Abstract
DNA looping is important for gene repression and activation in Escherichia coli and is necessary for some kinds of gene regulation and recombination in eukaryotes. We are interested in sequence-nonspecific architectural DNA-binding proteins that alter the apparent flexibility of DNA by producing transient bends or kinks in DNA. The bacterial heat unstable (HU) and eukaryotic high-mobility group B (HMGB) proteins fall into this category. We have exploited a sensitive genetic assay of DNA looping in living E. coli cells to explore the extent to which HMGB proteins and derivatives can complement a DNA looping defect in E. coli lacking HU protein. Here, we show that derivatives of the yeast HMGB protein Nhp6A rescue DNA looping in E. coli lacking HU, in some cases facilitating looping to a greater extent than is observed in E. coli expressing normal levels of HU protein. Nhp6A-induced changes in the DNA length-dependence of repression efficiency suggest that Nhp6A alters DNA twist in vivo. In contrast, human HMGB2-box A derivatives did not rescue looping.
INTRODUCTION
The intrinsic stiffness of double-stranded DNA with respect to bending and twisting (1,2) must be managed in vivo (3–6). This is particularly true because of the need to compact very long genomes into the prokaryotic nucleoid (7–9) or eukaryotic nucleus. Enhancing the apparent flexibility of DNA is apparently also essential to facilitate locally deformed nucleoprotein structures such as loops, required for certain kinds of gene control and recombination (10–13). Architectural DNA-binding proteins appear to play important roles in modifying the apparent flexibility of DNA (5,6,14–21).
The abundant heat unstable (HU) protein of Escherichia coli binds DNA with little or no sequence specificity, and induces a sharp DNA bend. X-ray crystallography and biochemical experiments suggest HU can induce hinge-like DNA flexibility (22–29). This protein appears to play a direct role in facilitating DNA loop formation in repression of the E. coli gal operon (30,31). In this case, an A/T-rich sequence element may be the preferred HU-binding site in the DNA loop, and direct GalR–HU interactions have been proposed. Though HU had not been previously implicated in facilitating repression by DNA looping in the lac operon, our previous work (14,15) has shown that lac repression becomes leaky in the absence of HU. Deletion of the genes encoding HU results in nucleoid instability and growth defects in E. coli (24,32,33).
In eukaryotes the abundant sequence-nonspecific high-mobility group B (HMGB) proteins appear to perform roles analogous to HU in enhancing apparent DNA flexibility (6,34). HMGB proteins contain one or two ∼80-amino acid domains that fold into ‘L’-shaped structures composed of three α-helices [Figure 1A and B; (35–43)]. In the case of sequence-nonspecific DNA binding by HMGB proteins such as mammalian HMGB1 and HMGB2 or yeast Nhp6A, DNA bending of 90° or more results from van der Waals and electrostatic interactions of the globular domains with the minor groove and also amino acid intercalation to force local kinking (44). Recent studies (40,44,45) have estimated that an individual HMG box (B) from HMGB1 binds DNA with an equilibrium dissociation constant of ∼10 nM and a 40° induced bend angle. The single-box Nhp6A protein from yeast has been estimated to bend DNA by 60–90°. Our optical tweezer studies of human HMGB2 box A (16) showed that the protein reduced the apparent persistence length of DNA from its intrinsic value of 50 nm (∼150 bp) to 10 nm (∼30 bp) at 100 mM salt and even to 5 nm (∼15 bp) at 50 mM salt. At saturation, protein binding increased the DNA contour length by 16% due to intercalation and unwinding (16). A binding site size of 5–6 bp was estimated with Kd ∼50–100 nM depending on the salt concentration. Average DNA bend angles of 114° and 87° were measured at 50 and 100 mM monovalent salt, respectively. More recent work using similar methods estimated DNA bending of 99° for the single HMG box A derived from human HMGB2 (17). Interestingly, a two-box HMGB protein may bind DNA with the protein domains out of alignment (42), and the NMR structure of such a protein bound to DNA gave an estimated DNA bend angle of 77°. Besides DNA bending, the degree of DNA untwisting by HMGB proteins depends on the protein variant.
Figure 1.
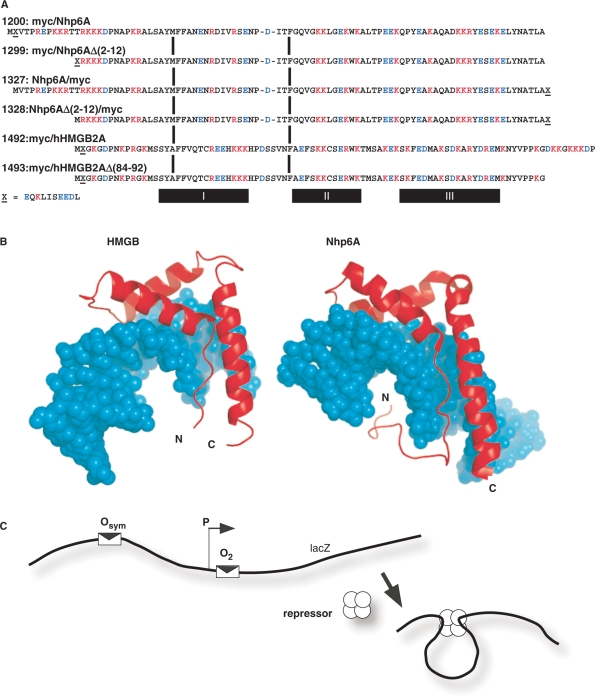
Proteins tested in E. coli in vivo DNA-looping assay. (A) HMGB proteins tested. The plasmid number and a description of protein domains (amino-to-carboxyl order) are indicated, along with the complete amino acid sequence aligned at conserved HMG box intercalating residues (vertical bars). The c-Myc epitope tag is indicated by ‘X’ and its sequence is given below the figure. Deletion derivatives removed portions of cationic tails at the amino (constructs 1299, 1328) or carboxyl (construct 1493) termini. The α-helical domains I, II and III are indicated below the diagram. (B) Molecular models of single HMG boxes bound to DNA. Left, HMG box A from mammalian HMGB1 (37,38,41), PDB code 1CKT, which is highly similar to the corresponding box from HMGB2. Right, S. cerevisiae Nhp6A (40), pdb code 1J5N. The strongly bent DNA molecules are shown as space-filling models in cyan. HMGB domains are shown in red. Note that both the amino and carboxyl termini (N and C, respectively) are positioned so that basic tail extensions can neutralize crowded DNA backbone phosphates in the compressed major groove. (C) Schematic representation of episomal lac looping assay constructs (14,15). Weak (O2) and strong (Osym) lac operators are positioned at various separation distances flanking an E. coli promoter (P) upstream of the lacZ reporter gene encoding β-galactosidase. In the presence of tetrameric lac repressor protein, DNA looping causes repression of the test promoter. The dependence of DNA looping on operator separation in a series of strains where this distance is varied in base pair increments provides information about the apparent bending and torsional stiffness of DNA in vivo. When experiments are performed in ΔHU cells, the effect of apparent DNA flexibility of exogenous eukaryotic HMGB protein expression can be monitored.
