Abstract
Quantification of a plasmid DNA (pDNA) and investigation of its polymer-associated state in the nucleus are crucial to evaluate the effectiveness of a gene-delivery system. This study was conducted with p3NF-luc-3NF, a pDNA-bearing optimized κB motif to favour NFκB-driven nuclear import. Here, a quantification of pDNA copies in the nucleus was performed by real-time confocal laser scanning microscopy in HeLa and C2C12 cells transfected with linear polyethylenimine or histidylated polylysine. Förster Resonance Energy Transfer (FRET) from the fluorescein-p3NF-luc-3NF donor to the co-localized rhodamine-polymer acceptor was carried out to investigate whether the pDNA was still condensed with the polymer in the nucleus. Upon 5 h of transfection, the nuclear amount of p3NF-luc3NF was ∼1500 copies in both cell lines whereas that of pTAL-luc, a 3NF-free counterpart pDNA, was less than 250. This quantity of p3NF-luc-3NF dropped dramatically to that of pTAL-luc in the presence of the BAY 11-7085, an inhibitor of NFκB activation. These data strongly support a nuclear import of p3NF-luc3NF mediated by NFκB. Moreover, FRET experiments clearly revealed that most of nuclear pDNA were still condensed with the polymer raising the question of their passage through the nuclear pore complex and their impact on the gene-expression efficiency.
INTRODUCTION
Plasmid DNA (pDNA)/cationic polymer complexes (so-called polyplexes) are attractive non-viral gene delivery systems for gene-therapy applications (1,2). Although the fundamental mechanisms of transfection by polyplexes are still not completely understood, it is assumed that the delivery of plasmid DNA (pDNA) in the nucleus of the targeted cells for gene expression requires polyplex capture, internalization, pDNA endosome escape into the cytosol and pDNA nuclear import. Considerable efforts have been made to overcome these important intracellular barriers. For instance, polyethylenimines (3) and histidylated polymers (4) contain primary amines that allow pDNA condensation, protecting it against nuclease digestion and enhancing its cellular uptake. They also possess protonable secondary amines or imidazole groups that induce membrane destabilization in an acidic medium favouring pDNA endosome escape and delivery in the cytosol (5). A nuclear localization signal (NLS) peptide such as that of the SV40 large T antigen has also been used to help the pDNA import into the nucleus of non-dividing cells (6). This peptide has been directly coupled with polymers or at the end of a linear plasmid. It has also been associated with pDNA either via a specific peptide nucleic acid or a padlock oligonucleotide. Unfortunately, the gain obtained with the addition of this device in terms of gene expression and the number of transfected cells has not been as high as expected.
Recently, the capacity of the NFκB transcription factor to shuttle between the cytoplasm and the nucleus under specific conditions has been exploited to increase pDNA nuclear import (7–9). In the cytoplasm, NFκB interacts with the IκB subunit masking the NLS moiety of NFκB, thus preventing its translocation into the nucleus (10). Upon stimulation by, e.g. TNFα, a cascade of events leads to IκB phosphorylation and its ubiquitinylation results in a rapid degradation. Owing to the NLS now exposed, NFκB rapidly penetrates into the nucleus where it binds to specific DNA elements (κB sites of 10 bp) in the promoter/enhancer regions with a high affinity (kD = 10−10–10−13 M) (11). Gene expression and fluorescence microscopy measurements have revealed that pDNA bearing a 50 bp fragment made of five κB motifs inserted upstream SV40 or CMV promoters driving the expression of the luciferase gene increased both the luciferase activity and the amount of fluorescent plasmids inside the nucleus of various TNFα-treated cell lines transfected with polyplexes (8). The nuclear accumulation of κB-pDNA mediated by NFκB has been demonstrated by confocal microscopy imaging and pDNA microinjection. The nuclear-associated fluorescence intensity inside the nucleus of HeLa cells upon cytoplasm microinjection of κB-pDNA/p50 suggested that 12% of pDNA were inside the nucleus within 8 h versus 2% for κB-free pDNA or κB-pDNA associated with a NLS-deleted p50 (9). In our study, we designed a specific sequence termed 3NF comprising three 10 bp κB sites separated by a 5 bp spacer optimizing NFκB binding. We have found that plasmids encoding the luciferase gene and containing 3NF sequences (3NF plasmids) bound to NFκB inside the cells and were more efficient in terms of gene expression than the ones mentioned above (Gonçalves et al., submitted). Fluorescent in situ hybridization (FISH) experiments have shown that 3NF-pDNA was imported into the nucleus more than 3NF-free pDNA. Nevertheless, the lack of an adequate method to determine the number of pDNA copies in the nuclear compartment limits the evaluation of formulations containing κB-pDNA in terms of nuclear uptake. Indeed, the determination of nuclear plasmid copies is usually done by real-time PCR or by flow cytometry on cell nuclei purified upon subcellular fractionation. It is hard to prevent artefacts due for instance to the absorption of cytosolic pDNA on the nuclear envelope upon cell homogenization (12,13).
The aim of the present study was to determine the number of pDNA copies in the nucleus of polyplex-transfected cells and to investigate whether nuclear pDNA are free of polymers or still condensed with polymers. We used real-time confocal laser scanning microscopy (CLSM) because this method offers a three-dimensional (3D) analysis on sequential z-series images allowing true quantitative determination of the copy number of a fluorescent plasmid in the nucleus (14). A comparative study was conducted in HeLa and C2C12 cells transfected with three kinds of plasmids encoding the luciferase (luc) gene: p3NF-luc-3NF (a 3NF-pDNA), pTAL-luc (a 3NF-free pDNA) or pCMV-luc. We compared the nuclear accumulation upon transfection with the pDNA complexed with linear polyethylenimine (LPEI) and histidylated polylysine (HIS). Furthermore, it is still unclear whether pDNA is or is not packaged with the polymers in the nucleus. Förster Resonance Energy Transfer (FRET) was used to measure plasmid/polymer interactions in the living cells and to address whether plasmids accumulated in the nucleus were still condensed with the polymers. This fluorescence method deals with a transfer from an energy donor (i.e. fluorescein bound to pDNA) towards an energy acceptor (i.e. rhodamine bound to polymer) only if the distance between fluorophores is close (10–100 Å). Overall, our data indicate that for both cell lines, p3NF-luc-3NF was rapidly and much more accumulated in the nucleus (∼1500) than pTAL-luc and pCMV-luc (<500) upon 5 h post-transfection. Moreover, pDNA in the nucleus was mostly condensed with the polymers.
MATERIALS AND METHODS
Reagents unless otherwise stated were purchased from Sigma (St Quentin Fallavier, France).
Cell culture
Human epithelial ovarian carcinoma (HeLa; CCL2, ATCC, Rockville MD, USA) and mouse skeletal muscle (C2C12; CRL1772, ATCC) cell lines were routinely grown at 37°C in a humidified atmosphere of 5% CO2. C2C12 and HeLa cells were maintained by regular passage in Dulbecco's modified Eagle's medium (DMEM) and minimum essential medium (MEM; Gibco, Invitrogen SARL, Cergy Pontoise, France), respectively. These media were supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine and 100 U/ml penicillin and 50 U/ml streptomycin each (Gibco). The HeLa cell medium was supplemented with 1% of a 100× non-essential amino acid solution (Gibco). Cells were free from mycoplasma as evidenced by bis-benzimidazole staining (15).
