Abstract
The Escherichia coli RutR protein is the master regulator of genes involved in pyrimidine catabolism. Here we have used chromatin immunoprecipitation in combination with DNA microarrays to measure the binding of RutR across the chromosome of exponentially growing E. coli cells. Twenty RutR-binding targets were identified and analysis of these targets generated a DNA consensus logo for RutR binding. Complementary in vitro binding assays showed high-affinity RutR binding to 16 of the 20 targets, with the four low-affinity RutR targets lacking predicted key binding determinants. Surprisingly, most of the DNA targets for RutR are located within coding segments of the genome and appear to have little or no effect on transcript levels in the conditions tested. This contrasts sharply with other E. coli transcription factors whose binding sites are primarily located in intergenic regions. We suggest that either RutR has yet undiscovered function or that evolution has been slow to eliminate non-functional DNA sites for RutR because they do not have an adverse effect on cell fitness.
INTRODUCTION
A cohort of over 250 transcription factors controls gene expression in Escherichia coli in response to specific environmental cues. Most factors are operon specific and regulate the transcription of a small number of genes, while a small number of ‘global’ regulators coordinate transcription from a large number of promoters (1–3). Newly developed whole genome technologies now enable us to catalogue binding targets for each factor. These studies confirm that most regulators bind to intergenic DNA sequences near the 5′ end of a gene to regulate transcription.
The E. coli rutABCDEFG operon, that encodes genes for the catabolism of pyrimidines, is regulated by RutR, a TetR family factor whose DNA binding is modulated by uracil (4,5). RutR is transcribed divergently from rutA and, using genomic SELEX, Shimada and co-workers (5) identified six DNA targets for RutR, including the rutR-rutABCDEFG intergenic region. Interestingly, this study reported that two of the targets are located within open reading frames (ORFs) and failed to detect any RutR-dependent modulation of transcription at one of the targets (ygiF/glnE). To investigate RutR further, here we have used chromatin immunoprecipitation (ChIP), in combination with DNA microarrays (ChIP-chip), to measure the chromosome-wide DNA-binding profile of RutR in vivo. We compared the binding profile of RutR in cells growing exponentially in media either with or without uracil, which inhibits DNA binding by RutR. Our study identifies 20 different binding targets for RutR. Surprisingly, 14 of these targets are located within genes and, for all but one of these targets, we were unable to measure any RutR-dependent effects on RNA levels.
MATERIALS AND METHODS
E. coli strains and oligonucleotides
Bacterial strains and synthetic oligodeoxynucleotides used in this work are listed in Supplementary Table 1. In all experiments we used E. coli strain BW25113 (6) or the ΔrutR derivative JW0998 (7). BW25113 expresses normal levels of RutR from a chromosomal copy of the rutR gene. Other than the rutR mutation, BW25113 and JW098 are isogenic. Cells were grown in M9 minimal medium supplemented with 0.4% glucose in either the presence or absence of 0.1 mM uracil. For experiments with exponentially growing cells, overnight cultures of E. coli strain BW25113 or JW0998 were diluted 1:100 into fresh medium either with or without uracil, and grown for ∼4 hours to an OD650 of 0.3–0.4.
ChIP and DNA microarray analysis
ChIP assays were used to measure the chromosome-wide DNA-binding profile of RutR in the presence and absence of uracil, using experimental protocols described in detail by Efromovich et al. (8). Assays were done either in triplicate (−uracil) or in duplicate (+uracil). Briefly, cultures of E. coli BW25113 and, as a control, JW0998 ΔrutR were grown to mid-log phase at 37°C. Cells were then treated with 1% formaldehyde and broken open by sonication which also fragments cross-linked nucleoprotein. Cross-linked RutR–DNA complexes were immunoprecipiated from cleared lysates of BW25113 using anti-RutR rabbit polyclonal anti-serum, and parallel samples were isolated from control JW0998 ΔrutR cells. Cross-links were then reversed and immunoprecipitated DNA was purified. DNA samples isolated from BW25113 cells and the control ΔrutR cells were labelled with Cy5 and Cy3, respectively. To identify segments of DNA specifically associated with RutR, the two labelled samples were combined and hybridised to a 22 000 feature DNA microarray (Oxford Gene Technology, Oxford, UK). For each probe, the Cy5/Cy3 ratio was measured and this was plotted against the corresponding position on the E. coli BW25113 chromosome, creating a profile of RutR binding (Figure 1). We then selected ‘peaks’, formed by two or more consecutive probes, with a Cy5/Cy3 ratio of >2.5. To increase the stringency of our search, we discarded the small number of peaks where the RutR-binding signal was not reduced at least two-fold when uracil was added to cultures. The centre of each peak is defined as the centre of the probe within the peak that had the highest Cy5/Cy3 signal.