Of particular interest here are additional DNA-bending forces attributed to cationic tails at the amino or carboxy termini of HMGB proteins. Either terminus is positioned to interact with crowded phosphates in the compressed major groove of bent DNA (Figure 1B). These tails may enhance bending by asymmetric charge neutralization (2,44,46,47). An estimate of 4–5 degrees of DNA bending per major groove salt bridge has been suggested (44). Removal of six cationic residues from the cationic carboxy-terminal tail of the sequence-specific HMGB protein LEF1 reduced DNA bending from 117° to 88° (44). In this work, we test whether this potential change in DNA bending has functional consequences by asking whether the highly cationic tail domains are essential for HMGB function in a heterologous assay system where the protein facilitates DNA looping by lac repressor protein in E. coli, as described below. We compare derivatives of the yeast Nhp6A protein to charge variants of human HMGB2 box A. The latter domain was selected for comparison with our detailed examination of its properties in single molecule experiments in vitro (16). Future studies will extend these approaches to the more conserved and robust box B of mammalian HMGB1.
The in vivo lac repression looping assay (14,15,48–53) was developed to understand the constraints on DNA repression loops in living bacteria due to inflexibility in the repressor protein and looped DNA. As shown in Figure 1C, the lacZ reporter gene is placed downstream from a simple promoter that overlaps a very weak binding site (O2 operator) for lac repressor. This gene is poorly repressed. Repression is dramatically enhanced by placement of a strong operator (Osym) upstream of the promoter. Analysis of lacZ repression as a function of operator spacing provides an extremely sensitive assay of DNA bending and twisting flexibility in vivo (4,14,15,48,54,55). In particular, fitting the oscillating plots of the repression dependence on operator spacing (14,15,48) suggests that the in vivo apparent DNA torsional modulus is ∼4-fold lower than the accepted range of ∼2–4 × 10−19 erg cm for naked DNA in dilute solution (56). DNA bending stiffness did not appear to affect looping for loop sizes of 60–90 bp (14,15). Such short loops are highly unfavorable in naked DNA (19), though exactly how unfavorable has been the subject of recent debate (57–59).
We have previously shown that in vivo lac repressor looping assays can be applied to understand the role of endogenous E. coli nucleoid proteins in facilitating DNA looping (14,15). In particular, cells disrupted for the genes encoding HU subunits were significantly disabled with respect to formation of repression loops, while cells lacking the H-NS nucleoid protein showed facilitated DNA looping (15). The detailed DNA length dependence of looping efficiency can be interpreted in terms of protein-induced changes in DNA structure or flexibility.
The compromised repression looping in ΔHU E. coli provides an opportunity to test the ability of heterologous eukaryotic HMGB proteins and mutants to complement this looping defect. Such complementation has been suggested by previous studies. Johnson and co-workers (18) showed that expression of the yeast HMGB protein Nhp6A in E. coli cells deleted for the HU and Fis nucleoid proteins restored normal nucleoid morphology and cell growth. Similarly, HMGB proteins substitute for bacterial HU in reconstituted DNA recombination reactions in vitro (5). It has also been shown that HU and the yeast mitochondrial HM protein can functionally replace each other in both bacteria and yeast (60). Our own initial studies of DNA looping by lac repressor in ΔHU E. coli showed that heterologous expression of the two-box rat HMGB1 protein could partially restore facilitated DNA looping (14).
We now extend the study of HMGB complementation of DNA looping in ΔHU E. coli by testing single box HMGB domains derived from yeast and human proteins. We further explore the importance of cationic HMGB tail domains by comparing complementation by constructs containing or lacking these extensions.
MATERIALS AND METHODS
Bacterial strains and gene disruption
The bacterial strains in this study are indicated in Table 1. The hupA and hupB genes were disrupted in parental E. coli strain FW102 (61) as described (14,62). Strain deletion status and the presence of looping assay episomes were confirmed by diagnostic PCR amplification following conjugation and selection.
Table 1.
Bacterial strains in this study
| Strain | Relevant genotype | Designation | Comment |
|---|---|---|---|
| FW102a | StrepR derivative of CSH142 [araD(gpt-lac)5] | WT | |
| BL643 | FW102 ΔhupA ΔhupB | ΔHU | Loss of both HU-1 and HU-2 subunits of HU heterodimer |
aFW102 was the kind gift of F. Whipple.
Cloning and bacterial expression of HMGB proteins
HMG protein expression constructs were created by inserting purified PCR products into plasmid pJ1035, which is a modified version of pLX20 containing a promoter for a moderate level of protein expression and an inactive lac operator (14,61). Both full length (pJ1200 and pJ1327) and deletion (pJ1299 and pJ1328) versions of Nhp6A were created for protein expression. In addition, a 10-amino acid c-Myc epitope tag (EQKLISEEDL) was added either to the N-terminus (pJ1200 and pJ1299), or the C-terminus (pJ1327 and pJ1328) of proteins to monitor expression. Nhp6A constructs were amplified from purified yeast genomic DNA. Briefly, pJ1200 contains the product of PCR with primer pair LJM-2498 (GCTCTAGA2TGGA2CA5CT2AT3CTGA3GA3GATCTGGTCAC4A2GAGA2C2TA2G) and LJM-2473 (CGAC2G2TCGCAGTC3TA2GC2A3GTG2CG), which amplify the coding region of Nhp6A from Met1 to Ala92. The encoded c-Myc tag is indicated in bold italics. Upper primer LJM-2498 also installs an XbaI site and lower primer LJM-2473 installs both a stop codon and an AgeI site for cloning. Nhp6A deletion construct pJ1299, missing amino acids 2–12, was created using upper primer LJM-2564 (GCTCTAGA2TG2A2CA5CT2AT3CTGA2GA2GATCTGAGA3GA2GA2G2AC3) and lower primer LJM-2473. Full length Nhp6A construct pJ1327 was created using primer pair LJM-2701 (GCTCTAGA2TG2TCAC4A2GAGA2C2TA2G) and lower primer LJM-2704 (CGA2GCT2GCAGCTACAGATCT2CT2CAGA3TA2GT5GT2CAGC2A3GTG2CGT2ATATA2C). Upper primer LJM-2701 installs a XbaI site. Lower primer LJM-2704 installs the c-Myc tag, a TAG stop codon and a Hind III restriction site for cloning. Deletion construct pJ1328 was created with upper primer LJM-2702 (GCTCTAGA2TGAGA3GA2GA2G2AC3A3TG) and lower primer LJM-2704.
