attB x attB (Figures 1F and 2).
Figure 2.
E449K is hyperactive in synapsis. A catalytically inactive double mutant of integrase, S12A, E449K was used to assay synapsis between all combinations of att sites. Annealed oligonucleotides were labelled to provide probes with identical lengths; attB (A), attP (B) attR (C) and attL (D). The formation of synaptic complexes was assayed by adding integrase S12A, E449K (125 nM) and partner fragments encoding a second att site. To demonstrate the presence of the partner fragments in the synaptic complexes, different sizes (104, 142, 235 bp) of partner were used in each assay that shift the mobility of the synapse. A control reaction containing probe, S12A integrase (125 nM) and a 235 bp partner was run in each panel. The position of the probe (attB) is only shown in (A); in (B–D) only the integrase:DNA complexes are shown.
A possible explanation for the ability of Int E449K to synapse most pairs of recombination sites is that this protein is tetrameric in solution. The oligomeric state of the hyperactive integrases was analysed by gel filtration. Wt Int, Int L460P, Int Y475H, Int E449G and Int E449K were all shown to have retention times consistent with dimers in solution (data not shown; 27). Thus, like wt integrase, Int E449K probably binds its substrates as a dimer and then forms the synaptic interface by protein–protein interactions within a tetramer.
The double mutants Int S12A,Y475H and Int S12A,L460P were prepared and used in synapsis assays. Neither was able to form a detectable synapse with attP x attB nor attL x attR (data not shown). In addition, DNA-binding affinities by the hyperactive integrases were not significantly different from wt Int (Supplementary Material Figure S1). Int Y475H was defective in recombination compared to wt Int but had higher DNA-binding affinities for all four att sites than wt Int.
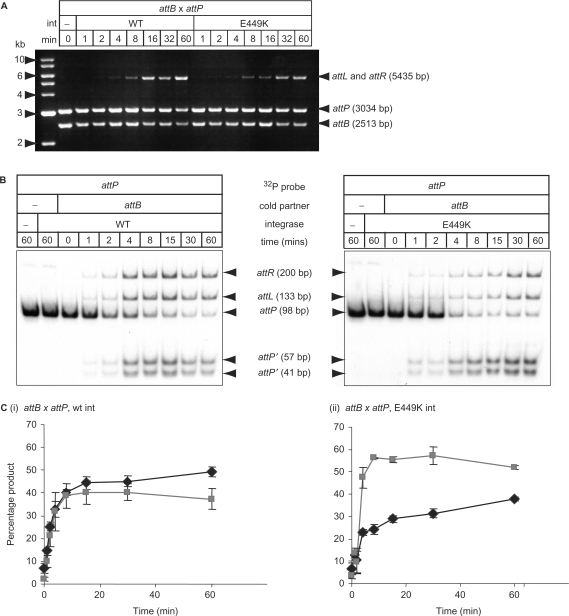
The hyperactive integrase, Int E449K, has gained the ability to recombine attL x attR without affecting integration
If the integration and excision reactions are in a reversible equilibrium and the rate of attP x attB recombination is much greater than attL x attR recombination, then integration by wt Int might appear to be irreversible. The observed attL x attR recombination in the hyperactive integrases could therefore be a consequence of a decrease in the overall rate of attB x attP recombination (Figure 3). When the supercoiled plasmid substrates encoding either attP (pRT702) or attB (pRT600) were mixed with either wt Int or Int E449K (at 15°C to slow the reaction), the initial rates of recombination appeared similar up to the 8 min time point. After this time wt Int consistently yielded more products. To test this quantitatively, a recombination assay was performed using a radio-labelled DNA fragment encoding attP and an unlabelled fragment encoding attB (Figure 3B and C). This assay confirmed that the initial rates of recombination by wt Int and Int E449K were similar in the attP x attB assay, but after the first few minutes the relative rate of product formation in the assay with Int E449K was significantly reduced. The level of cleaved attP present with Int E449K was considerably higher than that seen with wt integrase (Figures 3B and 4C). This is likely to be due to the ability of Int E449K to synapse and cleave the products of attP x attB recombination (attL and attR) upon formation. The same assay used with attL x attR as the initial substrates showed that the rate of excision was much lower than the rate of integration under these conditions and the level of cleaved substrate was about three times the amount of product (Supplementary Figure S2).
Figure 3.