Figure 1.
Distribution of RutR binding across the E. coli chromosome. (A) The figure shows an overview of results from ChIP-chip experiments that measure the profile of RutR binding across the E. coli chromosome during exponential growth in the absence of added uracil. Binding signals (y-axis) are plotted against their location on the 4.64 Mbp E. coli chromosome (x-axis). The locations of selected signals are lablelled. A complete list of the targets is presented in Table 1. Data shown in all panels are average values from replicate experiments. (B) The figure shows RutR binding to intergenic segments of the chromosome in either the absence (blue) or presence (red) of added uracil. (C) The figure shows RutR binding to coding segments of the chromosome in either the absence (blue) or presence (red) of added uracil.
DNA sequence analysis
The RutR-binding motif was extracted from 500-bp DNA sequences centred around the binding peaks using BioProspector (http://ai.stanford.edu/~xsliu/BioProspector/). A DNA sequence logo describing the binding motif was then generated using WebLogo (http://weblogo.berkeley.edu/). For RutR, each of the 20 binding sites identified by ChIP-chip were aligned in PREDetector (9) and a position weight matrix (PWM) was generated to describe the information content of the binding site. Each target was then assigned a score depending on how it matched the PWM. The average score was used as a cut-off when searching genome sequences for RutR-binding sites. The same approach was used for the other transcription factors shown in Figure 4 except that we used the binding site alignments of Robison et al. (10) to generate the PWM.
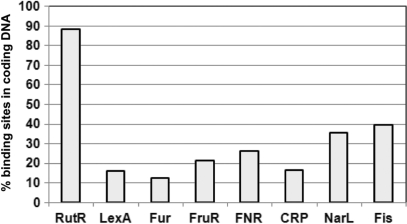
Figure 4.
Distribution of sites for global DNA-binding proteins between coding and non-coding DNA in the E. coli chromosome. Using equally stringent search conditions for each factor, we screened the E. coli MG1655 genome for DNA sequences that resembled the DNA-binding sites of RutR, LexA, Fur, FruR, FNR, CRP, NarL and Fis. The results show that, while predicted high-affinity-binding sites for most factors are found in non-coding DNA, most sequences resembling the RutR-binding site were found in coding DNA.
RutR purification and electrophoretic mobility shift assays
His-tagged RutR was over-produced and purified exactly as described by Shimada et al. (5) using plasmid pYcdC and E. coli strain BL21(λDE3). Purified RutR was more than 95% pure as analysed by SDS PAGE. DNA fragments for EMSA experiments were generated by PCR amplification using the appropriate DNA primers and chromosomal BW25113 DNA as a template (Supplementary Table 1). PCR products were purified, cut with HindIII and end labelled using [γ-32P]-ATP and polynucleotide kinase. DNA fragments were then incubated with purified RutR in buffer containing 20 mM Tris pH 7, 10 mM MgCl2, 100 μM EDTA, 120 mM KCl and 12.5 μg/ml herring sperm DNA. Reactions were loaded under tension onto a 5% polyacrylamide gel, run in 0.5× TBE at 160 V for 2–4 hours and analysed as described above.
RESULTS AND DISCUSSION
ChIP-chip analysis of RutR binding in mid-log phase E. coli
Our aim was to use ChIP to measure the binding of RutR across the chromosome of growing E. coli cells. Thus, strains BW25113 and the ΔrutR derivative JW0998 were grown aerobically, in M9 minimal medium supplemented with 0.4% glucose, to an OD650 of 0.3–0.4. Cells were then treated with formaldehyde, and cellular DNA was extracted and sonicated, yielding DNA fragments of ∼500–1000 bp. After immunoprecipitation with anti-RutR antibodies, DNA fragments from BW25113 or control JW0998ΔrutR cells were purified and labelled with Cy5 and Cy3, respectively, mixed and hybridised to the microarray. After washing and scanning, the Cy5/Cy3 signal intensity ratio was calculated for each probe. In parallel, the experiment was repeated using cells grown in the presence of 0.1 mM uracil. Complete data sets are shown in Supplementary Table 2. Figure 1A gives an overview of the profile for RutR binding. Most peaks for RutR binding are discrete, easily distinguishable from the background signal and sensitive to uracil (examples are shown in Figure 1B and C).