Plasmid constructs encoding box A of human HMGB2 were amplified from plasmid pJ583 (a gift from Phillip Sharp), with the addition of N-terminal c-Myc tags. A variant construct deletes the basic linker region (amino acids 84–92: DKKGKKKDP). Plasmid pJ1492 was prepared using upper primer LJM-3315 (GCTCTAGA2TGGA2CA5CT2AT3CTGA2GA2GATCTGG2TA3G2AGAC4A2C), with the c-Myc tag in bold italics, and lower primer LJM-3317 (CGA2GCT2GCAGCTACG3TC2T5CT2C), which amplify the human HMGB2 coding region from Met1 to Pro92. Plasmid pJ1493 was created using upper primer LJM-3315 and lower primer LJM-3327 (CGA2GCT2GCAGCTA2C2T3G3AG2A2CGTA2T5C), which amplify from Met1 to Gly83 of HMGB2A.
In vivo DNA-looping assay
DNA-looping constructs were based on plasmid pJ992, created by modifications of pFW11-null (61) as previously described (14). Constructs contained a strong distal Osym operator and a weak proximal O2 operator. The O2 operator normally present within the lacZ coding region was destroyed by site-directed mutagenesis (14). A construct with a proximal O2 but lacking upstream Osym was used as a normalization control. Test promoters did not contain promoter CAP-binding sites. The lacZ looping constructs were placed on the single copy F128 episome by homologous recombination between the constructed plasmids and bacterial episome. F128 carries the lacI gene producing wild-type levels of repressor. Maintaining both the lacI gene and the target promoter in single copy ensures that the protein and DNA concentrations are as close to wild-type as possible. Bacterial conjugation and selections were performed as previously described (61). After mating and selection, correct recombinants were confirmed by PCR amplification.
Reporter assay
All chemicals were obtained from Sigma (St Louis, MO, USA). The lacZ expression was measured by a liquid β-galactosidase colorimetric enzyme assay as described (63). The assay was modified to increase efficiency. Cultures were grown in 1.1 ml LB/antibiotic in 96-well boxes (2 ml capacity per well) with shaking (250 r.p.m.) at 37°C. Fresh 1.1 ml aliquots of media were then inoculated with 30 μl of overnight culture in the presence or absence of 2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Subcultures were grown with shaking at 37°C until OD600 reached ∼0.3. For samples with low β-galactosidase activity, 800 μl of bacterial culture was assayed after centrifugation and resuspension in 1 ml Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, pH 7.0, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). For samples with high levels of β-galactosidase activity, 100 μl of bacterial culture was diluted with 900 μl of Z-buffer before analysis. Cells were lyzed by addition of 50 μl chloroform and 25 μl 0.1% sodium dodecyl sulfate (SDS), followed by repeated pipetting (10–12 times) with a 12-channel pipettor. Samples were equilibrated at 30°C for 5 min, followed by the addition of 200 μl of 4 mg/ml O-nitrophenylpyranogalactoside (ONPG) in Z-buffer. Incubation at 30°C continued with accurate timing until OD420 reached ∼0.5. Reactions were stopped with 500 μl 1 M Na2CO3 and the reaction time was recorded. Cell debris was pelleted by centrifugation of the 96-well box for 10 min at 4000g. A total of 350 μl of cleared samples were transferred to 96-well plates. Sample OD readings were measured on a Molecular Devices SpectraMax 340 microtiter plate reader. The β-galactosidase activity (E, in Miller units) was calculated according to the following equation:
| 1 |
where ODx refers to optical density at wavelength x, t is the reaction time (min), and v is the assay culture volume (ml). Assays were performed with a total of six colonies from each independent strain repeated on 2 different days.
Looping data analysis and modeling
The enhancement of repression due to specific DNA looping is expressed in terms of the normalized expression parameter E′, according to:
| 2 |
where EOsymO2 is the raw β−galactosidase activity (induced or uninduced) from test constructs with both Osym and O2 operators, and EO2 is the corresponding raw β−galactosidase activity (induced or uninduced) from test constructs with only the proximal O2 operator. Note that Equation (2) is corrected from the original report (14).
The conventional repression ratio, RR, is given by
| 3 |
where E is the raw β-galactosidase activity under the indicated conditions.
An extended version of a previously described statistical weights/DNA mechanics model (14,15) was used for simultaneous fitting of experimental E′ and RR data. The model describes the distribution of looped, singly bound and free states of the O2 operator under repressed and induced conditions. The experimentally derived fraction of O2 that is bound by repressor (fbound) is modeled as a function of DNA spacer length (sp) with six adjustable parameters. Three parameters describe properties mainly of the LacI:DNA complex: the optimal operator spacing in base pairs (spoptimal), the equilibrium constant for specific Osym−O2 loop formation when operators are perfectly phased (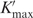 , replacing our previous Kmax as discussed below), and an equilibrium constant for nonspecific looping (KNSL). The KNSL term describes all forms of Osym-dependent enhanced binding to O2 other than the specific loop; for example, it could include looping between Osym and a pseudooperator overlapping O2, or enhanced O2 binding via sliding or hopping from Osym. Three fitting parameters describe mainly properties of the intervening DNA; the first two, as in our previous work, are the helical repeat (hr) and the apparent torsional modulus of the DNA loop (Capp). The qualitatively obvious length dependence of some of the data sets obtained here prompted us to add a third fitting parameter denoted Papp.
, replacing our previous Kmax as discussed below), and an equilibrium constant for nonspecific looping (KNSL). The KNSL term describes all forms of Osym-dependent enhanced binding to O2 other than the specific loop; for example, it could include looping between Osym and a pseudooperator overlapping O2, or enhanced O2 binding via sliding or hopping from Osym. Three fitting parameters describe mainly properties of the intervening DNA; the first two, as in our previous work, are the helical repeat (hr) and the apparent torsional modulus of the DNA loop (Capp). The qualitatively obvious length dependence of some of the data sets obtained here prompted us to add a third fitting parameter denoted Papp.