Plasmids
pCMV-luc (pTG11033, Trangène S.A., Strasbourg, France) and pTAL-luc (Clontech; Takara Bio Europe S.A.S, Saint-Germain-en-Laye, France) were plasmid DNA encoding the firefly luciferase (luc) gene under the control of the strong human cytomegalovirus (CMV) promoter and a weak promoter, i.e. the TATA-like promoter region (TAL) from Herpes simplex virus thymidine kinase (HSV-TK), respectively. p3NF-luc-3NF was a plasmid DNA with NFκB-binding sequence encoding the firefly luciferase gene under the control of the TAL from HSV-TK (Gonçalves et al., submitted). Plasmids DNA were prepared with the Plasmid Mega Kit (QIAGEN, Courtaboeuf, France).
p3NF-luc3NF construction
Two 3NF sequences containing three repeats of the κB 5′-GGGACTTTCC-3′ site were inserted in pTAL-luc, one placed upstream the promoter region and the other downstream the luciferase gene. Plasmids DNA were prepared with the Plasmid Mega Kit (Qiagen, Courtaboeuf, France). The 5′-CTGGGGACTTTCCAGCTGGGGACTTTCCAGCTGGGGACTTTCCAGG-3′ double strand (3NF fragment) was synthesized by Eurogentec S.A. (Seraing, Belgium). After digestion by NheI and BgLII restriction enzymes (MBI-Fermentas–Euromedex, Souffelweyersheim, France), the 3NF fragment was cloned into the same restriction sites of the pTAL-Luc cloning vector using the T4-DNA ligase (MBI-Fermentas-Euromedex). The ligation mixture was transformed into the TOP10 Escherichia Coli strain that has a tetracycline resistance. The recombinant colonies were selected on agar-LB medium containing ampicillin and tetracycline antibiotics. The recombinant plasmids were analysed by PCR on colonies using the following two specific primers purchased from Eurogentec: NFUP (5′-GCTGTCCCCAGTGCAAGTGC-3′) and NFLP (5′-TGCTCTCCAGCGGTTCCATC-3′) corresponding to 4910-4929 and 299-280 regions of pTAL-Luc vector, respectively. The PCR products were amplified using PCR-Ready-to-go beads (Amersham-Pharmacia BioTech, Saclay, France) with a Techne Genius-Avantec thermocycler. PCR conditions for amplifications were 45 s at 94°C, 30 s at 57°C, 50 s at 72°C for 33 cycles, preceded by 2.15 min at 94°C, and followed by 5 min at 72°C. The recombinant plasmid (p3NF-luc) was analysed upon extraction by digestion with various restriction enzymes. A second 3NF fragment was inserted into p3NF-luc downstream the luciferase gene. The presence of the insert was determined by PCR amplification using 3NFU (5′-GGCCGCTTCGAGCAGACATG-3′) and 3NFL (5′-CTGCCGGCACCTGTCCTACG-3′) primers (Eurogentec) hybridizing with 1898-1917 and 2273-2254 regions of p3NF-luc, respectively.
Fluorescent pDNA labelling
Each plasmid DNA was labelled with the Label IT nucleic acid labelling kit (MIRUS, Madison, WI, USA) at 1:2 reagent/pDNA weight ratio according to manufacturer's instructions. p3NF-luc-3NF, pTAL-luc and pCMV-luc were labelled either with fluorescein (fluo-p3NF-luc-3NF, fluo-pTAL-luc, fluo-pCMV-luc) or cyanine 3 (Cy3-p3NF-luc-3NF, Cy3-pCMV-luc). The labelled DNA was purified by ethanol precipitation. The labelling densities determined by absorbance according to the manufacturer's protocol were 1 fluorescein/90 bp and 1 cyanin 3/80 bp. We have verified that the gene expression was not affected by this type of plasmid labelling.
Cationic polymers
HIS (MW = 70.4 kDa) and LPEI (MW = 22 kDa) were prepared as previously described (16,17). Polymers (0.4 µmol) were labelled in 1 ml DMSO for 12 h at 20°C with 4 µmol TAMRA-5-SE (Molecular Probes, Invitrogen, France) in the presence of DIEA (100 µmol). LPEI and HIS were purified by precipitation in 20 volumes of ethyl acetate/HCl (10/0.05; v:v) and 20 volumes of 2-propanol, respectively. The precipitate was solubilized in distilled water and freeze-dried. The fluorescent polymers were free of unbound fluorophore as evidenced by thin layer chromatography in chloroform/methanol/water (6/6/1; v:v:v).
Transfection and luciferase gene expression measurements
Two days prior to transfection, cells were seeded at 2 × 105 cells per well in 1 ml of culture medium in a 24-well plate. Cells were washed 3 times in serum-free medium before transfection. LPEI (5 µg) or HIS (7.5 µg) in 5 µl or 7.5 µl of 10 mM Hepes pH 7.4 was added to 2.5 µg DNA in 37.5 µl of 10 mM Hepes and vortexed for 4 s. The mixtures were left for 30 min at 20°C and made up to 500 µl of serum-supplemented medium before being incubated with cells for 4 h at 37°C in a humidified atmosphere of 5% CO2. HeLa and C2C12 cells were transfected either in the absence or in the presence of 25 ng/ml of human TNFα (hTNFα) or murine TNFα (mTNFα), respectively. After 4 h transfection, the medium containing plasmids was removed, replaced with fresh medium and cells were incubated in the absence of TNFα. Luciferase gene expression was measured by monitoring its luminescence activity as previously described (17).
Uptake of polyplexes and flow cytometry
Two days prior to transfection, cells were seeded as above. At 80% confluent, cells were washed 3 times in serum-free medium and incubated for 5 h at 37°C in serum-supplemented medium with fluo-pDNA/polymer complexes. Cells were then washed twice with ice-cold PBS (pH 7.4), harvested by trypsin, centrifuged (1500 r.p.m. for 5 min at 4°C) and suspended in PBS. The cell-associated fluorescence was analysed by flow cytometry (FACSort, Becton-Dickinson, Grenoble, France) before and after a post-incubation for 30 min at 4°C in the presence of 50 µM monensin (18). The monensin treatment restores the fluorescein fluorescence partially quenched in acidic vesicles (endosomes and lysosomes). The cell-associated fluorescence was recorded at 520 nm after excitation at 488 nm and was expressed as the mean value of the fluorescence intensity of 104 cells. The cell-associated fluorescence was expressed relative to that of cells transfected with fluo-p3NF-luc-3NF/LPEI polyplexes to take into account fluorescence intensity variations of each fluo-pDNA/polymer complex.
CLSM
CLSM experiments were performed using a Zeiss Axiovert 200 M microscope coupled with a Zeiss LSM 510 scanning device (Carl Zeiss Co. Ltd., Iena, Germany). The inverted microscope was equipped with a Plan-Apochromat 63× objective (NA = 1.4) and with a temperature-controlled stage. In each experiment, the cell nuclei were labelled with 1 µM Draq5, a far-red fluorescent DNA dye (Biostatus Limited, Shepshed, UK). Images were recorded with Carl Zeiss's LSM Image Browser software and were calculated with the public-domain ImageJ software (19). Each image was represented with 512 × 512 pixels measuring 0.28 × 0.28 µm2 each, and recorded with a line mode to reduce background noise (average on two scanning images). Image acquisition was performed with the CLSM's meta mode selecting specific domains of the emission spectrum, i.e. Draq5 was excited at 633 nm with a He/Ne laser and its fluorescence emission was collected between 670 and 745 nm.
Plasmid quantitation in cell nucleus
Two days prior to the experiment, 104 C2C12 and HeLa cells were seeded in a 4-well Lab-Tek chambered coverglass (Nunc, Dutsher S.A., Brumath, France). Fluo-pTAL-luc (0.5 µg) and Cy3-p3NF-luc-3NF (0.5 µg) in 70 µl of 10 mM Hepes (pH 7.4) were separately mixed with 1 µg unlabelled LPEI in 10 mM Hepes (pH 7.4). After 30 min at 20°C, fluo-pTAL-luc/LPEI polyplexes were mixed with Cy3-p3NF-luc-3NF/LPEI polyplexes and the solution adjusted to 500 µl with a CO2-independent medium (Gibco) supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U/ml penicillin and 50 U/ml streptomycin. Then, HeLa and C2C12 cells were incubated with the polyplex mixture in the absence or in the presence of 25 ng/ml of hTNFα or mTNFα, respectively. Similar experiments were also carried out with the mixture of 0.5 µg fluo-pTAL-luc/1 µg LPEI and 0.5 µg Cy3-pCMV-luc/1 µg LPEI polyplexes. The same protocol was carried out with 0.5 µg of plasmid complexed with 1.5 µg HIS. For NFκB inhibition, HeLa cells were pre-treated with 1 µM BAY 11-7085 (Calbiochem – Merks, VWR, Strasbourg, France) for 1 h at 37°C in medium supplemented with 10% FBS, prior to incubation with 25 ng/ml hTNFα and 1 µM BAY 11-7085 at 37°C in the presence of fluo-pTAL-luc/LPEI and Cy3-p3NF-luc-3NF/LPEI polyplexes as described before. The cells were then observed under confocal microscopy in real-time after 1 h, 3 h and 5 h incubation. As a control, cells were incubated for 5 h at 37°C with a mixture of fluo-pTAL-luc polyplexes and Cy3-pTAL-luc polyplexes. Fluorescein (fluo) and cyanine 3 (Cy3) fluorophores were excited with Ar and He/Ne laser lines at 488 and 543 nm, respectively. Each fluorescence emission was collected with the CLSM's meta mode in two distinct channels limited to 500–542 and 560–605 nm, respectively.
