Abstract
Interactions between proteins bound to distant sites along a DNA molecule require bending and twisting deformations in the intervening DNA. In certain systems, the sterically allowed protein–DNA and protein–protein interactions are hypothesized to produce loops with distinct geometries that may also be thermodynamically and biologically distinct. For example, theoretical models of Gal repressor/HU-mediated DNA-looping suggest that the antiparallel DNA loops, A1 and A2, are thermodynamically quite different. They are also biologically different, since in experiments using DNA molecules engineered to form only one of the two loops, the A2 loop failed to repress in vitro transcription. Surprisingly, single molecule measurements show that both loop trajectories form and that they appear to be quite similar energetically and kinetically.
INTRODUCTION
The activity of promoters is often regulated by the interaction between proteins that are simultaneously bound to distant DNA segments to form a loop. Such complexes may be called enhanceosomes (1–3) or repressosomes (1,4–6), depending on their effect on transcription. The loop of DNA might, in principle, follow either a parallel or an antiparallel trajectory (7), and the particular trajectory can be influenced by requirements of the protein–DNA and protein–protein interactions, flexibility of protein–protein interfaces, binding of architectural proteins and length of the intervening DNA. In some systems, the scheme of protein–DNA and protein–protein interactions would allow more than one parallel or antiparallel geometry (7–9).
The Gal repressosome is a ternary nucleoprotein complex that represses transcription of the gal operon in Escherichia coli. Assembly of the Gal repressosome requires direct interaction of GalR dimers bound to two operator sites (OE and OI) separated by 113 bp. This long-range interaction is mediated by the transcriptional cofactor HU and negative DNA supercoiling (10). GalR dimers form a V-shaped, stacked tetramer in the repressosome (6,11,12). Binding of the symmetric GalR dimers to the operators could lead to four different DNA trajectories with respect to the DNA sequence, two of which are parallel (P1 and P2), while two are antiparallel (A1 and A2) (4,7). The relative stacking arrangements of the two operator-bound GalR dimers are different within each trajectory, and elastic energy calculations suggest that the A1 antiparallel GalR/HU-DNA loop (Figure 2a) is more stable than either of the parallel loops or the A2 antiparallel loop (4,8). The major difference between the two antiparallel trajectories results from the stacking of the operator-bound dimers. As a consequence of the 60° angle between the two dimers in the GalR tetramer, the DNA was calculated to be under-twisted (by slightly different amounts) in both antiparallel trajectories (8).
Figure 2.
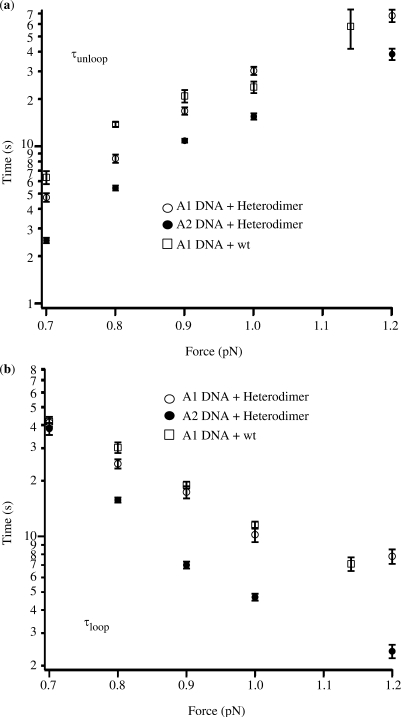
Dependence of the mean lifetime on the externally applied force for the (a) unlooped and (b) looped configuration.
The HU protein stabilizes the GalR-mediated DNA loop by bending the DNA near the apex of the loop (1,10). Both single-molecule manipulations using magnetic tweezers (10) and AFM visualization of DNA loops (13) predicted an antiparallel DNA trajectory in the repressosome, suggesting that HU binding does not assist formation of repressosomes containing a parallel DNA loop. These measurements, however, were unable to distinguish between the two alternative antiparallel configurations. Recently instead, only one (A1) of the two alternative antiparallel loops was found to repress gal transcription (4). Two explanations were proposed for this observation: (i) the A2 loop is thermodynamically unfavored such that it either does not form or forms with such thermodynamics and/or kinetics that it fails to repress transcription, or (ii) the A2 and A1 loops are geometrically/topologically different. Here, we test these hypotheses using magnetic tweezers to detect and characterize loop formation in DNA molecules in which engineered operator sequences and Gal repressor proteins formed either the A1 or A2 antiparallel loop.