Papp is an empirical parameter motivated by the expected decrease in DNA-bending free energy as operator spacing sp increases, as given by the expression below:
| 4 |
where we have absorbed the DNA persistence length P, the thermal energy RT, and the extent of bending (ΔΘ) into a single constant; for simplicity, we make the doubtful assumption of constant loop geometry for all sp. We do not have an independent measurement of absolute bending free energy at any length, but the bending energy above will contribute an e−Papp/sp factor in the looping equilibrium constant Kmax. Therefore, the functional form of Equation (4) is combined with normalization of the length dependence by replacing Kmax in the fitting routines with the following expression:
| 5 |
where spavg is the mean spacing over the data set. Kmax, the value of 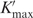 at sp = spavg can be compared directly to the Kmax obtained using our previous fitting routines. For the data sets here, values of Kmax determined as previously described without the use of Papp agree to within their estimated uncertainties with
at sp = spavg can be compared directly to the Kmax obtained using our previous fitting routines. For the data sets here, values of Kmax determined as previously described without the use of Papp agree to within their estimated uncertainties with 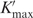 evaluated at spavg. None of the interpretations given here depend on the values of Papp except those that concern Papp itself. The relationship of Papp to a true in vivo DNA persistence length is complicated because the amount of curvature in the loop probably changes with spacing.
evaluated at spavg. None of the interpretations given here depend on the values of Papp except those that concern Papp itself. The relationship of Papp to a true in vivo DNA persistence length is complicated because the amount of curvature in the loop probably changes with spacing.
A second important motivation for the Papp parameter is that none of the other physical processes that we modeled can reproduce the observed increase in repression with increasing length. We were concerned that the multiparameter fitting procedure might lead to marked inaccuracies in the other values if the routines changed them to try to fit the length dependence. The results showed that this was not a serious problem, apparently because variations in the other parameters are simply unable to model the observations (not shown). However, we retained the Papp parameter in part because of the interesting trends in its values, as discussed subsequently.
Assay of protein expression
Derivatives of HMGB protein expression plasmid pJ1035 were transformed into BL643 (ΔHU) bacterial cells and grown on LB-Cb plates. Individual colonies were grown in 5 ml LB-Cb liquid cultures overnight at 37°C with aeration. Saturated overnight culture (1.5 ml) was pelleted by centrifugation, resuspended in 300 µl 2-(N-morpholino) ethanesulfonic acid (MES) buffer (50 mM MES, 50 mM Tris base, 3 mM SDS, 1 mM EDTA, pH 7.5) and cells were lyzed by sonication in bursts of 3 s. Samples were clarified by centrifugation and analyzed on 10% bis–Tris gels containing SDS by electrophoresis at 145 V for 45 min in 1× MES buffer. Protein was transferred to a nitrocellulose membrane using NuPAGE transfer buffer containing 20% methanol (30 V, 3 h, 4°C). The primary antibody for western blotting was specific for the c-Myc tag (op10T, CalBiochem, San Diego, CA, USA). Blots were blocked for 1 h in blocking buffer, incubated with primary antibody overnight at 4°C in blocking buffer and then incubated with secondary anti-mouse antibody at room temperature for 1 h. Imaging was performed with ECL Plus western blot detection (Amersham, Piscataway, NJ, USA).
RESULTS AND DISCUSSION
Experimental design
Our primary goals in this work were (i) to determine to what extent the eukaryotic HMGB proteins yeast Nhp6A and human HMGB2A can complement the repression looping defect observed in ΔHU E. coli cells, and (ii) to determine the importance of cationic amino acid terminal sequences of the HMGB proteins to any observed complementation. The experimental HMGB protein sequences are shown in Figure 1A. They contain decameric c-Myc epitope tags at the N-terminus (constructs 1200, 1299, 1492 and 1493) or C-terminus (constructs 1327 and 1328) as indicated in the designations. The initial tagged yeast Nhp6A construct is denoted 1200, and in 1299 there is a deletion of 11 of the native N-terminal amino acids of 1200. This deletion removes three of seven cationic amino acids from the Nhp6A N-terminal leader, adjacent to an N-terminal myc epitope tag that is anionic. To determine if the tag and its electrostatic character were important, Nhp6A constructs 1327 and 1328 were prepared with a C-terminal myc tag, with 1328 bearing the 11 amino acid N-terminal deletion relative to 1327. Constructs 1492 and 1493 contain human HMGB2 box A sequences with an N-terminal myc tag. The 1493 denotes a protein with a truncation of nine C-terminal residues (removing +3 net charge).
Both the Nhp6A and HMGB proteins have been shown to fold into L-shaped clusters of three helical domains with N- and C-terminal extensions (Figure 1B). In the case of Nhp6A, the cationic N-terminal extension appears to neutralize crowded phosphates in the compressed major groove of the bent DNA. Truncation of a similar segment from the sequence-specific LEF-1 HMGB protein significantly reduces DNA bending (44). The C-terminal extension of HMGB appears to be disposed to serve a similar charge neutralization function.
The ability of HMGB proteins to enhance the apparent flexibility of DNA and complement the DNA looping defect observed in ΔHU E. coli was assayed as in our previous work, as diagrammed in Figure 1C. Efficient repression by lac repressor tetramer requires DNA looping between weak (O2) and strong (Osym) operators that flank the promoter (P). Experimental measurement of the extent of DNA repression (compared to constructs where no Osym was present) was used to test the hypothesis that nonhomologous prokaryotic HU and eukaryotic HMGB proteins can increase the apparent flexibility of DNA similarly in vivo.
Expression of epitope-tagged HMGB proteins in E. coli
We measured the accumulation of HMGB deletion derivatives in E. coli using western blots. Surprisingly, preliminary experiments showed that any truncation of Nhp6A that removed amino acids of the RK3 sequence near the N-terminus, regardless of the presence or absence of an N-terminal myc epitope tag, completely eliminated protein accumulation. This result suggests that proper folding of Nhp6A requires these cationic residues. The western blot analysis of E. coli extracts in Figure 2 shows the relative levels of accumulation of those proteins that could be expressed and tested, normalized to constant total protein. The Nhp6A derivatives were expressed at comparable levels except that construct 1327 (Figure 2A, lane 3) accumulated to a much lower although clearly detectable level. The full box A of human HMGB2 and its C-terminal truncated derivative accumulated to high levels compared to Nhp6A construct 1328 (Figure 2B, compare lanes 2–3 with lane 1).
Figure 2.