Rate of recombination by wt versus E449K integrases. (A) A time course of attP x attB recombination and direct detection of recombinants by restriction and agarose gel electrophoresis. The time course was performed at 15°C to slow recombination with 733 nM integrase and reactions stopped by incubation at 80°C, 10 min. Reactions were digested with HindIII. The product of integration, pRT602700, gave two fragments, 5435 bp (attL) and 91 bp (attR; data not shown), resulting from the fusion of the substrates, 3034 bp (pRT700, attP) and 2513 bp (pRT602, attB). (B) Time course of attP x attB recombination using linear DNA fragments. Purified integrases (733 nM) were incubated with radio-labelled attP (98 bp; 1.5 nM) and a 235 bp fragment encoding attB (13 nM) for a time series at 30°C. The reactions were stopped by heating (80°C, 10 min) and then digested with subtilisin before loading onto a 5% polyacrylamide gel. The gel was dried and the radioactivity assayed using a phosphorimager. The positions of free probe (attP) the products of recombination (attL and attR) and the cleaved attP fragments (attP′ 57 bp and attP′ 41 bp) are indicated. (C) The amount of radioactivity in the products (black lines) versus the cleaved substrates (grey lines) was quantified and plotted against time for (i) attB x attP with wt integrase, (ii) attB x attP E449K integrase.
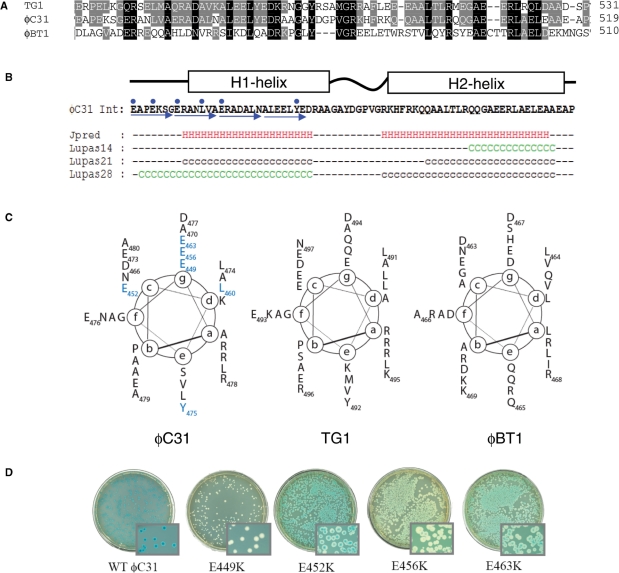
Figure 4.
Structure prediction for the hyperactive region of ϕC31 integrase. (A) An alignment of the sequences of the most similar homologues to ϕC31 integrase (ϕBT1 and TG1 integrases) is shown with the residues conserved between all three proteins shaded black and between two shaded grey. (B) The output from JPRED and COILS predictions are shown below the sequence of the hyperactive region from ϕC31 integrase (41,42). The blue arrows show the heptad repeat and the blue dots are the positions of the mutations conferring hyperactivity. H indicates a residue in a predicted α-helix, C (green) and c (black) indicate residues with coiled-coil forming probability of >90% and between 50% and 90%, respectively. (C) Helical wheel representations of the hyperactive region of ϕC31 integrase and comparable regions from TG1 and ϕBT1 integrases. The (a)–(g) are the positions of amino acids in the heptad repeat. The residues that confer hyperactivity are in blue. (D) Activities of wt and hyperactive integrases in the in vivo recombination assay using pPAR1000 as a reporter of attL x attR recombination. Blue colonies indicate no recombination. White, light blue or sectored colonies indicate recombination between attL x attR to excise the lacZ gene.
This assay revealed that about 40% of the radio-labelled attP remained cleaved after 1 h incubation with wt Int. This suggests that under these conditions, post-cleavage events (i.e. strand exchange and rejoining) were very slow compared to the rates of cleavage. In previous experiments using supercoiled substrates containing attP and attB, no accumulated cleaved intermediates were observed with wt Int (25,27,36). Recombination assays were performed specifically to detect the presence of any DNA cleaved at the crossover site in supercoiled substrates but none was observed (data not shown). The accumulation of cleaved substrates appears to be a feature of using linear DNAs as recombination substrates. When ϕC31 integrase is used in eukaryotic cell lines there is a significantly high level of micro-deletion that destroys the recombination sites, possibly due to non-homologous end-joining (NHEJ) machinery (7,40). It seems likely that integrase is also inhibited post-cleavage in the eukaryotic nucleus, allowing time for the NHEJ proteins to find and act on the recombination intermediates.