MATERIALS AND METHODS
Preparation of DNA
Linear DNA fragments of plasmid pSA580, ∼3.6 kb in length, were used as tethers. The original plasmid had been modified to contain properly arranged hybrid operator sites for oriented binding of the GalRA16T/GalRV15T,T322R heterodimers (6). The arrangement of the hybrid operator sites determined the mutual orientation of the active tetramerization interfaces of the operator-bound GalR heterodimers, allowing only either A1- or A2-type loop formation. The resulting plasmids were linearized by digestion with KpnI and SacI. Two ‘tails’ were synthesized with biotin or digoxigenin-labeled nucleotides by PCR of the multiple cloning site of a pBS plasmid comprising the restriction sites for KpnI and SacI. After restriction of these labeled tails, they were ligated to the complementary ends of the linear fragment of interest.
Stretching and twisting single DNA molecules
All looping experiments were performed in the presence of 25 nM GalR and 50 nM HU. One end of a single molecule of DNA was attached to the glass surface of a microscope flow-chamber (previously coated with antidigoxigenin) and a paramagnetic bead 2.4 μm in diameter (DYNAL MyOne beads coated with streptavidin) was attached to the other end. A pair of permanent magnets, above the microscope stage were used to gently attract the tethered, magnetic bead and effectively stretch the DNA with molecular-scale forces (14). Furthermore, rotation of the magnets causes synchronous rotation of the bead to enable twisting of the DNA tether, which does not swivel at either the glass or bead surfaces due to multiple attachments. The extension, l = <z>, of the molecule of the DNA was monitored with an error of ∼10 nm with 1 s averaging using 3D, video-rate tracking of the bead (15). The horizontal motion of the bead <▵x2> allowed the determination of the tension in the molecule via the equipartition theorem: F = kBT l/<▵x2> with 10% accuracy. Mechanical drift in the data was eliminated using differential tracking of a second bead stuck on the surface.
Loop detection in length versus time data
Data were analyzed as described previously (10). In brief: traces with transitions between longer (unlooped) and shorter (looped) lengths were best fitted to the raw data l(t) (filtered using a 1 s window) using a sliding Heaviside (step) function: lstep(t) = sθ(t−t1) + l1 defined over a time window of size Tav. In other words, for every data point, t, of the data set, the parameters of the step function, s, t1 and l1, were fitted such as to minimize the error (l(t)− lstep(t))2 in the time window t0 < t < t0 + Tav, where only one transition is expected. Finally, the parameters, that consistently scored best (x2-test), were selected as steps. The time intervals between successive looped and unlooped steps were included in histograms of τunlooped (or τlooped) corresponding to the time spent in the longer (or shorter) state.
RESULTS
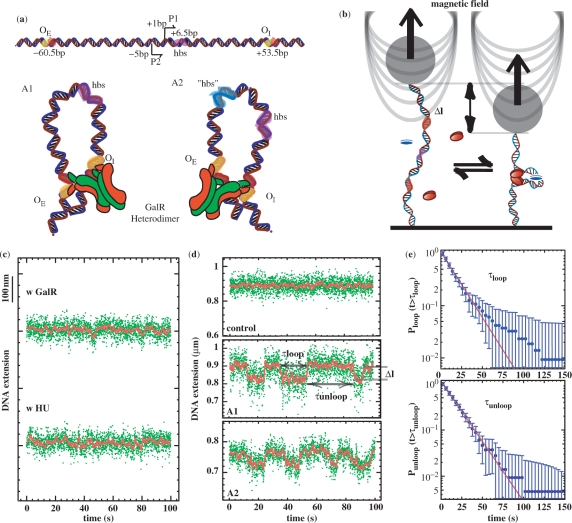
In order to characterize differences and similarities between the two antiparallel loops, A1 and A2 illustrated in Figure 1a, we used previously engineered A1 and A2 DNA molecules (4; see also Materials and methods section). These sequences contain hybrid GalR operators formed of half-sites, which determine the orientation of binding by a GalR hybrid (GalRA16T/GalRV15T,T322R mutant; GalR heterodimer, for brevity). Since this heterodimer also contains only one active surface for tetramerization, the operator-bound GalR heterodimers can only form either A1- or A2-type loops (Figure 1a). This oriented heterodimer loop formation strategy was a modification of the principle of Zhou et al. (16–18). Measurements were then performed using magnetic tweezers to stretch and twist a single DNA molecule between a paramagnetic microsphere and the glass surface of a microscope flow-chamber. In this pendulum-like system, fluctuations of the x or y positions of the microsphere allowed determination of the tension in the DNA (19). In addition, time-resolved records of the position of the microsphere along the tension axis revealed a telegraphic-like signal with alternating looped (short) and unlooped (long) configurations and their lifetimes (Figure 1d). The GalR or HU protein alone did not induce loop formation (Figure 1c).