Identification and sequence analysis of RutR targets
To select peaks for RutR binding, a cut-off was applied to the dataset (see Materials and Methods section). A total of 77 probes passed this cut-off, corresponding to 20 separate peak locations (listed in Table 1). The targets we identified for RutR included four of the six RutR-binding targets reported previously (carA, rutA, hyi and ygiF/glnE). Our failure to find all of the targets identified by Shimada et al. (5) may be due to differences in the in vivo and in vitro DNA-binding properties of RutR or the high false-negative rate of ChIP-chip analysis. Surprisingly, although RutR is bound to some targets in intergenic regions (Figure 1B) most targets are located within genes (Figure 1C).
Table 1.
RutR-binding sites identified by ChIP-chip analysis
| Peak centre | Sequence (5′–3′) | Match to consensus | Position of site with respect to nearest start codon | ORF or intergenic |
|---|---|---|---|---|
| A. DNA/RNA related | ||||
| 3211536 | TTGACTACCTGGTCAA | 13/14 | +804.5 | ORF (rpoD) |
| 3994092 | TTGACTGGCTGGTCAG | 11/14 | +258.5 | ORF (xerC) |
| B. Metabolism | ||||
| 29360 | TTGACCATTTGGTCCA | 13/14 | −284.5 | intergenic (carA)a |
| 139594 | CACACCAGTTGGTCAA | 11/14 | +1667.5 | ORF (gcd) |
| 331599 | No consensus site | N/A | intergenic (yahA) | |
| 535353 | TTAACTGTCTGGTCGG | 9/14 | +312.5 | ORF (hyi)a |
| 1015645 | TTGACCACACGGTCCA | 12/14 | +117.5 | ORF (fabA) |
| 1073323 | TGGACTAAACGGTCAA | 11/14 | −114.5 | intergenic (rutA)a |
| 2718517 | TGGACCAAACAGTCTG | 9/14 | +373.5 | ORF (yfiQ) |
| 3197797 | CCAACCATTCGGTCAA | 10/14 | −373.5 | ORF (ygiF)a |
| 3221219 | TTTACCATCTGGTCAT | 12/14 | +885.5 | ORF (ebgA) |
| 3578836 | ATGACCATGATTTCGT | 8/14 | −0.5 | overlapping (yhhX) |
| C. Function unknown | ||||
| 1272801 | CTGACCAATCGGTCAC | 11/14 | −69.5 | intergenic (ychN) |
| 1724133 | TAGACCGACTGGTCTA | 11/14 | −22.5 | intergenic (ydhM) |
| 1822567 | TTTACCACCTGGTCCG | 11/14 | +273.5 | ORF (ves) |
| D. Drug resistance/sensitivity | ||||
| 2371490 | TTGACCAGCCATTCCA | 10/14 | +9.5 | ORF (pmrD) |
| E. Transport | ||||
| 624085 | ATGACAAATTCGACAA | 10/14 | −45.5 | intergenic (fepB) |
| 2510573 | ATCGCCATCAGGTTGG | 7/14 | +183.5 | ORF (mntH) |
| 4419024 | TTGACCATACGGTAAA | 12/14 | +102.5 | ORF (ulaB) |
| 4603338 | ATGACCATTGCGCCAG | 9/14 | +176.5 | ORF (fhuF) |
aPreviously identified targets.
RutR binding sites are shown in bold and bases that match the consensus RutR binding sequence are underlined.
To pinpoint the precise RutR-binding sequences, we used BioProspector (http://ai.stanford.edu/~xsliu/BioProspector/) to search for short, over-represented DNA sequences in 500-bp segments centred on each peak. This identified the 16-bp sequence motif shown in Figure 2, which matches the previously proposed consensus RutR-binding sequence, 5′-TTGACCAnnTGGTCAA-3′, and predicts that positions 4 and 13 of the binding site are the most important for RutR binding. Following this, we located RutR-binding sites at 19 of the 20 targets identified by our ChIP-chip analysis (no motif was found at the yahA promoter). For the previously identified RutR targets, carA, rutA, hyi and ygiF/glnE (5), the motif that we identified corresponds exactly to the location of the known RutR-binding site (Table 1).