Up to 20 optical slices of 0.6 µm in depth each were recorded for every sample. Each 12-bit TIFF image was transferred to ImageJ to visualize every z-series of images in x–z side view with the Volume Viewer plug-in (20). The total number of nuclear spots was determined for 12 nuclei and converted into the number of pDNA copies per nucleus assuming that one spot contained 18 pDNA copies on average. This value took into account the average number of plasmid per polyplex molecule and the average number of polyplexes per fluorescent spot. By fluorescence correlation spectroscopy (21) and transmission electron microscopy (22), the number of plasmids per PEI or HIS polyplexes is 3.5 ± 1 and 4, respectively. Then, one can assume that four plasmids are complexed per polyplex on the bases of these previous results. Moreover, the number of polyplexes per fluorescent spot was determined from the total number of 3D pixels (voxel) of spots (35.3 ± 14.9 voxels) and the voxel number of a polyplex (8 voxels). This latter number was determined from fluorescence of polyplexes observed at the bottom of a cell-free Lab-Tek chambered coverglass poured with the polyplexes in culture medium supplemented with 10% heat-inactivated FBS. Only voxels having at least half of the fluorescence intensity of the brighter voxels in a given spot were considered.
pDNA condensation and FRET
One microgram fluo-p3NF-luc-3NF was complexed with either 2 µg Rho-LPEI or 3 µg Rho-HIS in 10 mM Hepes (pH 7.4) as described above. HeLa and C2C12 cells were incubated for 5 h at 37°C with 500 µl of polyplexes in CO2-independent medium supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U/ml penicillin and 50 U/ml streptomycin containing either in the presence of 25 ng/ml of hTNFα or mTNFα. Cells were washed in a polyplex-free medium and observed by confocal microscopy. Sensitized emission FRET experiments were performed with a 488 nm Argon laser excitation wavelength for fluo and rho, and fluorescence emissions were collected with the CLSM's multitracking mode limited to 500–530 nm for fluo, 580–628 nm for rho and 542–570 nm for FRET. Optical section thickness was 0.8 μm. For the acceptor photobleaching experiment, fluo, rho and Draq5 fluorophores were excited at 488, 543 and 633 nm in a multitracking mode, respectively. Bleaching of rho was performed at 543 nm with 800 pulses. One set of images was recorded every 5 s during 10 acquisitions.
FRET calculations
Spectral overlap and Förster distance between donor and acceptor
The J overlap integral (in M−1 cm3) expressing the degree of spectral overlap between the fluo emission and the rho absorption can be written in the alternative form on the wavelength (λ) scale (between 450 and 650 nm):
| 1 |
Where FD corresponds to the total fluorescence intensity normalized to unity of the fluo donor and εA is the extinction coefficient of the rho acceptor. The distance for which the energy transfer efficiency is equal to 50%, is called the Förster distance (R0) as:
| 2 |
Where φD is the quantum yield of fluo coupled with plasmid DNA (φD = 0.27), n is the refractive index of the medium (n = 1.33 in distillated water). κ2 is the factor describing the relative orientation in space of the transition dipoles of the donor and acceptor. A κ2-value of 2/3 was generally assumed. In our system, the J overlap integral was 3.64 × 10−13 M−1 cm3 (spectra not shown). Thus, R0 allowing an energy transfer efficiency of 50% was 51.3 Å. These values were in agreement with those previously reported by Wu and Brand (23) and indicated that an energy transfer between fluo and rho could occur in our cellular system.
Sensitized emission
We applied three methods to correct FRET images from changes in donor and acceptor emission. The calculations were performed from the variation in pixel response with the PixFRET plug-in of ImageJ in each region of interest (ROI) (24).
- The method according to Youvan et al. (25) determines only a FRET intensity image (Fc) corrected by the cross-talk and background intensity, but is not normalized for the concentration of the donor and acceptor:
IFRET, ID, and IA were intensities (subtracted from their background intensity) in ROIs under the distinct FRET, fluo and rho channels, respectively. A and B were the percentage of fluo and rho bleed-through in the FRET channel, respectively. They were determined by quantifying the relative intensity ratio in the FRET, fluo and rho channels of image analysis from cells transfected with fluo-pDNA/unlabelled polymer and unlabelled pDNA/rho-polymer complexes, respectively. No bleed-through signals from fluo in the rho channel and vice versa were observed.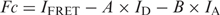
With Gordon et al.'s correction (26), Fc is normalized to the direct donor and acceptor signal according to the nF equation: nF = Fc/(ID × IA).
According to Xia and Liu (27), Fc is divided by the square root of the donor and acceptor product (
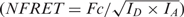 ) and should be constant and independent of the local concentration of donor and acceptor.
) and should be constant and independent of the local concentration of donor and acceptor.
Acceptor photobleaching
Alternatively, the absolute FRET efficiency (%F) between the donor (fluo) and the acceptor (rho) was obtained by measuring the donor emission variation before [(ID)Pre-bleaching] and after [(ID)Post-bleaching] selective photobleaching of the acceptor according to Siegel et al. (28):
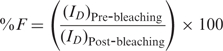 |
The FRET efficiency (%E = 1−%F) was also calculated from fluorescence recovery after the photobleaching kinetic of the donor emission in the presence of the acceptor in ROIs. This procedure used each cell or sub-region as its own internal standard. In this way, values as low as a few percent could be quantified with reasonable confidence.
RESULTS
Influence of 3NF sequences on gene expression
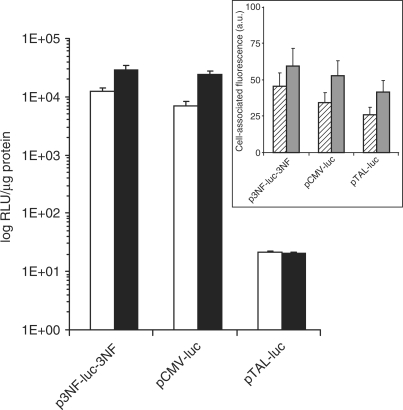
Two 3NF sequences composed of three 10 bp κB sites separated by a 5 bp spacer maximizing the binding of NFκB proteins were put both upstream the TATA-like promoter region from the HSV-TK promoter and downstream the luciferase gene. The luciferase gene expression of p3NF-luc-3NF upon transfection of HeLa cells with LPEI was compared to that obtained in cells transfected with either pCMV-luc or κB-deleted pDNA (pTAL-luc) (Figure 1). In the absence of TNFα-stimulation, the luciferase gene expression with p3NF-luc-3NF was 1600-fold higher than with pTAL-luc and not significantly different than with pCMV-luc in which the luciferase gene expression was driven under the strong CMV promoter. Upon TNFα stimulation to maximize NFκB availability, the luciferase activity with p3NF-luc-3NF was 8-fold higher than in the absence of stimulation whereas it did not change with pTAL-luc. Interestingly, gene expression with p3NF-luc-3NF reached a similar value as with pCMV-luc (29 400 versus 23 900 RLU/µg of protein, respectively). TNFα stimulation also improved CMV-luc expression. This was likely due to a transcriptional activation of the luciferase gene by NFκB. Indeed, three NFκB-binding consensus sequences separated by 142 and 152 bp elements are present in the CMV promoter. Similar results were obtained upon transfection with HIS (data not shown). This result underlined the influence of the 3NF sequences on the luciferase gene expression.
Figure 1.