Figure 1.
Loop formation by the GalR and HU proteins on supercoiled DNA. (a, top) Graphic representation of the gal regulatory region. Top, the two promoters, P1 and P2, are flanked by the gal operators, OE and OI. Using the transcriptional start site of the P1 promoter as a reference for numbering, the HU-Binding Site (hbs) is located downstream of the promoters, at position +6.5 (42) in A1. For the A1 and A2 constructs, functional GalR tetramerization interfaces are marked in green; the inactivated ones are marked red. Arrowheads indicate directions of transcription. (a, bottom) GalR heterodimer-mediated A1 and A2 DNA loops. The major difference between the two trajectories results from the interaction of the operator-bound dimers. As a consequence of the 60° angle between the two dimers in the GalR tetramer, the DNA is thought to be unwound with respect to relaxed DNA in both antiparallel conformations (8). The site for HU binding in the A1 construct is colored pink. The putative site for HU binding “hbs” in the A2 construct is colored blue (see Discussion section). (b) Scheme of the experimental set-up. A single DNA molecule containing the GalR and HU-binding sites is anchored at one end to the glass surface and at the other end to a paramagnetic bead. In response to small magnets placed above the sample, the bead can be used to stretch and twist the DNA. Loop formation by GalR (red ovals) and HU (blue oval) reduces the extension by an amount, Δl. (c) Control experiments performed in the presence of only GalR or only HU. (d, top) Typical signal from an A1 DNA molecule in the absence of proteins. This is indistinguishable from that of an A2 DNA molecule. (d, center and bottom) Typical telegraph-like signal observed for A1 or A2 DNA molecules, respectively, at 0.9 pN in the presence of proteins. The green dots are raw data and the red line is the averaged signal (1 s). In all experiments, molecules were unwound by 3% (σ = −0.03). From the trace, it is possible to measure the transition time (τloop and τunloop) between the looped and unlooped state, as well the loop size. (e) Cumulative probability distribution of τloop and τunloop for all the transitions observed at 0.9 pN in the A1 DNA (error bars are statistical errors). The distributions are fitted by a single exponential giving a mean lifetime: <τloop> = 17.3 ± 1.3 s and <τunloop> = 16.7 ± 0.9 s.
We monitored looping mediated by GalR heterodimer or wt GalR and HU proteins in DNA molecules maintained at a constant negative supercoiling of 3% (σ = −0.03) (10) and constant tension. We repeated these assays in the range of forces between 0.7 and 1.2 pN. Loop formation was undetectable at lower forces due to low signal-to-noise ratios and was prevented by higher tension. Wt GalR can interact with both A1 and A2 DNA with no orientation specificity; in this case, loops with either trajectory can in principle form. The distribution of the dwell times in the looped or the unlooped state was fit with an exponential decay function to determine the average lifetimes, τloop and τunloop, at a particular force (Figure 1d). In all cases, the dependence was exponential, and the lifetimes obtained from measurements carried out on A1 and A2 DNA in the presence of HU and heterodimer or wild-type GalR are reported in Table 1. For each protein/DNA combination, increased tension diminished the loop lifetime and increased the unloop lifetime (Table 1 and Figure 2). This was true both for heterodimeric or wild-type Gal repressor, and lifetimes were similar for loops formed in A1 DNA molecules by heterodimeric or wild-type protein (Figure 2). However, we found that the average lifetime of the unlooped configuration in the presence of heterodimeric Gal repressor was shorter for A2 DNA with respect to that measured for A1 DNA molecules (Figure 2a). In addition, the lifetime of loops formed by heterodimeric Gal repressor in A2 and A1 DNA were commensurate at 0.7 pN of tension, but A2 loops endure half as long as A1 loops with 0.9 or higher tension. Extrapolation from the data in Figure 2b indicates that at tensions below 0.7 pN the A2 loops may last longer than the A1 loops. Unfortunately, the small loop was undetectable at tensions lower than 0.7 pN due to the lower signal-to-noise ratio.