Expression of eukaryotic HMGB derivatives in E. coli. Western blotting with anti-myc epitope antibodies showing protein extracts from E. coli cells carrying HMGB expression plasmids. (A) Proteins expressed at lower levels. Lanes 1–4 correspond to proteins 1200 (myc/Nhp6A), 1299 (myc/Nhp6AΔ(2–12), 1327 (Nhp6A/myc) and 1328 (Nhp6AΔ(2–12)/myc), respectively. (B) Proteins expressed at higher levels. Lanes 1–3 correspond to proteins 1328 [Nhp6AΔ(2–12)/myc; repeated from panel A for comparison], 1492 (myc/hHMGB2A) and 1493 [myc/hHMGB2AΔ(84–92)], respectively.
Complementation of ΔHU phenotype by HMGB2 proteins and truncation mutants
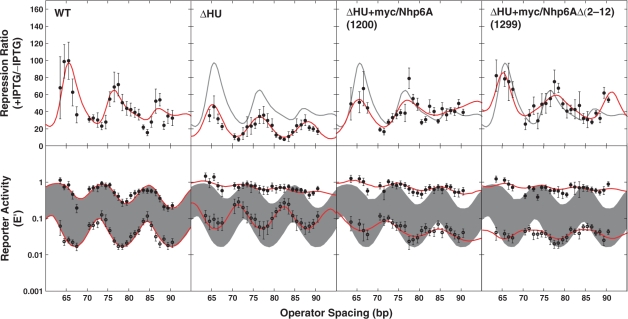
We measured DNA loop-dependent repression of lacZ in E. coli as reporter activity (expressed as E′, the activity normalized to controls with no upstream Osym operator) in the presence and absence of IPTG (Figures 3 and 4, bottom row). The ratio of induced to uninduced raw activities is reported as the repression ratio (Figures 3 and 4, top row). The results for wild-type E. coli (Figure 3, ‘WT’ column) are entirely consistent with our previous studies, showing oscillating repression dependent on operator spacing, even in the presence of IPTG, with the ±IPTG oscillations slightly shifted in phase and helical repeat so that the repression ratio is not sinusoidal (14,15). Looping was observed to be substantially disabled in ΔHU cells (Figure 3, ΔHU column), as we have previously reported (14,15). The resulting strain shows leaky repression of the lacZ reporter. Fitting parameters from the thermodynamic model of loop-dependent gene repression are given in Table 2, reflecting behavior consistent with our earlier work.
Figure 3.
DNA looping in vivo. Top row: the repression ratio (ratio of β-galactosidase activity under inducing versus noninducing conditions) is shown for E. coli WT, ΔHU and ΔHU strains expressing heterologous HMGB proteins. WT behavior is indicated by a shaded line in the three columns to the right. Bottom row: β-galactosidase activity normalized to controls lacking an upstream operator under inducing (upper curves: filled circles) or noninducing (lower curves: open circles) conditions. Error bars represent standard deviations based on replicate measurements. Curve fitting to the thermodynamic model is shown. Shading indicates uninduced and induced reporter expression from WT cells. Raw data are available at NAR Online.
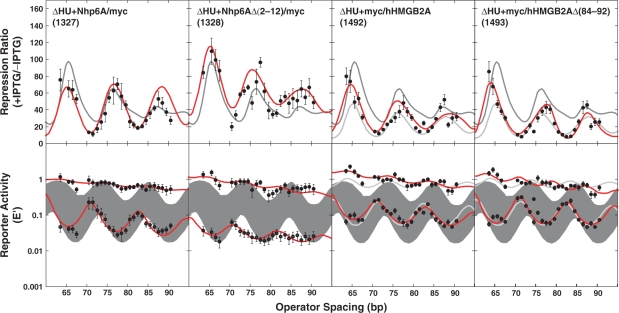
Figure 4.
DNA looping in vivo. The repression ratio and normalized activities are shown as in Figure 3 for additional tested strains. In columns three and four, light colored curves show ΔHU behavior for comparison. Considering length dependence explicitly clearly improves the fitting to the +IPTG E ′ data. Raw data are available at NAR Online.
Table 2.
Fitting parameters from thermodynamic DNA looping modela
| Strain |
spoptimal (bp) |
Capp (×10−19 erg cm) |
hr (bp/turn)b |
Kmax |
KNSLb |
Papp (bp)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | −IPTG | +IPTG | |
| WT+vec | 78.7 ± 0.3 | 79.3 ± 0.4 | 0.69 ± 0.13 | 0.89 ± 0.38 | 11.6 ± 0.3 | 11.0 ± 0.5 | 211 ± 103 | 2.5 ± 0.7 | (20.2) ± 21.0 | (0) ± 0.4 | (0) ± 188 | (238) ± 205 |
| ΔHU+vec | 76.8 ± 0.2 | 77.2 ± 1.0 | 0.66 ± 0.10 | 0.23 ± 0.08 | 11.1 ± 0.3 | (9.1) ± 1.2 | 65 ± 12 | 1.4 ± 0.4 | 8.8 ± 5.6 | (0) ± 0.8 | (10.4) ± 91.6 | (219) ± 304 |
| ΔHU+1327 | 77.0 ± 0.2 | 79.1 ± 2.3 | 0.53 ± 0.04 | 0.21 ± 0.13 | 11.6 ± 0.4 | (10.3) ± 2.5 | 113 ± 32 | 0.9 ± 0.3 | (0) ± 15 | (0) ± 0.7 | (160) ± 160 | (276) ± 376 |
| ΔHU+1328 | 76.7 ± 0.5 | 78.0 ± 1.4 | 0.30 ± 0.04 | 0.22 ± 0.09 | 11.0 ± 0.7 | (8.9) ± 1.5 | 171 ± 35 | 1.8 ± 0.7 | (0) ± 166 | (0) ± 0.8 | (114) ± 229 | (359) ± 398 |
| ΔHU+1200 | 78.7 ± 0.6 | 80.2 ± 0.9 | 0.32 ± 0.04 | 0.33 ± 0.12 | 12.9 ± 0.9 | 10.5 ± 1.1 | 87 ± 23 | 1.2 ± 0.4 | (0) ± 80 | (0) ± 0.5 | (168) ± 314 | (252) ± 330 |
| ΔHU+1299 | 78.2 ± 0.6 | 78.5 ± 1.2 | 1.21 ± 1.00 | 0.24 ± 0.11 | 13.2 ± 1.0 | (8.5) ± 1.3 | 112 ± 33 | 1.2 ± 0.5 | 62.5 ± 12.3 | (0) ± 2.4 | (8.1) ± 346 | (85.5) ± 497 |
| ΔHU+1492 | 77.1 ± 0.4 | 79.0 ± 1.5 | 0.42 ± 0.05 | 0.24 ± 0.10 | 12.0 ± 0.6 | (10.0) ± 1.5 | 108 ± 25 | 2.2 ± 0.7 | (0) ± 41 | (0) ± 0.8 | (90.4) ± 154 | (284) ± 320 |
| ΔHU+1493 | 76.7 ± 0.3 | 83.0 ± 0.9 | 0.55 ± 0.06 | 0.29 ± 0.08 | 11.9 ± 0.4 | (8.1) ± 0.6 | 103 ± 23 | 2.2 ± 1.0 | (3) ± 21 | (0) ± 1.2 | (22.0) ± 121 | (307) ± 442 |
aIndicated errors are 95% confidence limits. The values and ranges for Kmax, KNSL and Papp are fit only to expression data E+IPTG and E − IPTG. The values and ranges for spoptimal, Capp, and hr are fit to E+IPTG, E − IPTG and the repression ratio RR simultaneously, with Kmax, KNSL, and Papp held fixed.