Influence of 3NF sequences on the luciferase gene expression. HeLa cells were transfected with LPEI complexed with the indicated plasmids in the absence (open bar) or the presence (solid bar) of 25 ng/ml TNFα. The luciferase activity expressed as relative light units per microgram protein (RLU/µg of protein) was measured upon 24 h of culture. The values shown are averages of three independent experiments. Insert: polyplex uptake. HeLa cells were incubated at 37°C for 5 h in the presence of 2.5 µg of the indicated fluorescein-labelled plasmids complexed either with LPEI (hatched bar) or HIS (grey bar). Cells were then harvested with trypsin and their fluorescence intensity was measured by flow cytometry after a post-treatment with monensin. The cell-associated fluorescence (MFI) in arbitrary units was expressed relative to that of cells incubated with fluo-p3NF-luc-3NF/LPEI polyplexes.
The uptake by HeLa cells of fluorescein-labelled pDNA complexed with LPEI or HIS revealed that the cell fluorescence associated with p3NF-luc-3NF and pCMV-luc were quite comparable after 5 h incubation whatever the polymer used (Figure 1 insert). Compared to pCMV-luc, the remarkable effect on the luciferase gene observed in Figure 1 with p3NF-luc-3NF may be likely due to a larger number of pDNA copies imported into the nucleus. This could imply that the weakness of the TATA-like promoter in p3NF-luc-3NF was counterbalanced by the higher number of pDNA copies in the nucleus. The cell fluorescence associated with pTAL-luc was a third (with LPEI) to a fifth (with HIS) less than that with p3NF-luc-3NF, suggesting that pTAL-luc polyplexes were slightly less taken up by the cells. Nevertheless, this uptake difference cannot explain the 1300-fold higher luciferase activity observed with p3NF-luc-3NF compared to pTAL-luc (Figure 1). This could be consecutive to an enhancement of the 3NF-plasmids penetration in the nucleus. Similar results were obtained with C2C12 cells except that the cell-associated fluorescence was 2- to 3-fold higher than with HeLa cells (data not shown).
FRET imaging of polyplexes inside the cells
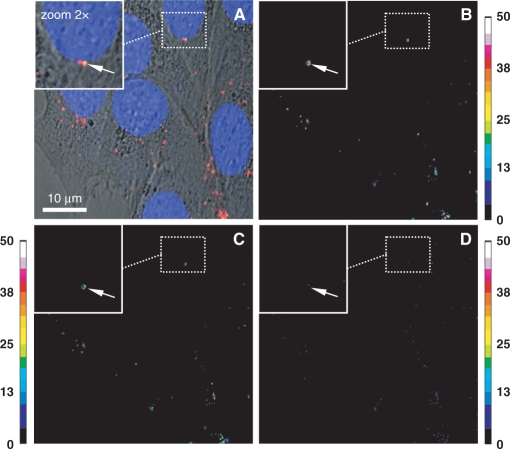
The condensation status of endocyted polyplexes in cells was investigated by FRET imaging. C2C12 cells were transfected for 1 h with double-labelled fluo-p3NF-luc-3NF/rho-LPEI polyplexes. Nuclei were stained with 1 µM Draq5 to visualize the nuclei size easily. The merged image with fluo, rho, Draq5 and transmission channels showed a majority of red spots (rho-polymer) in the cytoplasm close to the nucleus (Figure 2A). The absence of green fluorescence indicates a probable energy transfer between rho-polymer and fluo-p3NF-luc-3NF due to the presence of highly condensed pDNA in polyplexes. Conversely, the presence of yellow spots (white arrows on Figure 2A insert) corresponding to co-localization of fluo-p3NF-luc-3NF and rho-LPEI revealed that some pDNA were weakly condensed in polyplexes. FRET experiments undergo a fluorescence bleed-through of the direct excitement of the acceptor at donor excitation wavelengths, the dependence of FRET on the concentration of the acceptor, and background noise. We therefore calculated corrected FRET ratio images according to either Youvan (Figure 2B), Xia (Figure 2C) or Gordon (Figure 2D) and compared them to the polyplex location in Figure 2A. For a direct comparison with Figure 2A, we chose the FRET ratio from the image in Figure 2C obtained by the Xia method that normalized the FRET ratio apart from the donor and acceptor concentrations. In contrast, the image in Figure 2B obtained with the Youvan method that did not take into account the concentration of the two fluorophores and the image in Figure 2D obtained with the Gordon method that overcompensated for the concentration did not fit very well with the image in Figure 2A. The comparison of Figure 2A and C shows that FRET ratio values varied with the fluorescent spot considered, indicating that the strength of interaction between LPEI and pDNA was not equally distributed. In fact, many red spots in Figure 2A displaying a high FRET ratio ranging from 18 to 35 revealed the presence of well-condensed pDNA in polyplexes in this area. On the other hand, the absence of FRET for other spots presumes either polyplex dissociation with pDNA separation from polymer in different z-sections or the presence of rho-LPEI alone. Sometimes, one can notice the presence of red and green overlay with a low FRET ratio (from 10 to 17) indicating that pDNA was weakly condensed in those polyplexes (white arrow on Figures 2A and C insert).
Figure 2.
Sensitized emission measurements of double-labelled LPEI-polyplexes by CLSM. HeLa cells were incubated with fluo-p3NF-luc-3NF/rho-LPEI polyplexes in the presence of TNFα. Fluorescein (fluo), rhodamine (rho) and Draq5 were excited at 488, 543 and 633 nm, respectively. (A) Transmitted light images merged with recorded images for the fluorescence emission of fluo-p3NF-luc-3NF (in green), rho-LPEI (in red) and Draq5 (in blue) in a multi-tracking mode. (B–D) FRET ratio images calculated according to the method of Youvan (B), Xia (C) and Gordon (D). Sidebars: colour pixels scoring for FRET level from 0 (dark blue) to 50 (white). Each image corresponds to 512 × 512 pixels measuring 0.28 × 0.28 µm2 each.
Polyplexes are present in the nucleus
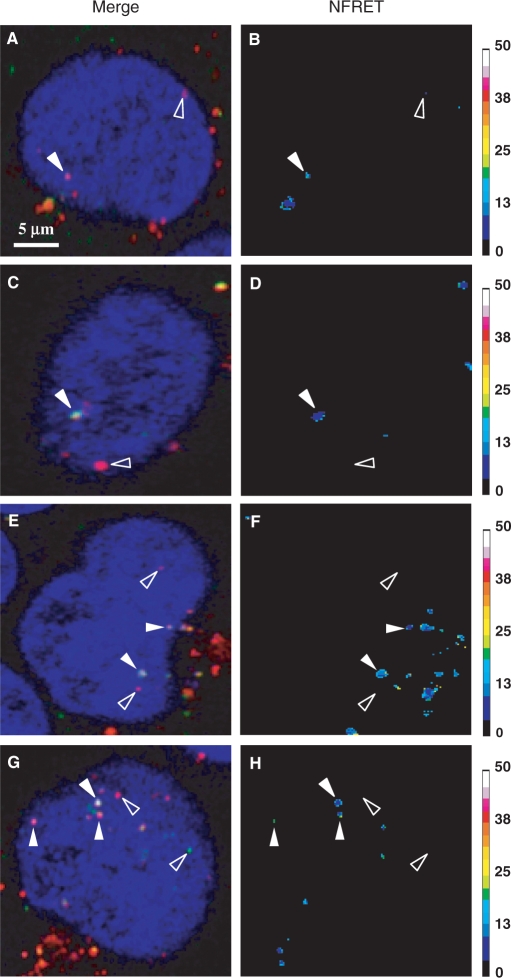
As aforementioned, FRET imaging according to Xia's normalization was used to achieve analysis of the condensation status of pDNA in polyplexes in the nucleus. C2C12 and HeLa cells were transfected for 5 h either with fluo-p3NF-luc-3NF/rho-LPEI or fluo-p3NF-luc-3NF/rho-HIS polyplexes (Figure 3). In merged images performed with fluo, rho and Draq5 emissions, a majority of red spots was clearly observed in the nucleus revealing at least the distribution of rho-LPEI (or rho-HIS) (open arrows in Figures 3A, C, E and G). In contrast, only a few green spots corresponding to the presence of free fluo-p3NF-luc-3NF were detected in this area (right open arrow in Figure 3G). Three types of DNA statuses were observed when comparing the merged images to their respective FRET ratio images. Several red spots exhibiting a FRET ratio higher than 18 (solid arrow in Figure 3B, G and H) correspond to well-condensed pDNA in this ROI. Yellow spots with a significant FRET ratio ranging from 10 to 17 are detected in certain ROIs indicating a subsequent pDNA unpackaging (some of the solid arrows in Figure 3D, F and H). Some green and red spots displaying no significant FRET ratio reveal some pDNA molecules free of polymer and polymer molecules free of pDNA, respectively (open arrows in Figure 3B, D, F and H). A proper statistical analysis carried out in ten transfected HeLa cells after 5 h indicated that 58 spots displayed a FRET ratio higher than 10 and 18 green spots displayed no FRET. These results indicated that the major part (76%) of p3NF-luc-3NF was still highly condensed with the polymer in the nucleus.