Table 1.
Kinetic and thermodynamic values (mean ± SD) measured and calculated respectively from magnetic tweezers assays
| Force (pN) | Event number | τloop (S) | τunloop (S) | τunloop/τloop | ΔGl,F (kBT) | Δl (nm) |
|---|---|---|---|---|---|---|
| A1 DNA heterodimer | ||||||
| 0.7 | 100 | 41.1 ± 2.9 | 4.7 ± 0.3 | 0.1 ± 0.01 | −2.3 ± 0.1 | 79.9 ± 2.5 |
| 0.8 | 152 | 24.6 ± 1.4 | 8.3 ± 0.5 | 0.3 ± 0.03 | −1.2 ± 0.1 | 78.9 ± 2.3 |
| 0.9 | 215 | 17.3 ± 1.3 | 16.7 ± 0.9 | 1.0 ± 0.1 | 0 ± 0.1 | 75.0 ± 1.8 |
| 1.0 | 196 | 10.2 ± 0.9 | 30.1 ± 1.8 | 3.0 ± 0.3 | 1.1 ± 0.1 | 57.9 ± 1.9 |
| 1.2 | 136 | 7.8 ± 0.7 | 67.5 ± 6.1 | 8.6 ± 1.1 | 2.2 ± 0.1 | 60.1 ± 2.9 |
| A2 DNA heterodimer | ||||||
| 0.7 | 165 | 38.0 ± 2.8 | 2.5 ± 0.1 | 0.1 ± 0.01 | −2.3 ± 0.1 | 72.4 ± 1.3 |
| 0.8 | 152 | 15.7 ± 0.5 | 5.4 ± 0.2 | 0.3 ± 0.02 | −1.2 ± 0.1 | 69.3 ± 1.5 |
| 0.9 | 103 | 7.0 ± 0.3 | 10.8 ± 0.3 | 1.5 ± 0.1 | 0.4 ± 0.1 | 66.6 ± 1.9 |
| 1.0 | 280 | 4.7 ± 0.2 | 15.4 ± 0.7 | 3.3 ± 0.2 | 1.2 ± 0.1 | 66.1 ± 1.8 |
| 1.2 | 100 | 2.4 ± 0.2 | 38.4 ± 3.2 | 16.0 ± 1.9 | 2.8 ± 0.1 | 56.2 ± 2.0 |
| A1 DNA wt | ||||||
| 0.7 | 183 | 41.5 ± 2.5 | 6.3 ± 0.6 | 0.2 ± 0.02 | −1.6 ± 0.1 | 81.5 ± 1.4 |
| 0.8 | 277 | 30.1 ± 2.0 | 13.7 ± 0.5 | 0.5 ± 0.04 | −0.7 ± 0.1 | 82.0 ± 1.4 |
| 0.9 | 208 | 18.9 ± 0.8 | 20.8 ± 1.9 | 1.1 ± 0.1 | 0.1 ± 0.1 | 57.4 ± 0.7 |
| 1.0 | 251 | 11.5 ± 0.5 | 23.7 ± 2.0 | 2.1 ± 0.2 | 0.7 ± 0.2 | 53.8 ± 1.2 |
| 1.1 | 135 | 7.1 ± 0.6 | 57.7 ± 16.2 | 8.1 ± 2.4 | 2.1 ± 0.3 | 51.0 ± 1.5 |
τloop and τunloop are the average lifetimes for the looped and unlooped configurations, respectively, calculated from the dwell time distributions in each case. From the data, it is possible to directly extract the free energy for the looping reaction at a given force, A Gl,F, using the following equation: Gl,F = kBT ln(τunloop/τloop). Δl is the average change in the DNA length associated with looping.