The hyperactive mutations lie within a putative coiled-coil motif
The ϕC31 Int and the integrases from two other Streptomyces phages, ϕBT1 and TG1 were subjected to structure prediction algorithms. The ϕBT1 and TG1 integrases have 23 and 49% identity to ϕC31 Int, respectively. Sequence alignments between protein homologues can be used to train secondary structure prediction algorithms such as JPRED (41). In the region where the hyperactive mutations mapped in ϕC31 Int, JPRED predicted two α-helices, H1 and H2, separated by a short linker (Figure 4). The equivalent regions in ϕBT1 Int and TG1 Int were also predicted to contain two α-helices (not shown). Another structure prediction programme, COILS (42), searches for probable coiled-coil regions in proteins. The ϕC31 Int, ϕBT1 Int and TG1 Int were all predicted to contain a region of coiled-coil that in ϕC31 Int spans the hyperactive region (Figure 4, Table 2). Coiled-coil motifs are normally identified by the presence of a canonically repeating heptad of amino acids, abcdefg, occupying positions (a)–(g) that can be displayed on a helical wheel (Figure 4C). Typically, within a coiled-coil domain amino acids at positions (a) and (d) are hydrophobic and (b), (c), (e), (f) and (g) are polar (43). The predicted coiled-coils in ϕC31, ϕBT1 and TG1 all appeared to have a hydrophobic position (d). However, the (a) position in all three of these proteins had two or more positively charged residues that would normally consist of hydrophobic amino acids. Most striking was the abundance of negatively charged amino acids at position (g). The COILS program compares the output from ‘weighted’ and ‘unweighted’ scans. The unweighted scan gives equal importance to the presence of the polar residues as to the hydrophobic residues but this can lead to false positives when the sequence is rich in polar residues. The weighted scan gives the same importance to the two hydrophobic positions as to the five polar positions thus reducing the impact of an abundantly polar sequence. If the difference between the two scans is >20–30% the prediction is likely to be a false positive. In addition, the COILS prediction uses three scanning windows; 14, 21 and 28 aa. A 28 residue window that displays the heptad repeat is highly likely to have a genuine coiled-coil motif, while the smaller scanning windows are more prone to false positives that would arise from a random sequence of amino acids. The ϕC31 integrase H1-helix maintained a high probability of forming a coiled-coil in both the weighted and unweighted scans using the 28 aa window (Figure 4; ‘Lupas28’ prediction). The H2-helix, however had a >40% drop in the probability of coiled-coil in the weighted versus the unweighted scans with the 28 and 21 residue windows. Only the smaller 14-residue scanning window maintained a >90% probability of forming a coiled-coil (Figure 4). E449 is located at the start of the longest predicted coiled-coil region and so has been included within the schematic representation despite the presence of the probable helix-breaker, P451. Y475 and L460 are located in the predicted coiled-coil domain. ϕBT1 H1-helix and TG1 H1- and H2-helices were predicted to form coiled-coils using the COIL algorithm (Figure 4, Table 2).
Table 2.
Location of putative coiled-coil regions in phage serine integrases
| Origin of integrase | Size (aa) | Predicted coiled-coil region(s)a | Region size (aa) | Accession number |
|---|---|---|---|---|
| Phage ϕC31 | 605 | 452–480/489–518 | 28/29 | NP047974 |
| Phage TG1 | 606 | 463–497/506–533 | 34/27 | BAF03600 |
| Phage Bcep22 | 518 | 350–387/390–423 | 37/33 | AAQ54941 |
| Phage U153 | 453 | 337–366/374–424 | 29/50 | CAD10283 |
| Prophage CP-933X | 508 | 367–398 400–427 | 31/27 | NP287356 |
| Phage ϕFC1 | 464 | 343–370/385–423 | 27/35 | AAD26564 |
| Phage ϕ105 | 477 | 358–385/388–415 | 27/27 | NP690785 |
| Phage TP91 | 398 | 284–314/316–350 | 30/34 | CAA72268 |
| Phage SPBc2 | 549 | 398–426/444–475 | 28/31 | NP046553 |
| Phage 370 | 476 | 385–412 | 27 | NP606888 |
| Phage TP901-1 | 487 | 405–434 | 29 | NP112664 |
| Phage ϕK381 | 552 | 427–455 | 28 | BAF03598 |
| Phage ϕBT1 | 594 | 435–469 | 34 | NP813744 |
| Phage ϕRv1 | 469 | 346–380 | 34 | NP216102 |
aBold values indicates a high probability of forming a coiled-coil structure based on the COILS programme (42).
Other large-serine recombinases also showed the presence of predicted α-helix in the C-terminal domain, many of which had a high probability of forming coiled-coils (Table 2). A coiled-coil domain has also been predicted in the large serine transposase TnpX from Tn4451 (44).
Residues that confer hyperactivity lie on one face of the putative coiled-coil motif
A spontaneous mutant of pHS62 was isolated that appeared to be active in attL x attR recombination using the DH5α (pPAR1000) reporter strain. Sequencing of the int gene from this spontaneous mutant revealed a mutation that caused the E463K substitution. E463K was able to give 100% recombinants when introduced into DH5α (pPAR1000) or DH5α (pRT504). E463K, like E449K resides on the (g) face of the predicted canonical coiled-coil (Figure 4C and D). Because of the location and nature of the mutations conferring hyperactivity, SDM was used to change all the acidic residues from E449 to D477 to lysine with the hope of isolating more hyperactive mutants. Two additional mutant integrases were isolated that could recombine attL x attR in vivo; E452K and E456K (Figure 4D). E449K, E456K and E463K all reside at the (g) position of the proposed coiled-coil and E452K lies in the adjacent (c) position (Figure 4C). Mutations of the other residues were either as wt (D466K, A470K, E473K, E476K and A479K) or inactive (D477K) (Figure 4C and data not shown). These data are consistent with this region presenting an interaction surface, which is altered in the hyperactive proteins.