Figure 3.
FRET imaging of double-labelled polyplexes in living cells. C2C12 (A–D) and HeLa (E–H) cells were transfected for 5 h with fluo-p3NF-luc-3NF/rho-LPEI (A, B and E, F) or fluo-p3NF-luc-3NF/rho-HIS (C, D and G, H). NFRET ratio images were calculated according to Xia's normalization. Fluo, rho and Draq5 were excited at 488, 543 and 633 nm, respectively. Sidebars: colour pixels scoring for FRET level from 0 (dark blue) to 50 (white). Each image corresponds to 512 × 512 pixels measuring 0.28 × 0.28 µm2 each.
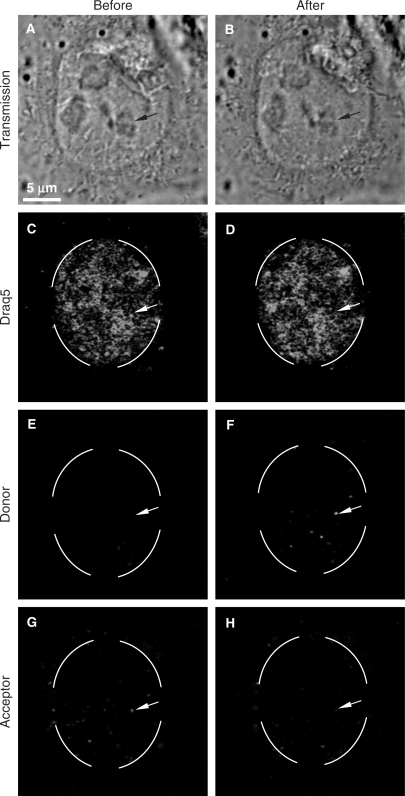
In a second step, the presence of FRET in the nucleus was validated by acceptor photobleaching experiments (Figure 4). Prior to the rho photobleaching, many red spots were observed around and inside the nucleus of HeLa cells transfected with fluo-p3NF-luc-3NF/rho-HIS polyplexes (Figure 4G). No green spot of fluo-p3NF-luc-3NF was visible indicating the presence of FRET between rho-HIS and fluo-p3NF-luc-3NF (Figure 4E). After the rho photobleaching, a clear fluorescence recovery of the donor (fluo) was observed in the bleached ROI (Figure 4F in circle), whereas a decrease in the fluorescence intensity of the corresponding red spots occurred (Figure 4H, white arrow). The fluorescence variation of the donor in intracellular polyplexes was determined from pixel intensities in two ROIs (n = 10 spots for each): a first ROI submitted to the rho photobleaching and a second area taken as control ROI. A third one recorded outside of cells was used to determine the background. Compared to the control ROI, a high increase (from 27.9% to 98.5%) in the fluo fluorescence efficiency was observed immediately after acceptor photobleaching. According to Siegel et al. (28), the calculated FRET efficiency was about 72.1 and 1.5% before and after the rho phothobleaching, respectively. No significant fluorescence variation was observed inside the control ROI. Thus, a recovery of the fluo intensity was clearly measured after the rho photobleaching. This FRET result characterized via the rho photobleaching combined with the direct FRET measurements clearly demonstrated that the majority of pDNA in the nucleus were still in a highly condensed form with the polymer 5 h post-transfection.
Figure 4.
Acceptor photobleaching of double-labelled HIS-polyplexes in living cells. HeLa cells were transfected for 5 h with fluo-p3NF-luc-3NF/rho-HIS polyplexes. Fluo, rho and Draq5 were excited at 488, 543 and 633 nm, respectively. Before and after the rhodamine photobleaching, three images were simultaneously recorded by CLSM in a multi-tracking mode. The broken circle on images corresponds to the bleached area. Experimental conditions: acceptor bleaching was processed with 800 pulses at 543 nm in a time series (one image recorded every 5 s); each image corresponds to 512 × 512 pixels measuring 0.28 × 0.28 µm2 each.
Nuclear quantitation of pDNA
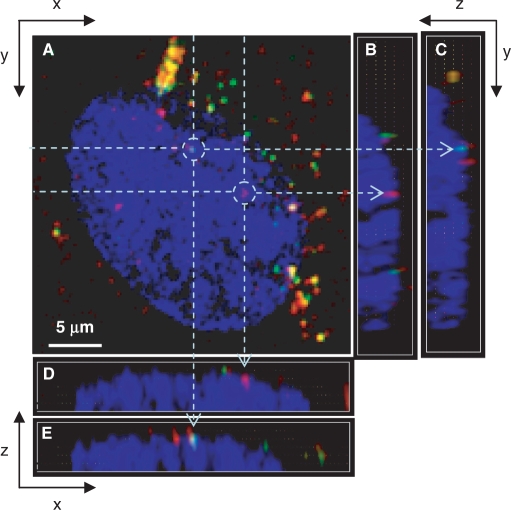
Transfection was performed both with a 3NF-free pDNA (fluo-pTAL-luc) and 3NF-pDNA (Cy3-p3NF-luc-3NF) to visualize the influence of 3NF sequences on the nuclear localization inside the same cell and to determine simultaneously the amount of each plasmid in the same nucleus. In this way, we avoided artefacts due to the cell cycle heterogeneity. Figure 5 shows a typical confocal z-series image of TNFα-stimulated HeLa cells upon 2 h incubation at 37°C with a mixture of fluo-pTAL-luc/LPEI and Cy3-p3NF-luc-3NF/LPEI polyplexes. Many red or green intracellular spots were clearly observed, confirming the cellular uptake of these two plasmids. Fluo-pTAL-luc and Cy3-p3NF-luc-3NF were sometimes co-localized (yellow spots). Interestingly, the presence of several red spots in the nucleus clearly revealed that Cy3-p3NF-luc-3NF was efficiently imported. Fluo-pTAL-luc (green spots) was also observed in the nucleus but at much lower frequency than Cy3-p3NF-luc-3NF.
Figure 5.
CLSM images of transfected living cells. C2C12 cells were transfected for 2 h with a mixture of Cy3-p3NF-luc-3NF/LPEI and fluo-pTAL-luc/LPEI polyplexes. Red denotes Cy3-p3NF-luc-3NF; green, fluo-pTAL-luc; yellow, the overlap of the two denotes pDNA; blue, Draq5-labelled nucleus. For each set, (A) is the x–y top view at a given z; (B, C) and (D, E) are two examples on the respective y–z and x–z side views along dotted lines. Objects in drawn circles are indicated by arrows in the x–z and y–z plane side views. Fluo, Cyanin 3 (Cy3) and Draq5 were excited at 488, 543 and 633 nm, respectively. Fluorescence emissions were collected in a multi-tracking mode. Each confocal image corresponds to 512 × 512 pixels measuring 0.28 × 0.28 µm2 each.
A 3D analysis based on CLSM images was performed to evaluate the rate of pDNA nuclear accumulation in C2C12 and HeLa lines. Cells were co-incubated either with the mixture of fluo-pTAL-luc and Cy3-p3NF-luc-3NF polyplexes or of fluo-pTAL-luc and Cy3-pCMV-luc polyplexes, and observed in real-time.