Thermodynamic theory can be used to relate the average lifetime of each DNA configuration to the free-energy change involved in the looping reaction at given forces, ▵Gl,F, according to:
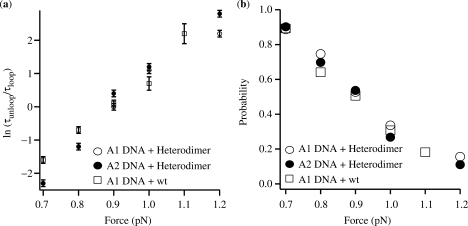
where τl(F) and τu(F) are the average lifetimes for the looped and unlooped configurations at a particular force, ▵Gl,F is the free-energy difference between the looped and unlooped states (column 6 in Table 1), kB is Boltzman's constant and T is the temperature. A plot of ▵Gl,F as a function of force is shown in Figure 3a. The related probability of loop formation Ploop = tl/(tl + tu), which was calculated from the aggregate time spent in the looped state as a fraction of the total observation, was practically indistinguishable across the range of forces employed as shown in Figure 3b.
Figure 3.
(a) Dependence of the free energy for loop formation, ΔGl,F, on the stretching force. (b) Probability of loop formation as a function of force. The probability was calculated as the ratio between the aggregate time spent in the looped configuration and the total observation time.
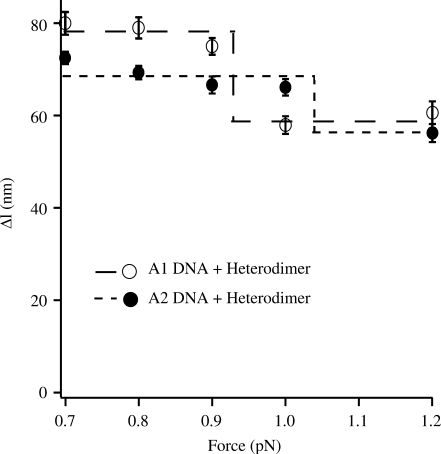
The DNA shortening due to loop formation (▵l, in Figure 1c) was observed to change with the applied force (Figure 4). This force dependence can be explained as follows. At lower forces, the additional DNA unwinding introduced by loop formation generates extra compensatory plectonemes outside the loop (20); as a consequence ▵l is large (14,19,21). At high forces, such plectonemes do not form and instead negatively supercoiled DNA denatures locally. This local, torque-induced melting absorbs any change in twist (22), due to loop formation, via a change in the amount of denaturation, thereby the overall change in extension, ▵l, is closer to the effective loop size.
Figure 4.
Force dependence of the change in DNA length (Δl) associated with heterodimer/HU-mediated loop formation of A1 and A2. The dashed/dotted lines show the amplitude of the total change in Δl observed in each case over the force range investigated.
DISCUSSION
DNA loop formation
Interactions between proteins bound to well-separated sites on a DNA molecule require bending and twisting deformations in the intervening DNA. Double-stranded DNA is a semi-flexible polymer, with a persistence length of ∼50 nm (∼150 bp) (23–25). DNA segments shorter than the persistence length do not easily bend. However, many DNA ‘transactions’ require formation of short DNA loops (150 bp or less).The feasibility of loop formation over distances shorter than its persistence length depends on the intrinsic shape and flexibility of the DNA sequence, the phasing of the binding sites being juxtaposed, supercoiling of the DNA, in concert with the effect of architectural proteins.
In many systems, protein–DNA and protein–protein interactions can produce loops with different geometries. However, not all these geometries are energetically equivalent and loops with a specific DNA trajectory may be preferred (1,8,26–28). For example, in the case of the Gal repressosome, which contains a 113-bp long DNA loop mediated by two Gal repressor dimers and the HU protein, there are four possible DNA trajectories. Two of these trajectories have antiparallel DNA at the entry/exit points (A1 and A2), while two other trajectories have parallel DNA (P1 and P2) (6,8). The DNA exiting an antiparallel loop must curve under tension and therefore formation of this kind of loop would be expected to cause a larger change in the overall end-to-end distance of the molecule (larger ▵l) than formation of a parallel loop where the exiting DNA is straight. However, parallel loops are generally more strained than antiparallel ones (9). Stereochemical models of GalR/HU-DNA loops confirm this and also predict that the A1 loop is much more stable than the A2 loop. In addition, they also predicted that, as a consequence of the 60° angle between the two dimers in the GalR tetramer, the DNA is similarly under-twisted in both conformations (8). These structural predictions might be relevant to in vitro transcription assays, in which the heterodimer and the HU protein repressed transcription from DNA engineered to form A1 but not A2 configurations. Note, however, that these twist calculations were performed on relaxed DNA.