Figure 2.
The RutR-binding site DNA sequence logo. The sequence logo was generated by aligning binding sites identified by ChIP-chip.
In vitro analysis of RutR binding
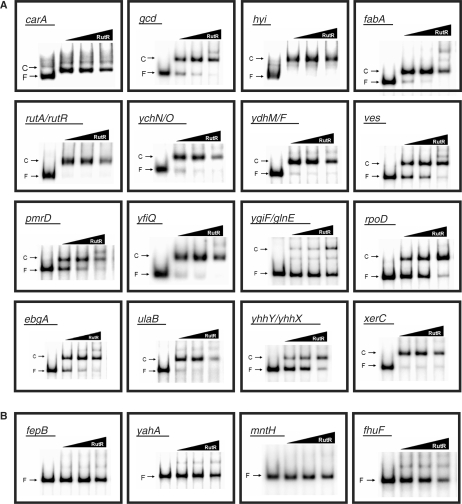
DNA fragments covering each of the 20 RutR targets were amplified, end-labelled and incubated with purified RutR protein in in vitro EMSA assays to measure the binding of RutR to each target (Figure 3). For 16 of the 20 targets, purified RutR clearly retarded the migration of purified DNA fragments (Figure 3A), while for four of the targets (fepB, yahA, mntH and fhuF), addition of purified RutR resulted in little or no retardation, indicating that RutR has a low affinity for these targets (Figure 3B). To understand this, we consulted the RutR-binding motifs shown in Table 1 and the RutR-binding site sequence logo shown in Figure 2. Interestingly, all 16 of the high-affinity RutR-binding sites matched the consensus at both of the strongly conserved positions 4 and 13. In contrast, the low-affinity fepB, mntH and fhuF targets all had non-consensus sequences at either position 4 or 13 and we could find no match to the RutR-binding site at the yahA target. In this latter case, RutR-binding specificity may be more relaxed in vivo or RutR might bind cooperatively with some other factor.
Figure 3.
Binding of RutR to its DNA targets in vitro. The figure shows the results of electrophoretic mobility shift assays in which the binding of RutR (10, 25 or 50 nM) to purified end-labelled PCR products was measured in vitro. Free DNA fragments (F) and RutR–DNA complexes (C) are labelled. High-affinity targets are shown in (A) and low-affinity targets are shown in (B).
Genome-wide distribution of binding sites for RutR and other global DNA-binding proteins
While 5 of the 20 RutR targets identified here fall in intergenic regions, 14 are in coding DNA and 1 target overlaps a translation start site. Many of the 14 targets in coding DNA are hundreds of base-pairs downstream of the nearest start codon. Since this was unexpected, we created a PWM from the 20 RutR targets and used this to identify other potential DNA sites for RutR in the E. coli genome. Of the 95 predicted targets, 84 fell within ORFs. We repeated the analysis, using equally stringent search criteria, for other global DNA-binding proteins (LexA, Fur, FruR, FNR, CRP, NarL and Fis). We found that the majority of predicted sites for these factors were within non-coding promoter DNA (Figure 4).
To determine if this phenomenon is peculiar to E. coli K-12, we used our PWM to search the genomes of other bacteria containing rutR homologues, for putative RutR-binding sites. The results of the analysis, presented in Table 2, show that these genomes have a similar density of RutR-binding sites and that these binding sites are mostly found in coding DNA. We note that the RutR-binding sites in coding DNA are found in a variety of ORFs and that all of the DNA targets for RutR listed in Table 1 would be translated into different amino acid sequences.
Table 2.