Structural asymmetry in the Int E449K synaptic complexes with attL and attR
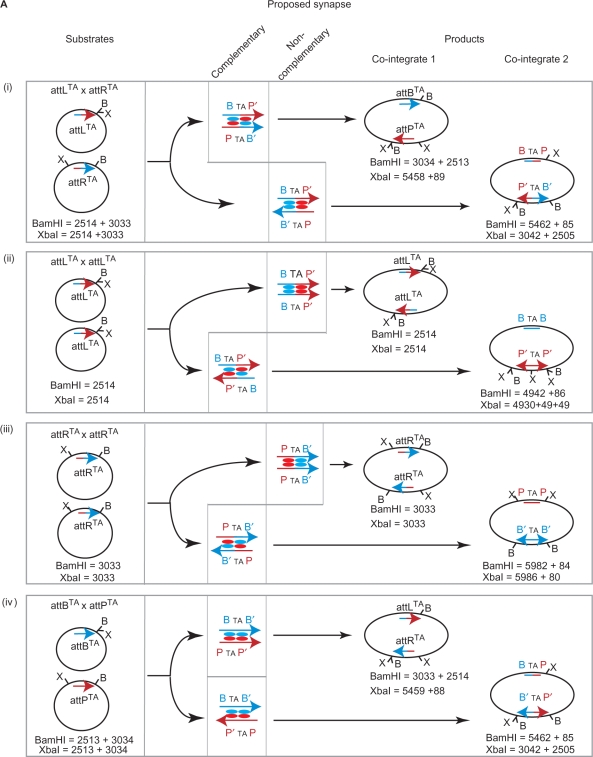
In the integration reaction, the polarity of the recombination sites is determined only by the nature of the 2 bp at the crossover site (26). In ϕC31 att sites, this is 5′TT and normally the left side of attB (B arm) joins to the right side of attP (P′arm) to make attL (BTTP′) and the reciprocal product, attR (PTTB′). If the crossover sequence is changed to 5′TA in both substrates or 5′AA in one substrate then polarity is either lost (5′TA) or switched (5′AA) (26). We showed previously that with wt Int, attBTA and attPTA recombines to give the normal products, BTAP′ (attLTA) and PTAB′ (attRTA) and aberrant products, BTAP and P′TAB′, in equal quantities (26). This was interpreted to mean that wt Int forms two synapse structures, differing by the spatial arrangements of P, P′ and B, B′ arms in the complex [Figure 5A(iv)]. Both synapse structures contain Int subunits bound to P or P′ arms making a synaptic contact with an Int subunit bound to the B or B′ arm. The premise is that integrase does not dictate a preference for the arrangement of the B and B′ arms with respect to the P or P′ arms in the integrative synapse [Figure 5A (iv)]. Where an Int subunit is bound to a B-type half site and synapses with an Int subunit bound to a P-type half site, we call this interaction ‘complementary’ (Figure 5A). Both synapse structures in attP x attB recombination involve complementary pairings of Int subunits [Figure 5A (iv)]. The ability of the hyperactive integrases to recombine attL x attR, attL x attL, attR x attR (Figure 1) provided the opportunity to interrogate the nature of the excisive synapse. When each pair of sites is brought together by Int E449K, we expect that two alternative synapse structures are possible that differ markedly from each other [Figure 5A (i) to (iii)]. One type of synapse involves complementary subunit interactions and in the other type of synapse the interactions between the Int subunits are non-complementary, i.e. both Int subunits contact a P-type arm or a B-type arm. If the interaction by an integrase subunit bound to P-type arms is different to that bound to B-type arms then the Int E449K synaptic complexes containing the hybrid sites, attL and attR, may be structurally biased.
Figure 5.