Instead, the magnetic tweezing assays reported here indicate that the A1 and A2 loops formed with similar energies in DNA, which was unwound by an amount similar to that found in the plasmids used in in vitro transcription assays and in vivo (Table 1 and Figure 3a). The two loops had nearly equivalent probabilities of formation, Ploop (Figure 3b), which in all cases studied was about 50% at F ∼0.9 pN (force at which τloop ∼ τunloop). As expected, the change in free energy for loop formation, ▵Gl,F, rose with increasing force (i.e. tension destabilizes the loop in the DNA). Therefore, there is no thermodynamic reason to expect a functional difference in transcriptional repression between A1 and A2 loops. In fact, the only significant difference is that the A2 loop forms and breaks down more frequently than the A1 loop having shorter loop and unloop lifetimes, at least for tensions above 0.7 pN (Figure 2). With the current understanding of repression, it is difficult to relate this observation to RNA polymerase activity.
Of course, in vitro transcription assays utilize supercoiled plasmids that are not under so much tension. However, it would not be rigorous to extrapolate the experimental lifetime data to zero force given the necessarily small range of forces investigated, the distance from the zero point, and the logarithmic scale of the y-axis of Figure 2. In addition, loop lifetimes in the absence of force may not be relevant. Evidence is accumulating in the literature (29) that DNA is under tension in vivo and several motor enzymes, such as RNA polymerase have been reported to exert large forces on the topologically constrained DNA (30). Furthermore, DNA molecules negatively supercoiled by 6% have a built-in entropic tension of about 0.5 pN (31).
Alternative DNA trajectories in transcriptional regulation
Thermodynamic stability plays a key role in the function of DNA loops that regulate transcription. Formation of a transient DNA loop may accompany transcriptional activation when a distant, DNA-bound transcription factor directly interacts with RNA polymerase to close the DNA loop. For transient loops, different DNA trajectories might support similar levels of transcriptional activation (32). Instead, DNA loops that repress transcription are generally quite stable. Repressors involved in DNA loop closure can inhibit RNA polymerase action directly (33,34) [e.g. sterically hindering RNA polymerase binding (35–37) or contacting the promoter-bound RNA polymerase to inhibit transcription initiation (38)] or indirectly by reducing the effective torsional flexibility of DNA (39,40) as in the case of loop formation. Loop formation by repressors may be enhanced by accessory DNA-binding proteins that bend the DNA at a critical segment or contribute stabilizing protein–protein interactions. Especially for the transcriptionally repressive cases, thermodynamic characterization of the macromolecular complex permits quantitative prediction of the probability of DNA loop formation (40,41), which might therefore also predict transcription efficiency.
However, we found that gal DNA loops with A1 and A2 trajectories form with similar energies and probabilities in the range of forces investigated, when DNA is untwisted to the level found in vivo and in in vitro transcriptional assays. Therefore, the failure of the A2 loop to repress transcription cannot be explained on the basis of thermodynamics alone. Previously it has been proposed that, failure of the A2 trajectory to repress in vitro transcription of naturally supercoiled DNA may result from destabilization of the DNA loop by RNA polymerase (6). Calculations performed on relaxed DNA show that, despite the fact that the A1 and A2 loops have similar overall structure, the direction of local DNA bending is different; the DNA surface that is inside the A1 loop apex is turned halfway outside in the A2 loop. One consequence is that the HU-binding site, which is experimentally observed at position +6.5 in the A1 loop, shifts to −14.5, a structurally equivalent position with respect to the loop apex in the A2 loop [Figure 1a and Figure 6 in (4)]. A structural- instead of a sequence-dependent binding site is consistent with the very high nonspecific binding affinity of HU for DNA. Thus, these calculations suggest that in the A2 trajectory, RNA polymerase may easily transition from closed to open complex, facilitating transcription (40), or evict HU from the −13.5/−14.5 site, which overlaps the −10 promoter element. Our data support this idea. In our experimental conditions, DNA may be already unwound by an amount sufficient to abrogate the energetic difference between the A1 and A2 loop and yet maintain the structural difference between A1 and A2 loops found in (4).