bParentheses indicate a large error estimate, making the parameter value unreliable. However, trends ±IPTG are robust: upon induction spoptimal increases, Capp decreases, hr decreases, Kmax decreases, KNSL decreases or is negligible and Papp increases. When Capp is less than about 0.3, torsional oscillations are so small that hr cannot be estimated reliably.
The oscillations in E′ even after induction provide strong evidence for residual lac repressor looping. The ratio of Kmax values provides an estimate of ∼100–200 for the relative Kdiss values for the lac repressor:DNA loop ±IPTG, roughly in accord with in vitro results (64). On the other hand, it is perhaps surprising that the helical phase dependences ±IPTG differ (as in our previous work). There are several possible explanations: the level of static or dynamic supercoiling, and therefore the helical repeat, may differ depending on the frequency of transcription initiation. The stable −IPTG loop may have a different structure, so it may organize the local topological domain differently than the unstable +IPTG loop. The ±IPTG loops might sequester different architectural proteins, so there could be an indirect effect on helical repeat. Note that the change in the optimal spacing parameter seems to be a consequence of the change in helical repeat, (Figure 7), although this does not necessarily mean that the ±IPTG loops have the same structure.
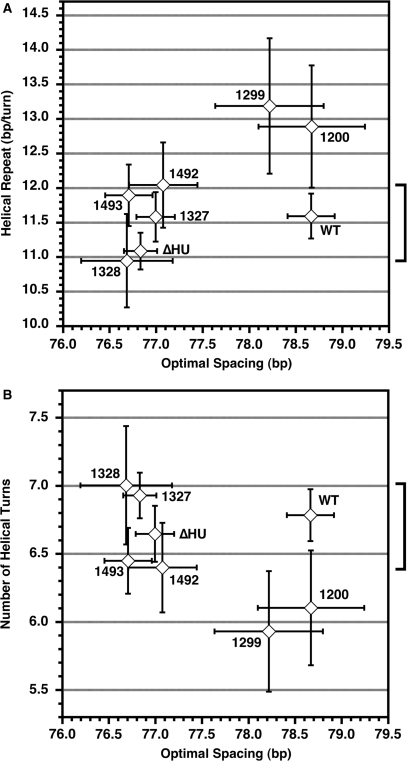
Figure 7.
Optimal spacing in terms of base pairs varies among bacterial strains but the optimal number of DNA helical repeats between operators is relatively consistent. (A) The range of helical repeat values from Table 2 (–IPTG only) versus the optimal loop spacing, with the bracket indicating that WT falls in the middle of the range of the well-determined helical repeats. (B) The number of helical turns between operators (calculated as optimal spacing/helical repeat) also clusters around the WT value. Error bars indicate 95% confidence limits on the parameters, with the estimated error in the number of helical turns being given by standard propagation of errors. The fitting parameters for constructs 1200 and 1299 are deemed unreliable, as discussed in the text.
The inclusion of a new length parameter Papp was motivated by the qualitatively obvious length dependence in several of the data sets reported here. This is especially clear under noninducing conditions. In every case, the fit value for Papp is larger for a given bacterial strain under inducing versus noninducing conditions, suggesting that the latter is more sensitive to length. This is consistent with the idea that because IPTG destabilizes DNA binding, bending free energy becomes relatively more important to overall loop stability. Note that this explicit consideration of the length (as opposed to torsion) dependence made little difference in the values of any of the other fit parameters. Also, this behavior contrasts with the decrease in Capp upon induction, which we have attributed to decreased LacI specificity or increased LacI flexibility.
Strong ΔHU complementation by four versions of yeast Nhp6A suggests twist flexibility
Expression of myc-tagged Nhp6A derivatives in ΔHU E. coli cells complemented the ΔHU deletion, showing dramatic rescue of DNA looping and lacZ repression. Improved repression is clearly seen for Nhp6A constructs 1200 and 1299 (Figure 3). Moreover, the presence of Nhp6A derivatives 1200 and 1299 strongly attenuated the face-of-the-helix dependence of repression, as reflected in the length dependence of both the E′ values in the absence of IPTG and the repression ratios (Figure 3, two right columns). This observation suggests that the ability of Nhp6A derivatives to enhance apparent DNA twist flexibility exceeds that of the E. coli HU protein. Physically, this could be caused by the binding of a variable number of Nhp6A proteins, which each induce about 30° of untwisting (40), or by variability in the extent of untwisting induced by protein binding; variability in bending and writhing has been demonstrated for HU (27) and suggested for TATA-binding protein (65). The HU protein stabilizes supercoiling via DNA bending and protein assembly leading to writhe (66). HU therefore appears to topologically unwind the DNA (note the increase in the apparent helical repeat), but does not damp torsional oscillations. The bending variability may be suppressed in the small loops found here or HU may bind with a defined stoichiometry. The HMGB protein structures in Figure 1B do not show obvious out of plane bending that would lead to writhe.
The fit parameters in Table 2 confirm the damping of torsional oscillations in 1200 and 1299 but, interestingly, the analysis suggests different root causes. For 1200, we find a dramatic decrease in the apparent torsional modulus Capp, consistent with variable extents of binding of an untwisting ligand. For 1299, the fit suggests a large value of KNSL, reflecting decreased binding specificity by LacI; this decreases torsional oscillations because the limits of the oscillations are KNSL and 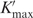 . In this case, we suspect that the KNSL value is an artifact of high uncertainties caused by very low absolute activities that may be influenced by background. In general, individual fit parameters are not precisely determined, so we seek to detect trends across the data sets. The fit values for helical repeat approach ∼13 residues/turn, consistent with untwisting, but we caution that these are highly uncertain because of the small amplitude of the torsional oscillations.