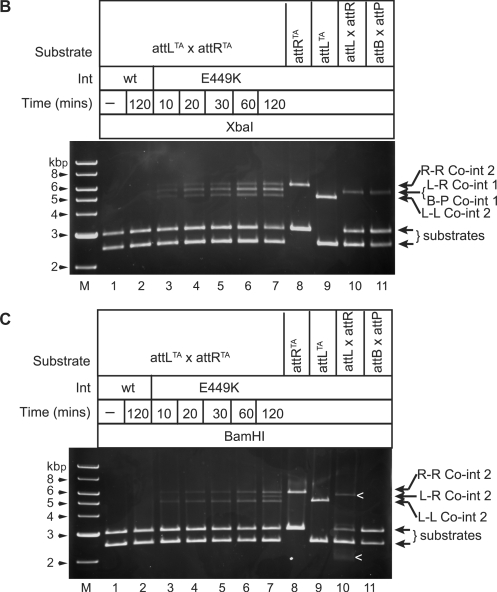
(B and C) Int E449K preferentially catalyses attLTA x attRTA recombination through a synapse, where Int interactions are complementary. A time course of Int E449K recombination using pMS92 (attLTA) and pMS93 (attRTA) as substrates (lanes 3–7) is shown. Control reactions also shown are pMS92 alone (lane 8), pMS93 alone (lane 9), pMS94 (attL) and pMS95 (attR), and pRT602 (attB) and pRT700 (attP). Reactions without integrase (lane 1), with wt Int (733 nM; lane 2) or with Int E449K (733 nM; lanes 3–11) were incubated at 15°C for the indicated times. Reactions were stopped by heat (80°C, 10 min), then divided equally and the appropriate restriction buffer and either XbaI (B) or BamHI (C) added. Arrows show the positions of products from attLTA x attRTA (L-R co-int 1 or L-R co-int2) or from attLTA x attLTA (L-L co-int 2) or from attRTA x attRTA (R-R co-int2). In (C), ‘<’ indicates that the bands shown were incomplete digests.
Figure 5.
Structural bias in the formation of the Int E449K synaptic complexes with attL and attR sites. (A) Recombination products from plasmids encoding attLTA (pMS92), attRTA (pMS93), attBTA (pRT603) and attPTA (pRT701). Products (co-integrates 1 and 2) are derived from reactions between (i) pMS92 (attLTA) and pMS93 (attRTA) (ii) pMS92 and pMS92 (iii) pMS93 and pMS93 and (iv) pRT603 (attBTA) and pRT701 (attPTA). Each attachment site is represented as an arrow. The halves without the arrow heads are the B or P arms, the halves with the arrowheads are the B′ or P′ arms. B-type arms are blue and P-type arms are red. The structures of the proposed synaptic complexes are indicated by the alternative alignments of P or P′ and B or B′ arms. The putative synaptic tetramer of integrase subunits is shown with each subunit coloured according to whether it is interacting with a B-type arm (blue) or a P-type (red). Int interactions at the synaptic interface are either complementary (Int bound to P-type arms interact with Int bound to B-type arms) or non-complementary (Int subunits that are both bound to P-type or B-type arms). The positions of relevant restriction sites are indicated (B, BamHI; X, XbaI) and the expected restriction fragment sizes of the parental plasmids and the co-integrate products are shown.
Plasmid substrates encoding attLTA (pMS92) and attRTA (pMS93) were mixed and incubated with Int E449K at 15°C (Figure 5A–C). The products of recombination reactions containing pMS92 and pMS93 are predicted to include recombinants between three different reactions; attLTA x attRTA, attLTA x attLTA and attRTA x attRTA [Figure 5A (i), (ii) and (iii)]. From each reaction two products (co-integrates 1 and 2) could, theoretically, be made, arising from alternative synapse structures that contain complementary or non-complementary Int interactions (Figure 5A). The two co-integrate products from the attLTA x attRTA recombination can be detected with XbaI (co-integrate 1; 5458 bp; Figure 5B) or BamHI (co-integrate 2; 5462 bp; Figure 5C). Co-integrate 1 appeared well before co-integrate 2 suggesting that when the sites come together in a synapse, complementary pairing of Int E449K subunits is preferred to the non-complementary pairing [Figure 5A(i), B and C]. As seen previously with wt Int (26), Int E449K did not show any bias towards generating either co-integrate 1 or 2 when the substrates were attPTA and attBTA (data not shown).