Furthermore, we speculate that, given the similarity between the lifetime and ▵l data relative to the interaction between heterodimer and wt GalR and A1, the A1 trajectory is preferred in the wild-type case. This is also to be expected given the similar transcriptional repression by A1 and wt loops but not by A2 loops.
In summary, our data show unequivocally that thermodynamic probabilities of Gal repressor/HU-induced alternate DNA loops failed to quantitatively predict their physiological effect. Therefore, in order to predict transcription modulations due to different DNA loop trajectories, one must carefully consider not only looping probabilities but also how DNA supercoiling affects the double helix topology and how this may impact the interactions of proteins associated with a given trajectory. Single molecule experiments such as those described here are very useful for the characterization of this effect. Furthermore, they emphasize how local tension in the DNA may alter the formation of repressive loops. A better understanding of this and the effect of supercoiling on macromolecular complex formation is emerging from such work.
ACKNOWLEDGEMENTS
We thank Dr Kim Sneppen at the Niels Bohr Institute for helpful discussion. We also thank our colleagues in our laboratories for their assistance. This work was supported by Human Frontier Science Programme Organization (RGP0050/2002-C to L.F. and S.A.); Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (1Z01BC010017 to S.A.); János Bolyai fellowship of the Hungarian Academy of Sciences (to S.S.). Funding to pay the Open Access publication charges for this article was provided by LF's Emory startup.
Conflict of interest statement. None declared.
REFERENCES
- 1.Courey AJ, Jia ST. Transcriptional repression: the long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- 2.Kim TK, Kim TH, Maniatis T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-beta enhanceosome in vitro. Proc. Natl Acad. Sci. USA. 1998;95:12191–12196. doi: 10.1073/pnas.95.21.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. doi: 10.1101/sqb.1998.63.609. (1998) Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol., 63, 609–620. [DOI] [PubMed] [Google Scholar]
- 4.Semsey S, Tolstorukov MY, Virnik K, Zhurkin VB, Adhya S. DNA trajectory in the Ga1 repressosome. Genes Dev. 2004;18:1898–1907. doi: 10.1101/gad.1209404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 6.Geanacopoulos M, Vasmatzis G, Lewis DEA, Roy S, Lee B, Adhya S. GalR mutants defective in repressosome formation. Genes Dev. 1999;13:1251–1262. doi: 10.1101/gad.13.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semsey S, Virnik K, Adhya S. A gamut of loops: meandering DNA. Trends Biochem. Sci. 2005;30:334–341. doi: 10.1016/j.tibs.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Geanacopoulos M, Vasmatzis G, Zhurkin VB, Adhya S. Gal repressosome contains an antiparallel DNA loop. Nat. Struct. Biol. 2001;8:432–436. doi: 10.1038/87595. [DOI] [PubMed] [Google Scholar]
- 9.Mehta RA, Kahn JD. Designed hyperstable lac repressor center dot DNA loop topologies suggest alternative loop geometries. J. Mol. Biol. 1999;294:67–77. doi: 10.1006/jmbi.1999.3244. [DOI] [PubMed] [Google Scholar]
- 10.Lia G, Bensimon D, Croquette V, Allemand JF, Dunlap D, Lewis DEA, Adhya SC, Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geanacopoulos M, Adhya S. Genetic analysis of GalR tetramerization in DNA looping during repressosome assembly. J. Biol. Chem. 2002;277:33148–33152. doi: 10.1074/jbc.M202445200. [DOI] [PubMed] [Google Scholar]
- 12.Semsey S, Geanacopoulos M, Lewis DEA, Adhya S. Operator-bound GalR dimers close DNA loops by direct interaction: tetramerization and inducer binding. EMBO J. 2002;21:4349–4356. doi: 10.1093/emboj/cdf431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virnik K, Lyubchenko YL, Karymov MA, Dahlgren P, Tolstorukov MY, Semsey S, Zhurkin VB, Adhya S. ‘Antiparallel’ DNA loop in gal repressosome visualized by atomic force microscopy. J. Mol. Biol. 2003;334:53–63. doi: 10.1016/j.jmb.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 14.Strick TR, Allemand JF, Bensimon D, Croquette V. Stress-induced structural transitions in DNA and proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:523–543. doi: 10.1146/annurev.biophys.29.1.523. [DOI] [PubMed] [Google Scholar]