HeLa cells
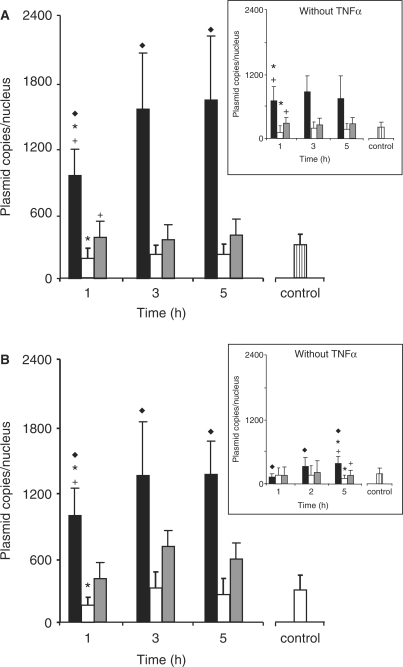
A rapid nuclear import of the three kinds of pDNA was clearly observed upon 1 h in TNFα-stimulated HeLa cells transfected with LPEI-polyplexes (Figure 6A). Nevertheless, a progressive accumulation of p3NF-luc-3NF occurred and reached a steady state after 3 h incubation. Upon 5 h incubation, the copy number of p3NF-luc-3NF (1493 ± 672 copies) was 4- and 6-fold higher than that of pCMV-luc (368 ± 116 copies) and pTAL-luc (223 ± 88 copies), respectively. In contrast, the nuclear copy number of these pDNA did not increase significantly upon 1 h incubation in the absence of TNFα (Figure 6A insert). These results indicated that TNFα stimulation increased the kinetic of pDNA accumulation into the nucleus. When cells were transfected with HIS-polyplexes in the presence of TNFα, a progressive increase in p3NF-luc-3NF also occurred in the nucleus and a plateau was observed after 3 h (Figure 6B). Moreover, the copy number of p3NF-luc-3NF (1235 ± 276 copies) was 2.5- and 5-fold higher than that of pCMV-luc (526 ± 211 copies) and pTAL-luc (252 ± 149 copies), respectively. In the absence of TNFα, the nuclear quantities of the three pDNA were 2- to 3-fold lower than that in TNFα-stimulated cells (Figure 6B insert).
Figure 6.
Quantitative measurement of the number of plasmid copies in the nucleus of HeLa cells. TNFα-stimulated cells were incubated with a mixture of Cy3-p3NF-luc-3NF polyplexes (solid bar) and fluo-pTAL-luc polyplexes (open bar) or a mixture of Cy3-pCMV-luc polyplexes (grey bar) and fluo-pTAL-luc polyplexes made with LPEI (A) or HIS (B). In control, cells were incubated for 5 h with a mixture of Cy3-pTAL-luc polyplexes (hatched bar) and fluo-pTAL-luc polyplexes. Insert: transfection in the absence of TNFα. Up to 20 optical slices 0.6 µm in depth were recorded by CLSM. Results are the mean value of 12 nuclei. Significant statistical difference between cells transfected with p3NF-luc-3NF and cells transfected with p3NF-luc-3NF (filled diamond), pTAL-luc (asterisk) or pCMV-luc (cross) according to a Student's test with P < 0.01.
C2C12 cells
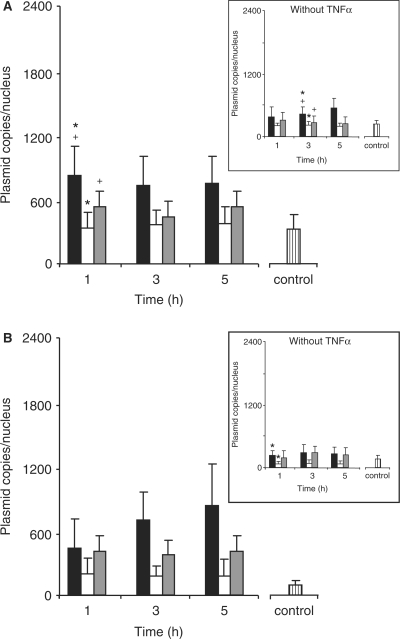
As compared to TNFα-stimulated HeLa cells, a rapid nuclear import of the three kinds of pDNA was clearly observed upon 1 h in stimulated C2C12 cells transfected with LPEI-polyplexes (Figure 7A). Subsequently, no change was observed at 3 and 5 h. However, the copy number of p3NF-luc-3NF (685 ± 234 copies) was 2- and 1.5-fold higher at any time than that of pTAL-luc (345 ± 146 copies) and pCMV-luc (489 ± 215 copies), respectively. In the absence of TNFα, these pDNA constructs were rarely present in the nucleus (Figure 7A insert). A progressive accumulation of p3NF-luc-3NF was observed over 5 h whereas no change was detected for pCMV-luc and pTAL-luc. Concerning transfection with HIS-polyplexes, the rate of the nuclear accumulation of p3NF-luc-3NF in TNFα-stimulated cells increased progressively compared to LPEI-polyplexes (Figure 7B). However, the number of p3NF-luc-3NF copies in cells transfected with HIS-polyplexes or LPEI-polyplexes reached an equivalent level at 5 h. Moreover, the number of p3NF-luc-3NF copies was 1.5–2 times lower in C2C12 cells than in HeLa cells. No change of pTAL-luc and pCMV-luc copy number was observed over 5 h. Interestingly, the number of p3NF-luc-3NF (765 ± 357 copies) was clearly 2-fold higher than that of pCMV-luc (375 ± 141 copies). Again, the nuclear copy number of pTAL-luc varied considerably (162 ± 142 copies). In the absence of TNFα, the nuclear amount of each pDNA was rapidly saturated. p3NF-luc-3NF (259 ± 129 copies) and pCMV-luc (243 ± 130 copies) was 3.5-fold higher than pTAL-luc (70 ± 58 copies) (Figure 7B insert). Nevertheless, the number of p3NF-luc-3NF in stimulated cells was 3 times higher than in non-stimulated cells.
Figure 7.
Quantitative measurement of the number of plasmid copies in the nucleus of C2C12 cells. Experimental conditions and legends are as in Figure 6. Results are the mean value of 12 nuclei. Significant statistical difference between cells transfected with p3NF-luc-3NF and cell transfected with p3NF-luc-3NF (filled Diamond), pTAL-luc (asterisk) or pCMV-luc (cross) according to a Student's test with P < 0.01.
As controls, HeLa and C2C12 cells were transfected with a mixture of Cy3-pTAL-luc polyplexes and fluo-pTAL-luc polyplexes in the absence of any other plasmids. We found that the amount of Cy3-pTAL-luc delivered in the nucleus was similar to that of fluo-pTAL-luc (data not shown). This evidenced that there was no influence of the fluorophore on the intracellular trafficking of plasmid. Moreover, the amount of Cy3-pTAL-luc (hatched bar in Figures 6 and 7) delivered in the nucleus after 5 h incubation was equal to that of fluo-pTAL-luc co-transfected either with Cy3-p3NF-luc-3NF or Cy3-pCMV-luc. This indicated that the nuclear accumulation of pTAL-luc upon co-transfection was not influenced by the presence of the other plasmids.