. In this case, we suspect that the KNSL value is an artifact of high uncertainties caused by very low absolute activities that may be influenced by background. In general, individual fit parameters are not precisely determined, so we seek to detect trends across the data sets. The fit values for helical repeat approach ∼13 residues/turn, consistent with untwisting, but we caution that these are highly uncertain because of the small amplitude of the torsional oscillations.
The 1299 results suggest that deletion of cationic residues in the 2–12 leader of Nhp6A does not negatively impact the function of Nhp6A in enhancing apparent DNA flexibility. As described earlier, it was impossible to assess Nhp6A deleted for the additional basic amino acid cluster at positions 13–16 because these variants did not accumulate in E. coli. Because positions 13–16 lie closest to the compressed major groove in the Nhp6A/DNA complex, it is these residues that may be important for full DNA bending (40,44).
To determine if ΔHU repression rescue by Nhp6A derivatives was influenced by the position of the short myc epitope tag, we compared constructs 1200 and 1299 with the corresponding Nhp6A derivatives 1327 and 1328, which have C-terminal rather than N-terminal tags. The repression results are shown in the first two columns of Figure 4 and in Table 2. Constructs 1327 and 1328 were remarkably effective in complementing the ΔHU phenotype of the E. coli test strain. Even though it was poorly expressed, 1327 nearly recapitulated WT behavior except for a marked shift in the optimum spacing for looping (discussed below). Repression was most dramatically enhanced by 1328, often being more efficient than in WT E. coli. Again the length-dependent oscillations in the repressed E′ and repression ratio values were strongly damped compared to WT (Figure 4, column 2, bottom). We note that the 1327 construct presumably has the highest N-terminal positive charge density of the four Nhp6A variants, whereas the 1299 construct has the lowest, so this charge density appears to correlate with the amplitude of torsional oscillations. It is possible that the loss of N-terminal charge neutralization leads to increased torsional flexibility in the binding site DNA. We have suggested a similar compensation mechanism for enhanced bending flexibility in the context of specific bHLH protein–DNA interaction (67).
Poor ΔHU complementation by two versions of human HMGB2 box A
Our recent single-molecule experiments with an isolated HMGB box domain (box A) derived from human HMGB2 (16,17) documented the ability of the protein to bind DNA in either a low-density mode or by saturating the DNA to form a filament. In the low-density mode, the protein profoundly increased the apparent DNA-bending flexibility. The ability of hHMGB2A to complement the DNA looping defect in vivo in ΔHU E. coli was compared to that of Nhp6A. The hHMGB2A constructs 1492 and 1493 (Figure 1A) were expressed with N-terminal myc epitope tags. Construct 1493 has a deletion of a C-terminal octapeptide containing five cationic residues (+4 net charge).
Interestingly, whereas Nhp6A derivatives effectively complemented the ΔHU looping defect, the hHMGB2 derivatives did not (Figure 4, last two columns; Table 2). The data points nearly overlap the behavior of the ΔHU mutant cells. We conclude that although the HMGB2 box A construct alters the apparent flexibility of DNA in vitro (16,17) and is expressed well in E. coli (Figure 2B), this protein differs from yeast Nhp6A in that it is unable to complement the E. coli ▵HU phenotype. This result is surprising in light of in vitro activity and the observation that expression of the rat HMGB1 protein (boxes A and B) partially complemented the E. coli ΔHU looping defect (14). Perhaps the human HMGB2A domain has some residual sequence specificity, interacts with other DNA-binding proteins, or is otherwise sequestered away from the lac promoter in vivo.
Other possible explanations for the lack of in vivo activity of HMGB2 box A derivatives involve amino acid sequence details. As shown in Figure 1A, box A derivatives of mammalian HMGB proteins lack the intercalating methionine residue in helix I found in box B derivatives and yeast Nhp6A (68). There is structural evidence from the Drosophila HMGD protein (69) and yeast Nhp6A protein (40) that this intercalating residue confers on box B domains the ability to more highly distort bound DNA, both by bending and twisting. In contrast, the box A domains appear to bind distorted DNA structures without causing additional distortion (41). HMG box B domains were reported to be superior to box A when substituting for HU in vitro (5). We therefore hypothesize that an intercalating residue in helix I is important for facilitating DNA flexibility in the E. coli in vivo DNA-looping assay.
The DNA looping parameter fits for 1492 and 1493 do show subtle differences from the ΔHU mutant (Table 2). The E′ values greater than 1 at short lengths accompanied by relatively large values of Papp suggest that the HMGB2A protein may destabilize short loops preferentially, and increased values for the helical repeat relative to WT and ΔHU suggest some DNA unwinding.
It is important to note that the low accumulation of C-terminally myc-tagged Nhp6A (Figure 2 compare lane 3 to lane 1) was nonetheless sufficient to complement the looping defect in ΔHU cells. This suggests that levels of expressed protein are not limiting for these HMGB proteins. This result further supports the conclusion that box A derivatives of HMGB2 show a qualitative, rather than a quantitative, failure to complement the looping defect. We have previously performed more rigorous quantitation of mammalian HMGB proteins expressed in E. coli from the same promoters in similar bacterial strains (14). In that study, we determined that the eukaryotic protein accumulated to ∼4000 copies per cell. The present recombinant protein levels appear to be comparable and sufficient. In contrast, endogenous HU protein is reported to accumulate to ∼50 000 copies per cell during log phase growth (8).
Promoter/operator spacing does not alter optimal loop lengths
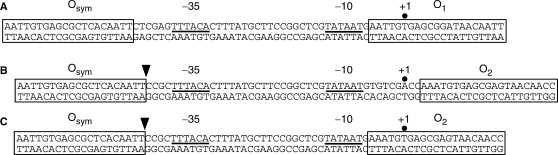
During the course of this work, we were again concerned by the discrepancy between operator spacing optima in our experiments versus those previously reported by Muller et al. (52) in their elegant study of a similar recombinant promoter. The earlier work reported maximal repression ratios for operator spacings of 59.5, 70.5, 81.5 and 92.5 bp, apparently counted as in our designs. This compares with maximal repression ratios at 65.5, 76 and 87 bp in our work (Figure 3, WT). This difference of ∼5 bp is puzzling. The maximal repression in the E′ data (Figure 3, WT, bottom panel) occurs at 67, 78 and 90 bp, a difference of ∼3 bp from the Muller work. We emphasize that the experiments of Muller et al. used a system in which repression resulted from two or five times the wild-type level of lac repressor and induction represented the complete absence of both lac repressor and residual looping, giving at least 20-fold greater repression ratios than we observe. Also, we have shown that IPTG-saturated repressor still participates in residual loops (14), and dephasing of the free and IPTG-bound repression curves contributes to the complex repression ratio trace that we observe. However, none of these differences provides an obvious explanation for the shift in operator spacings for optimal repression. One possible resolution might be a subtle difference in the promoter design between Muller et al. (52) and our system (14). The Muller work positioned the weak O1 sequence immediately flanking the −10 promoter sequence, with the reporter transcript predicted to initiate within O1 (Figure 5A). In contrast, our design places the weak O2 operator 9 bp further downstream, with transcription initiation just upstream of O2 (Figure 5B). We considered the possibility that different DNA loop regimes might be supported by our promoter design if, for example, RNA polymerase could be trapped between the lac operators in our design (Figure 5A), but not in the design of Muller et al. (Figure 5B).