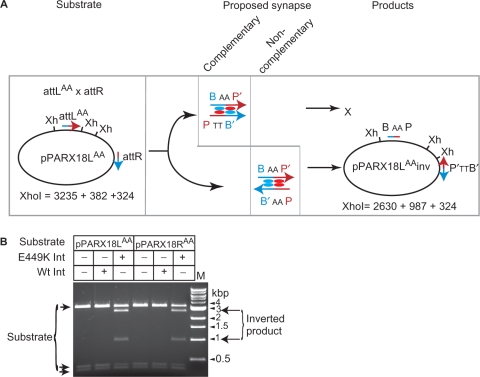
In these reactions, we also observed attLTA x attLTA and attRTA x attRTA recombination. The appearance of co-integrate 2-type products are indicated by the presence of the ∼4900 bp band for attLTA x attLTA and the ∼5980 bp band for the attRTA x attRTA in both the BamHI and XbaI digests. These appeared at the same time as the favoured product (co-integrate 1) from the attLTA x attRTA reaction [Figure 5A (ii), (iii), B, C] and well before the appearance of the attLTA x attRTA unfavoured (co-integrate 2) product. The proposed synapse structures that generated the co-integrate 2-type products from attLTA x attLTA or attRTA x attRTA recombinations are those where the bound Int E449K subunits interact in complementary pairings, i.e. a subunit bound to a P-type arm interacts with a B-type arm bound subunit (Figure 5A). Thus, in all three reactions, attLTA x attRTA, attLTA x attLTA and attRTA x attRTA products generated from synaptic complexes where the Int E449K interactions are complementary appear at the same time and well before products that arise from synaptic complexes containing non-complementary subunit pairings. It is possible that the unfavoured product of attLTA x attRTA recombination (co-integrate 2) could have arisen indirectly through the generation of co-integrate 1 followed by inversion of DNA between the attPTA and attBTA sites. To test whether products could be obtained at all via a synapse between attL and attR where the Int E449K subunits interact via non-complementary pairings, it was necessary to invert the polarity of one of the sites by changing the dinucleotides at the crossover site. Two new plasmids, pPARX18LAA and pPARX18RAA were generated by SDM of pMSX18 so that the crossover dinucleotides in attL were changed to attLAA in pPARX18LAA and those in attR to attRAA in pPARX18RAA. Due to the constraint provided by the crossover dinucleotides the only possible recombination products from pPARX18LAA and pPARX18RAA are inversion products arising through synaptic complexes in which Int subunits are in non-complementary pairings (Figure 6A). Inversion products were indeed detected, indicating that non-complementary pairings are possible in attL x attR recombination (Figure 6B). The outcome of this experiment was anticipated by the high number of deletion products from E449K Int acting on attL x attL and attR x attR (pMSX34 and pMSX36, respectively; Figure 1), reactions in which the Int subunit interactions were likely to be non-complementary.
Figure 6.
Recombination between attL and attR via the unfavoured synapse is not prevented. (A) Expected recombination products from pPARX18LAA encoding attLAA and attR in head to tail orientation. The att sites are represented as described in the legend to Figure 5A except that the central dinucleotide in the attL site has been changed to 5′AA. Synapsis of attLAA and attR in the complementary format will be permitted but formation of products is prevented (‘X’) by the mismatch at the cleavage sites. Synapsis with the attLAA and attR sites in the non-complementary arrangement will give rise to products where the DNA between the att sites is inverted. Recombination by attL x attRAA in pPARX18RAA is also expected to give an inverted product with the same restriction fragments as pPARX18LAA; Xh, XhoI. The fragment sizes for the substrate and the expected inversion products are indicated. (B) Inversion of pPARX18LAA and pPARX18RAA by Int E449K. Recombination reactions were performed under standard conditions and then digested with XhoI. The substrates and products are indicated. M, molecular weight markers.
These data show that the synaptic complexes between attL/attR and the hyperactive integrase, Int E449K, are structurally biased towards a synapse that proceeds through recombination to generate attP and attB rather than aberrant recombination products (Figure 5). This structural bias between two forms of synapse is not observed for attP x attB recombination (26).
DISCUSSION
The ϕC31 integrase is a unidirectional phage integrase that acts specifically on attP and attB to form the products attL and attR. In the absence of any accessory factors, the reverse reaction (attL x attR to form attP and attB) does not occur. Previous work has shown that the specific block to attL x attR recombination is the failure to form a stable synapse (25,27). Thus, integrase may bind to attP and attB with confomers that enable synapsis of these sites but prevent attL and attR synapsis. In order to understand directional control in integrase, we isolated and characterized mutants of integrase that were bi-directional, i.e. able to recombine attP x attB and attL x attR. The phenotypes of the hyperactive integrases resemble the naturally bi-directional large serine transposases, TnpX and TndX (20,21).
Mutations in integrase that conferred hyperactivity lay within a putative coiled-coil motif that spans from E449 to E463 in the large C-terminal domain (Figures 1 and 4). Many of the mutations lay on the same face of the predicted coiled-coil (Figure 4). By analogy with other proteins containing coiled-coil domains the putative coiled-coil motif in ϕC31 integrase may comprise a protein-protein interaction surface (43). In the resolvases, the synaptic interface occurs via the N-terminal domain and hyperactivity (in this case defined as an ability to recombine substrates in the absence of activation by resolvase subunits bound to accessory sites) results from mutations in this region (45,46). If the synaptic interface in ϕC31 integrase also occurs exclusively through the N-terminal domain, the hyperactive region may enable synapsis indirectly through communication between the C-terminal domain and the N-terminal catalytic domain. Thus, the C-terminal domain may control the formation of the synaptic interface in an analogous way to resolvase responding to the presence of accessory sites or Hin interacting with Fis (16,22). Alternatively, the putative coiled-coil region may act directly through inter-molecular interactions. Possibly, the coiled-coil domain is normally sequestered unless integrase is bound to attP and attB when the coiled-coil region becomes exposed. If the mutations conferring hyperactivity destabilize the sequestration then the coiled-coil domain may be exposed and able to form inter-molecular interactions even when integrase is bound to attL or attR. This is the first indication that synapsis might occur via the C-terminal domain. In both models, synapsis is the step which activates recombination.