NFκB inhibition
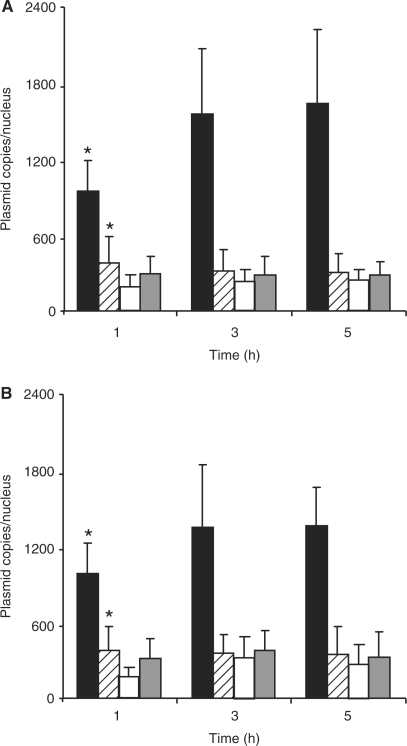
BAY 11-7085, an inhibitor of the NFκB activation by TNFα was used to demonstrate the involvement of NFκB in the nuclear accumulation of p3NF-luc-3NF (29). As shown in Figure 8A, the number of p3NF-luc-3NF copies in the nucleus at 1, 3 and 5 h clearly decreased and was 4-fold lower than in untreated cells. In contrast, the number of pTAL-luc copies did not statistically change in BAY 11-7085-treated cells. The same procedure carried out in cells transfected with HIS-polyplexes led to similar observations (Figure 8B). Thus, the dramatic inhibition of the pDNA nuclear import in the presence of BAY 11-7085 was indicative of the NFκB-mediated nuclear delivery of p3NF-luc-3NF. Moreover, we evaluated the influence of 1 µM BAY 11-7085 on the luciferase expression. Upon transfection with p3NF-luc-3NF in the presence of this inhibitor, the luciferase activity was 2.5 times lower than in its absence. In contrast, no significant change was observed when the cells were transfected with pCMV-luc in the absence and in the presence of the inhibitor. These results are in good correlation with the decrease of the nuclear import of p3NF-luc-3NF in the presence of the inhibitor of the NFκB activation.
Figure 8.
NFκB inhibition effect on the p3NF-luc-3NF nuclear import. HeLa cells were incubated with a mixture of Cy3-p3NF-luc-3NF polyplexes (hatched bar) and fluo-pTAL-luc polyplexes (grey bar) made with LPEI (A) or HIS (B) in the presence of BAY 11-7085 and TNFα. Cells were also transfected in the absence of BAY 11-7085 with Cy3-p3NF-luc-3NF polyplexes (solid bar) and fluo-pTAL-luc polyplexes (open bar). Results are the mean value of 12 nuclei. (asterisk) Significant statistical difference of values obtained between cells tranfected with p3NF-luc-3NF in the absence or the presence of BAY 11-7085 according to a Student's test with P < 0.01.
DISCUSSION
Although the transfer of pDNA into the nucleus of post-mitotic cells is a prerequisite for efficient transfection by polyplexes, quantification of pDNA molecules in the cell nucleus upon transfection is weakly documented. In this work, we determined the number of pDNA copies in the nucleus of two different cell lines transfected with pDNA/LPEI and pDNA/HIS polyplexes. Moreover, the state of nuclear pDNA was investigated by FRET and FRAP techniques. The study was conducted with three different pDNA constructs encoding the luciferase gene. Amongst them, p3NF-luc-3NF bore two 3NF optimized sequences for binding to NFκB to favour the pDNA import into the cell nucleus. In terms of luciferase gene expression, the transfection of HeLa cells with p3NF-luc-3NF was indeed several orders of magnitude higher than with the 3NF-free pDNA counterpart, pTAL-luc and reached a level close to that obtained with a luciferase gene expression driven by a strong promoter. We found that the amount of fluo-pDNA taken up by the cells was similar whatever the plasmid constructs. Confocal microscopy images showed that fluo-pDNA was present in the cell nucleus as fluorescent spots, the number of which was higher when cells were transfected with p3NF-luc-3NF than with the other pDNA. To determine whether spots corresponded to polyplexes or plasmid free of polymer, fluorescence-based techniques such as FRET and FRAP were applied to assess plasmid/polymer interaction in the nucleus. FRET imaging using Xia's normalization revealed the presence of many yellow spots that displayed significant FRET ratios, indicating the presence of plasmid–polymer interaction. In addition, few free p3NF-luc-3NF and free polymer were found in the nucleus. The presence of many condensed plasmids in the nucleus evidenced by the FRET phenomenon was confirmed by the clear fluorescence recovery of fluo-p3NF-luc-3NF observed upon acceptor (rho) photobleaching. These results are in agreement with those previously reported for other polyplexes. Indeed, Godbey et al. (30) have shown by microscopy observations that pDNA and branched PEI were also co-localized in the nucleus upon transfection of EA.hy 926 cells. Ho and others (31, 32) have shown by FRET microscopy that pDNA and chitosan were also co-localized in HEK293 cells transfected with red quantum dot-DNA/Cy5-chitosan polyplexes. According to Itaka et al.'s work (33), the FRET level of LPEI/pDNA polyplexes in the cytoplasm remained high upon 4 h transfection indicating that the majority of pDNA was still condensed in polyplexes. Then, a significant decrease of FRET occurred 16 h post-transfection indicating a pDNA decondensation. Our results are in agreements with Itaka's data since 76% of pDNA were condensed with polymer in the nucleus upon 5 h transfection.
The quantification of pDNA copies in the nucleus was achieved in living cells by fluorescent pDNA and a 3D analysis on sequential z-series of real-time confocal microscopy images. We found that the number of pDNA copies accumulated in the nucleus upon 5 h transfection with p3NF-luc-3NF was clearly higher than upon transfection with pTAL-luc or pCMV-luc. These results demonstrate that 3NF sequences favour pDNA nuclear import. We also observed a rapid nuclear accumulation of p3NF-luc-3NF within 1 h in both cell lines and saturation after 3 h incubation. After 5 h, the number of p3NF-luc-3NF copies in the nucleus of HeLa cells was 1493 ± 672 with LPEI and 1235 ± 276 with HIS, compared with 223 ± 88 to 526 ± 211 p3NF-free plasmid copies (pTAL-luc and pCMV-luc). In comparison, James and Giorgio (12) have found by quantitative flow cytometry measurements on isolated nuclei that 1250 pCMV-plasmid copies were delivered into nuclei of HeLa cells after 4 h transfection with cationic liposomes. Tachibana et al. (34) have estimated by real-time PCR that about 5840 copies of pGEM/SV2CAT were present in the nucleus of hepatic AH130 cells upon 36 h transfection with cationic liposomes. These values are higher than those that we have found for pTAL-luc (∼250 copies/nucleus) and pCMV-luc (∼500 copies/nucleus). This discrepancy could be related to subcellular fractionations performed to purify nuclei. Indeed, contamination of the nuclear fraction by cytoplasmic pDNA often leads to overestimation of plasmid numbers contained in the nuclei. The number of p3NF-luc-3NF copies (1493 ± 672 copies) found in HeLa nuclei was 6-fold more than that of pTAL-luc, resulting in a luciferase gene expression difference of three orders of magnitude. According to Pollard et al. (35), it seems that 103 pDNA copies in the nucleus is a threshold leading to a 100% chance of transfecting the cell. Indeed, intra-nuclear microinjection of 103 pDNA copies free or complexed with PEI led to the transfection of the cell while that of 102 copies had a 40–50% chance of transfecting the cell. In addition, the gene expression driven by the two p3NF-luc-3NF and pCMV-luc plasmids was similar, despite the stronger promoter present in the pCMV-luc plasmid. These results suggest that the 3-fold higher copy number of p3NF-luc-3NF into the nucleus balances the weakness of its promoter. The CMV promoter contains κB-binding consensus sequences that can be potentially recognized by NFκB, leading to a transcriptional activation upon TNFα stimulation. Thus, an improved expression of pCMV-luc occurs. These κB sequences are separated by 142 and 152 bp, an arrangement that is not optimal to favour a NFκB-mediated nuclear import of pCMV-luc in comparison with p3NF-luc-3NF plasmid.
According to our data, the amount of pTAL-luc inside the nucleus did not depend on either the cell type or on the polymer type. In contrast, the number of p3NF-DNA imported into the nucleus was almost 2-fold more in HeLa cells than in C2C12 cells. This could be due to different thresholds of NFκB activation between these two cell types. Similarly, we also noticed that in cells transfected with LPEI and HIS in the absence of TNFα, the amount of p3NF-luc-3NF in the nucleus was 3- and 5-fold more than that of pCMV-luc or pTAL-luc, respectively. It has been previously reported that many polyamines are able to increase the transcription of genes, in part, through the activation of NFκB pathway (36). Indeed, Shah et al. (36) have demonstrated that in transient transfection experiments using an NFκB-driven secreted alkaline phosphatase reporter, spermine analogues induced NFκB activity by ∼2-fold. Therefore, in the absence of TNFα, the higher number of p3NF-luc-3NF copies compared to pTAL-luc or pCMV-luc could be related to an activation of the NFκB pathway by cationic LPEI and HIS polymers.