Distribution of putative RutR-binding sequences in different bacteria
| Bacterium | Size of genome (megabasepairs) | RutR homologue | No. of putative binding sites | No. sites in coding DNA |
|---|---|---|---|---|
| E. coli K-12 | 4.64 | Yes | 95 | 84 |
| E. coli O157 H7 Sakai | 5.50 | Yes | 142 | 123 |
| Salmonella typhimurium LT2 | 4.86 | Yes | 107 | 96 |
| Shigella flexneri 2A | 4.60 | Yes | 100 | 82 |
| Citrobacter koseri ATCC BAA 859 | 4.72 | Yes | 95 | 85 |
| Enterobacter sp. 638 | 4.52 | Yes | 112 | 97 |
| Klebsiella pneumonia MGH 78578 | 5.32 | Yes | 141 | 125 |
CONCLUSIONS
To date, chromosome-wide DNA-binding profiles have been generated for only a small number of bacterial transcription factors. For E. coli, studies with LexA, CRP, FNR and MelR have shown that binding at high-affinity sites is largely restricted to non-coding DNA (11–15). Similar conclusions have been reached for some of the nucleoid-associated proteins (16,17) and large-scale bioinformatic screens for known transcription factor-binding sites also conclude that targets are mainly found in non-coding DNA (10). We show that this is not the case for the uracil responsive transcription factor RutR; binding sites are mainly located in genes and hundreds of base-pairs away from the nearest start codon. Our results may have implications, for example, bioinformatic screens tend to discard transcription factor-binding targets within genes as artefacts (9,18–21). Clearly, this conclusion is not applicable to RutR.
Although the RutR-binding sites identified here are undoubtedly recognised by the protein, we were able to detect effects of RutR on transcript levels only in the case of the ves gene (Supplementary Figure 1). Hence, it is possible that these sites play little or no role in the regulation of transcription. For example, they may be binding sites that occur by chance and have not been removed by evolution. Recently, Wade and co-workers (11) demonstrated that LexA binds to artificial sites introduced within coding DNA and concluded that, since few such LexA sites exist in the wild type E. coli genome sequence, they must have been removed as the genome evolved. Indeed, high-affinity-binding sites for most bacterial transcription factors are hugely biased towards intergenic DNA (Figure 4). However, by definition, an evolving genome can only ever be considered as a ‘work in progress’. Hence, to explain the sites we report here, we are obliged to suggest that, either many RutR targets have an unassigned function or they are evolutionary relics that are yet to be resolved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Tomoya Baba for providing E. coli strain BW25113 and the rutR derivative JW0998. This work was supported by a Wellcome Trust programme grant awarded to S.J.W.B. Funding to pay the Open Access publication charges for this article was provided by Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Babu MM, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Antonio A, Collado-Vides J. Indentifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Browning DF, Busby SJW. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 4.Loh DK, Gyaneshwar P, Papadimitriou EM, Fong R, Kim K-S, Parales R, Zhou Z, Inwood W, Kustu S. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl Acad. Sci. USA. 2006;103:5144–5199. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 2007;66:744–757. doi: 10.1111/j.1365-2958.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efromovich S, Grainger DC, Bodenmiller D, Spiro S. Genome-wide identification of binding sites for the nitric oxide-sensitive transcriptional regulator NsrR. Meth. Enzymol. 2008;437:209–231. doi: 10.1016/S0076-6879(07)37012-2. [DOI] [PubMed] [Google Scholar]
- 9.Hiard S, Maree R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Rigali S. PREDetector: a new tool to indentify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 2007;357:861–864. doi: 10.1016/j.bbrc.2007.03.180. [DOI] [PubMed] [Google Scholar]
- 10.Robison K, McGuire A, Church GA. Comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- 11.Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl Acad. Sci. USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grainger DC, Aiba H, Hurd D, Browning DF, Busby SJ. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 2007;35:269–278. doi: 10.1093/nar/gkl1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, Constantinidou C, Struhl K, Church G, Busby SJW. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2004;186:6938–6943. doi: 10.1128/JB.186.20.6938-6943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wade JT, Struhl K, Busby SJ, Grainger DC. Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 2007;65:21–26. doi: 10.1111/j.1365-2958.2007.05781.x. [DOI] [PubMed] [Google Scholar]
- 16.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome. Res. 2008 doi: 10.1101/gr.070276.107. [Epub ahead of print, Mar 13.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Rhodius V, Gross C, Siggia ED. Identification of the binding sites of regulatory proteins in bacterial genomes. Proc. Natl Acad. Sci. USA. 2002;99:11772–11777. doi: 10.1073/pnas.112341999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavesi G, Mauri G, Pesole G. In silico representation and discovery of transcription factor binding sites. Brief. Bioinform. 2004;5:217–236. doi: 10.1093/bib/5.3.217. [DOI] [PubMed] [Google Scholar]
- 20.Wei W, Yu X-D. Comparative analysis of regulatory motif discovery tools for transcription factor binding sites. Genomics Proteomics Bioinformatics. 2007;5:131–142. doi: 10.1016/S1672-0229(07)60023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babu MM, Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003;19:75–79. doi: 10.1016/S0168-9525(02)00039-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.