Additionally, several mutations within the C-terminal coiled-coil region (Int L460P, Int Y475H and Int D477K) were found to be defective in integration. Derivatives of Int L460P and Int Y475H were unable to form stable synaptic complexes. Clearly the putative coiled-coil domain has an important role in both integration and excision.
It is evident that the integration and excision reactions in the hyperactive mutants are different. Most striking is that Int E449K showed a bias towards one of two alternative synaptic complexes with respect to the spatial positioning of P, P′ and B, B′ arms in attL x attR recombination, whereas no bias was observed with attP x attB as substrates. The bias in the excision synapse is consistent with an Int subunit bound to a P-type arm preferring to synapse with an Int subunit bound to a B-type arm (Figure 5A). This implies that integrase has a mechanism of transmitting information, probably through protein conformation, from the DNA sequence to which it is bound to the synaptic interface. If this is the case, then Int complexes bound to attP or attB will be conformationally symmetrical but not the same. The symmetry arises from the sequences of attP and attB, which are imperfect inverted repeats. When attP and attB sequences are aligned (with the crossover dinucleotide in the centre), 15 out of 38 bases are identical (36). Most of the sequence divergence lies at the extremities and it is here that mutations in the attB site can have highly detrimental effects on the activity of attB in integration (36). Possibly, it is these unique sequences that specifically affect the conformations of integrase subunits bound to P-type arms or to B-type arms.
The excision reaction catalysed by ϕC31 Int E449K has similar features to Bxb1 integrase, catalysing attL x attR recombination in the presence of a recombination directionality factor (RDF; gp47) (38). In the presence of gp47, Bxb1 integrase also has a preference for one of the two possible synaptic complexes with respect to the arrangement of P- or B-type arms (38). Because the ϕC31 excision reaction with Int E449K occurs without the accessory protein it is possible to infer that the conformational ‘readout’ from DNA sequence interactions to the structure of the synaptic interface is inherent in the mechanism of the large serine recombinases generally. We emphasize however that Int subunits do not act independently of each other in adopting a conformation that will enable synapsis. No productive synapsis (and almost no recombination) was observed with Int E449K where the numbers of B to P-type arms deviated from 2:2, i.e. reactions such as attB x attR, attB x attL, attP x attL and attP x attR (3:1), as well as attP x attP and attB x attB (4:0). This selectivity indicates that prior to synapsis, conformations adopted by Int dimers when binding to pairs of att sites are still the primary step in controlling against unwanted DNA rearrangements. The discovery of a putative interaction motif in the C-terminal domain of ϕC31 integrase now provides a focus for understanding how different protein conformations mediate synapsis and the control of directionality in integration versus excision.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Andrew McEwan and Dr Milind Gupta for useful discussions and Dr John Walshaw for help with the coiled-coil analysis. This work was supported by the Biotechnology and Biological Science Research Council of the UK (grant reference: BB/D007836/1) and by a University of Aberdeen studentship to P.A.R. Funding to pay the Open Access publication charges for this article was provided by the BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Campbell A. In: The Bacteriophages. 2nd edn. Calendar R, editor. Oxford, New York: Oxford University Press; 2006. pp. 66–73. [Google Scholar]
- 2.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Ann. Rev. Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 3.Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29:2205–2216. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groth AC, Calos MP. Phage integrases: biology and applications. J. Mol. Biol. 2004;335:667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 5.Dafhnis-Calas F, Xu Z, Haines S, Malla SK, Smith MCM, Brown WR. Iterative in vivo assembly of large and complex transgenes by combining the activities of ϕC31 integrase and Cre recombinase. Nucleic Acids Res. 2005;33:e189. doi: 10.1093/nar/gni192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malla S, Dafhnis-Calas F, Brookfield JF, Smith MCM, Brown WR. Rearranging the centromere of the human Y chromosome with ϕC31 integrase. Nucleic Acids Res. 2005;33:6101–6113. doi: 10.1093/nar/gki922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 9.Calos MP. The ϕC31 integrase system for gene therapy. Curr. Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 10.Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using ϕC31 integrase. Nat. Methods. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc. Natl Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kittiwongwattana C, Lutz K, Clark M, Maliga P. Plastid marker gene excision by the ϕC31 phage site-specific recombinase. Plant Mol. Biol. 2007;64:137–143. doi: 10.1007/s11103-007-9140-4. [DOI] [PubMed] [Google Scholar]
- 13.Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA, Calos MP. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- 14.Thomason LC, Calendar R, Ow DW. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage ϕC31 site-specific recombination system. Mol. Genet. Genomics. 2001;265:1031–1038. doi: 10.1007/s004380100498. [DOI] [PubMed] [Google Scholar]