We have demonstrated that plasmids encoding the luciferase gene and containing 3NF sequences (3NF-plasmids) were (i) recognized by NFκB inside HeLa cells, (ii) more efficient than 3NF-free plasmids to transfect cells and (iii) more imported in the nucleus than 3NF-free plasmids on the basis of FISH experiments. Here, we demonstrated that the nuclear import of the p3NF-plasmid was mediated by NFκB. Indeed, the p3NF-luc-3NF copy number dropped dramatically in the nucleus in the presence of BAY 11-7085 and reached the amount of pTAL-luc.
Besides, FRET and FRAP measurements have evidenced that some polyplexes appeared intact and located within the nucleus at 4 h post-transfection. These findings raise the question of the plasmid passage into the nucleus through the nuclear pore complex (NPC) (37). Although small macromolecules (≤40 kDa) diffuse passively through NPCs, larger ones are actively transported by a process mediated by NLS signal recognized by a heterodimeric protein complex including α and β importins. Most importantly, nucleopores can only be expanded up to 39 nm (38), a diameter considerably smaller than the current size (70–300 nm) of LPEI or HIS-polyplexes. The following hypotheses can be considered to explain the presence of polyplexes in the nucleus. First, the size or the shape of polyplexes can undergo modifications near the nuclear envelope, allowing a passage through the nucleopores. Second, the plasmid is unpackaged on the cytosolic side of the nuclear envelope, and is repacked in the nucleus upon the separate passage of the polymer and the plasmid through the nucleopores. To date, no current data support these hypotheses. Further work to determine the polyplexes localization in the nucleopores as well as the plasmid and polymer interactions with NPCs on both sides of the nuclear envelope will contribute to elucidate this question.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants from the Association Française contre les Myopathies (AFM) and Vaincre la Mucoviscidose (VLM). G.B. received a fellowship from AFM. The English language of this manuscript has been edited by Elizabeth Rowly—PublishInEnglish® (France). Funding to pay the Open Access publication charges for this article was provided by CBMCNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fabre JW, Collins L. Synthetic peptides as non-viral DNA vectors. Curr. Gene. Ther. 2006;6:459–480. doi: 10.2174/156652306777934865. [DOI] [PubMed] [Google Scholar]
- 2.Wong SY, Pelet JM, Putman D. Polymer systems for gene delivery - past, present, and future. Prog. Poly. Sci. 2007;32:799–837. [Google Scholar]
- 3.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pichon C, Goncalves C, Midoux P. Histidine-rich peptides and polymers for nucleic acids delivery. Adv. Drug Deliv. Rev. 2001;53:75–94. doi: 10.1016/s0169-409x(01)00221-6. [DOI] [PubMed] [Google Scholar]
- 5.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 6.Wagstaff KM, Jans DA. Nucleocytoplasmic transport of DNA: enhancing non-viral gene transfer. Biochem. J. 2007;406:185–202. doi: 10.1042/BJ20070505. [DOI] [PubMed] [Google Scholar]
- 7.Kuramoto T, Nishikawa M, Thanaketpaisarn O, Okabe T, Yamashita F, Hashida M. Use of lipoplex-induced nuclear factor-kappaB activation to enhance transgene expression by lipoplex in mouse lung. J. Gene. Med. 2006;8:53–62. doi: 10.1002/jgm.812. [DOI] [PubMed] [Google Scholar]
- 8.Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol. Ther. 2001;3:653–657. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- 9.Mesika A, Kiss V, Brumfeld V, Ghosh G, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum. Gene. Ther. 2005;16:200–208. doi: 10.1089/hum.2005.16.200. [DOI] [PubMed] [Google Scholar]
- 10.Gilmore TD. Introduction to NFkappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 11.Carlotti F, Chapman R, Dower SK, Qwarnstrom EE. Activation of nuclear factor kappaB in single living cells. Dependence of nuclear translocation and anti-apoptotic function on EGFPRELA concentration. J. Biol. Chem. 1999;274:37941–37949. doi: 10.1074/jbc.274.53.37941. [DOI] [PubMed] [Google Scholar]
- 12.James MB, Giorgio TD. Nuclear-associated plasmid, but not cell-associated plasmid, is correlated with transgene expression in cultured mammalian cells. Mol. Ther. 2000;1:339–346. doi: 10.1006/mthe.2000.0054. [DOI] [PubMed] [Google Scholar]
- 13.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene. Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 14.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol. Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Chen TR. In situ detection of mycoplasma contamination in cell cultures by fluorescent Hoechst 33258 stain. Exp. Cell Res. 1977;104:255–262. doi: 10.1016/0014-4827(77)90089-1. [DOI] [PubMed] [Google Scholar]
- 16.Brissault B, Leborgne C, Guis C, Danos O, Cheradame H, Kichler A. Linear topology confers in vivo gene transfer activity to polyethylenimines. Bioconjug. Chem. 2006;17:759–765. doi: 10.1021/bc050287v. [DOI] [PubMed] [Google Scholar]
- 17.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjug. Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 18.Midoux P, Roche AC, Monsigny M. Quantitation of the binding, uptake, and degradation of fluoresceinylated neoglycoproteins by flow cytometry. Cytometry. 1987;8:327–334. doi: 10.1002/cyto.990080314. [DOI] [PubMed] [Google Scholar]
- 19.Rasband W. ImageJ. Bethesde, MA: US National Institues of Health; 1997–2007. http://rsb.info.nih.gov/ij/plugins.html. [Google Scholar]
- 20.Barthel KU. Volume Viewer. 2005. http://rsb.info.nih.gov/ij/plugins/volume-viewer.html.
- 21.Clamme JP, Azoulay J, Mely Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys. J. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Midoux P, LeCam E, Coulaud D, Delain E, Pichon C. Histidine containing peptides and polypeptides as nucleic acid vectors. In: Luo D, Saltzman WM, editors. Synthetic DNA Delivery Systems. London: Kluwer Academic/Plenum Publishers Hardbound; 2003. pp. 23–44. [Google Scholar]
- 23.Wu P, Brand L. Resonance energy transfer: methods and applications. Anal. Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 24.Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
- 25.Youvan D, Silva C, Bilina E, Coleman W, Dilworth M, Yang M. Calibration of fluorescence resonance energy transfer in microscopy using genetically engineered GFP derivatives on nickel chelating beads. Biotechnology. 1997;3:1–18. [Google Scholar]
- 26.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel RM, Chan FK, Zacharias DA, Swofford R, Holmes KL, Tsien RY, Lenardo MJ. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci. STKE. 2000;2000:PL1. doi: 10.1126/stke.2000.38.pl1. [DOI] [PubMed] [Google Scholar]
- 29.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J. Biol. Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 30.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl Acad. Sci. USA. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol. Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho YP, Chen HH, Leong KW, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J. Control. Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itaka K, Harada A, Yamasaki Y, Nakamura K, Kawaguchi H, Kataoka K. In situ single cell observation by fluorescence resonance energy transfer reveals fast intra-cytoplasmic delivery and easy release of plasmid DNA complexed with linear polyethylenimine. J. Gene. Med. 2004;6:76–84. doi: 10.1002/jgm.470. [DOI] [PubMed] [Google Scholar]
- 34.Tachibana R, Harashima H, Ide N, Ukitsu S, Ohta Y, Suzuki N, Kikuchi H, Shinohara Y, Kiwada H. Quantitative analysis of correlation between number of nuclear plasmids and gene expression activity after transfection with cationic liposomes. Pharm. Res. 2002;19:377–381. doi: 10.1023/a:1015162722295. [DOI] [PubMed] [Google Scholar]
- 35.Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 36.Shah N, Thomas T, Shirahata A, Sigal LH, Thomas TJ. Activation of nuclear factor kB by polyamines in breast cancer cells. Biochemistry. 1999;38:14763–14774. doi: 10.1021/bi991291v. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg MW, Allen TD. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J. Mol. Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- 38.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.