- 15.Strick TR, Charvin G, Dekker NH, Allemand J-F, Bensimon D, Croquette V. Tracking enzymatic steps of DNA topoisomerases using single-molecule micromanipulation. C. R. Physique. 2002;3:595–618. [Google Scholar]
- 16.Meibom KL, Kallipolitis BH, Ebright RH, Valentin-Hansen P. Identification of the subunit of cAMP receptor protein (CRP) that functionally interacts with CytR in CRP-CytR-mediated transcriptional repression. J. Biol. Chem. 2000;275:11951–11956. doi: 10.1074/jbc.275.16.11951. [DOI] [PubMed] [Google Scholar]
- 17.Zhou YH, Busby S, Ebright RH. Identification of the functional subunit of a dimeric transcription activator protein by use of oriented heterodimers. Cell. 1993;73:375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YH, Pendergrast PS, Bell A, Williams R, Busby S, Ebright RH. The functional subunit of a dimeric transcription activator protein depends on promoter architecture. EMBO J. 1994;13:4549–4557. doi: 10.1002/j.1460-2075.1994.tb06776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 20.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol. Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strick TR, Croquette V, Bensimon D. Homologous pairing in stretched supercoiled DNA. Proc. Natl Acad. Sci. USA. 1998;95:10579–10583. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagerman PJ. Investigation of the flexibility of DNA using transient electric birefringence. Biopolymers. 1981;20:1503–1535. doi: 10.1002/bip.1981.360200710. [DOI] [PubMed] [Google Scholar]
- 24.Shore D, Langowski J, Baldwin RL. DNA flexibility studied by covalent closure of short fragments into circles. Proc. Natl Acad. Sci. USA Biol. Sci. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA-molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Landy A. Lambda-Int protein bridges between higher-order complexes at 2 distant chromosomal loci Attl And Attr. Science. 1992;256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavoie BD, Shaw GS, Millner A, Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 28.Nunesduby SE, Smithmungo LI, Landy A. Single base-pair precision and structural rigidity in a small Ihf-induced DNA loop. J. Mol. Biol. 1995;253:228–242. doi: 10.1006/jmbi.1995.0548. [DOI] [PubMed] [Google Scholar]
- 29.Blumberg S, Pennington MW, Meiners JC. Do femto Newton forces affect genetic function? A review. J. Biol. Phys. 2006;32:73–95. doi: 10.1007/s10867-005-9002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 31.Charvin G, Allemand JF, Strick TR, Bensimon D, Croquette V. Twisting DNA: single molecule studies. Contemp. Phys. 2004;45:383–403. [Google Scholar]
- 32.Lilja AE, Jenssen JR, Kahn JD. Geometric and dynamic requirements for DNA looping, wrapping and unwrapping in the activation of E. coli glnAp2 transcription by NtrC. J. Mol. Biol. 2004;342:467–478. doi: 10.1016/j.jmb.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 33.Friedman AM, Fischmann TO, Steitz TA. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 34.Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob F, Monod J. Genetic regulatory mechanisms in synthesis of proteins. J. Mol. Biol. 1961;3:318–326. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 36.Johnson A, Meyer BJ, Ptashne M. Mechanism of action of Cro-protein of bacteriophage-lambda. Proc. Natl Acad. Sci. USA. 1978;75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlax PJ, Capp MW, Record MT. Inhibition of transcription initiation by lac repressor. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 38.Roy S, Semsey S, Liu MF, Gussin GN, Adhya S. GaIR represses gaIP1 by inhibiting the rate-determining open complex formation through RNA polymerase contact: a GaIR negative control mutant. J. Mol. Biol. 2004;344:609–618. doi: 10.1016/j.jmb.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 39.Choy HE, Adhya S. Control of Gal transcription through DNA looping - inhibition of the initial transcribing complex. Proc. Natl Acad. Sci. USA. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choy HE, Park SW, Parrack P, Adhya S. Transcription regulation by inflexibility of promoter DNA in a looped complex. Proc. Natl Acad. Sci. USA. 1995;92:7327–7331. doi: 10.1073/pnas.92.16.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilar JMG, Saiz L. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 2005;15:136–144. doi: 10.1016/j.gde.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]