- 15.Grindley NDF. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington: ASM Press; 2002. pp. 272–302. [Google Scholar]
- 16.Johnson RC. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington D.C.: ASM Press; 2002. pp. 230–271. [Google Scholar]
- 17.Smith MCM, Thorpe HM. Diversity in the serine recombinases. Mol. Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 18.Crellin PK, Rood JI. The resolvase/invertase domain of the site-specific recombinase TnpX is functional and recognizes a target sequence that resembles the junction of the circular form of the Clostridium perfringens transposon Tn4451. J. Bacteriol. 1997;179:5148–5156. doi: 10.1128/jb.179.16.5148-5156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullany P, Roberts AP, Wang H. Mechanism of integration and excision in conjugative transposons. Cell. Mol. Life Sci. 2002;59:2017–2022. doi: 10.1007/s000180200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Mullany P. The large resolvase TndX is required and sufficient for integration and excision of derivatives of the novel conjugative transposon Tn5397. J. Bacteriol. 2000;182:6577–6583. doi: 10.1128/jb.182.23.6577-6583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyras D, Adams V, Lucet I, Rood JI. The large resolvase TnpX is the only transposon-encoded protein required for transposition of the Tn4451/3 family of integrative mobilizable elements. Mol. Microbiol. 2004;51:1787–1800. doi: 10.1111/j.1365-2958.2003.03950.x. [DOI] [PubMed] [Google Scholar]
- 22.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Ann. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh P, Pannunzio NR, Hatfull GF. Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites. J. Mol. Biol. 2005;349:331–348. doi: 10.1016/j.jmb.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh P, Kim AI, Hatfull GF. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol. Cell. 2003;12:1101–1111. doi: 10.1016/s1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 25.Smith MC, Till R, Brady K, Soultanas P, Thorpe H, Smith MCM. Synapsis and DNA cleavage in ϕC31 integrase-mediated site-specific recombination. Nucleic Acids Res. 2004;32:2607–2617. doi: 10.1093/nar/gkh538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith MCA, Till R, Smith MCM. Switching the polarity of a bacteriophage integration system. Mol. Microbiol. 2004;51:1719–1728. doi: 10.1111/j.1365-2958.2003.03942.x. [DOI] [PubMed] [Google Scholar]
- 27.Thorpe HM, Wilson SE, Smith MCM. Control of directionality in the site-specific recombination system of the Streptomyces phage ϕC31. Mol. Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 28.Kamtekar S, Ho RS, Cocco MJ, Li W, Wenwieser SV, Boocock MR, Grindley NDF, Steitz TA. Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination. Proc. Natl Acad. Sci. USA. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley NDF, Steitz TA. Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 30.Stark WM, Boocock MR, Sherratt DJ. Catalysis by site-specific recombinases. Trends Genet. 1992;8:432–439. [PubMed] [Google Scholar]
- 31.Stark WM, Grindley NDF, Hatfull GF, Boocock MR. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J. 1991;10:3541–3548. doi: 10.1002/j.1460-2075.1991.tb04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson RC, Simon MI. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 33.Thorpe HM, Smith MCM. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl Acad. Sci. USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bibb LA, Hancox MI, Hatfull GF. Integration and excision by the large serine recombinase ϕRv1 integrase. Mol. Microbiol. 2005;55:1896–1910. doi: 10.1111/j.1365-2958.2005.04517.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S, Hatfull GF. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol. Microbiol. 2003;50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 36.Gupta M, Till R, Smith MCM. Sequences in attB that affect the ability of ϕC31 integrase to synapse and to activate DNA cleavage. Nucleic Acids Res. 2007;35:3407–3419. doi: 10.1093/nar/gkm206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh P, Wasil LR, Hatfull GF. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh P, Bibb LA, Hatfull GF. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc. Natl Acad. Sci. USA. 2008;105:3238–3243. doi: 10.1073/pnas.0711649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 40.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage ϕC31 integrase. Mol. Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 42.Lupas A. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 1997;7:388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- 43.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 44.Adams V, Lucet IS, Lyras D, Rood JI. DNA binding properties of TnpX indicate that different synapses are formed in the excision and integration of the Tn4451 family. Mol. Microbiol. 2004;53:1195–1207. doi: 10.1111/j.1365-2958.2004.04198.x. [DOI] [PubMed] [Google Scholar]
- 45.Burke ME, Arnold PH, He J, Wenwieser SV, Rowland SJ, Boocock MR, Stark WM. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol. Microbiol. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 46.Sarkis GJ, Murley LL, Leschziner AE, Boocock MR, Stark WM, Grindley NDF. A model for the γδ resolvase synaptic complex. Mol. Cell. 2001;8:623–631. doi: 10.1016/s1097-2765(01)00334-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.