Figure 5.
Alternate promoter/operator arrangements tested to explore whether promoter overlap with O2 influences optimal loop lengths. (A) Arrangement tested by Muller et al. (52) with 57.5 bp operator spacing shown. (B) Original promoter/operator spacing in our studies (14) with 63.5 bp operator spacing shown. (C) Design tested to simulate arrangement in (A), with 55.5 bp spacing shown. Arrowheads in (B) and (C) indicate locations of inserted sequences that alter operator spacing.
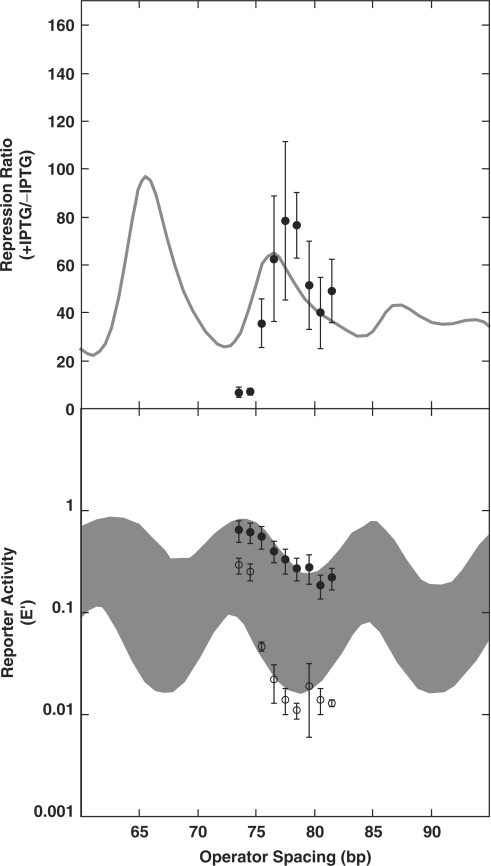
To test this idea, we created a series of DNA-looping constructs with the promoter design shown in Figure 5C. This design closely mimics the promoter/operator spacing of Muller et al., while retaining the weaker O2 operator in the proximal position. Figure 6 shows data for a series of operator spacings, compared to previous repression measurements. The maximal repression ratio is at an operator separation of 77 bp, and the maximal repression in the absence of IPTG occurs at a separation of 79 bp. These parameters are within ∼1 bp of our previous measurements. We conclude that changing the promoter/operator arrangement in the lac promoter does not influence optimal operator separation and cannot explain the discrepancy between the optima in our reports and that of Muller et al. In part, because of the unavailability of the earlier reporter constructs for sequence verification, it is unlikely that this discrepancy will be resolved definitively.
Figure 6.
Changing promoter/operator spacing does not alter optimal loop lengths for repression. Top: repression ratio for WT cells with spacing shown in (C) of Figure 5 (filled circles) versus spacing shown in (B) of Figure 5 (shaded line). Below: reporter gene expression E ′ levels for uninduced (open circles) and induced (filled circles) cells with the promoter/operator arrangements shown in Figure 5C versus those in Figure 5B (curves and filled region). Data are available at NAR Online.
Our current results do suggest that changes in optimal operator spacing may be quite common under different conditions. Figure 7 shows the optimal spacing for each of the constructs tested here versus the estimated helical repeat, yielding the calculated number of DNA helical turns between operators. The results show that the optimal spacing varies significantly depending on the different DNA-binding proteins, but that the number of helical turns at optimal spacing clusters around 6.75, both for WT and all of the variants where the number of helical turns could be measured with reasonable precision. It is clear that small changes in helical repeat can shift the optimal spacing, but the results suggest that the loop geometry is relatively constant in the different strains. The results of Muller et al. would give 70.5 bp/(11.0 bp/turn) = 6.4 turns, at the low end of the range of values observed here, but not an obvious outlier. It remains possible that there was a subtle difference in DNA supercoiling in E. coli strains tested in the earlier work. Thus, meaningful comparisons between fit parameters can only be made within sets of otherwise very similar strains and constructs.
SUMMARY AND IMPLICATIONS
The present experiments build upon the classic work of Müller–Hill (49,52) and Record (48,50,51), as well as our more recent studies (14,15). DNA repression looping in living E. coli provides a powerful system for observing the apparent physical properties of DNA in vivo. This work has suggested that some properties of the nucleoid confer on DNA enhanced apparent torsional and longitudinal flexibilities (14,48,50). We have argued that the sequence-nonspecific architectural protein HU plays an important role, though no defined HU-binding site is present in looped lac promoter DNA (14). Indeed, it has been suggested that the measured properties of DNA loops in ΔHU E. coli are close to the expectations for naked DNA in vitro (55), although there are other E. coli proteins present that can also affect looping.
The detailed fitting routines that we have developed are useful for identifying subtle trends in the data and for enabling numerical comparisons of qualitative trends. We suggest that in this case the analysis has contributed to three insights: (i) subtle increases in the helical repeat coupled with decreased torsional oscillations are consistent with the known untwisting induced by HMGB proteins; (ii) the relatively unstable +IPTG loop is much more sensitive to DNA persistence length than the more stable –IPTG repression loop and (iii) changes in the levels of DNA-binding proteins and the strain background can influence the optimal length of the LacI loop but perhaps not its geometry.
The results presented here also support and extend the previous observation that defects in E. coli nucleoid proteins can be complemented by eukaryotic HMGB proteins (14,18,60). Such complementation suggests HMGB restoration of some fundamental property of DNA packaging that does not depend upon protein homology, as HU and HMGB proteins employ very different DNA-bending mechanisms. We suggest that it is the ability of both HMGB proteins and HU to introduce sequence-nonspecific sites of DNA bending or flexure that explains their functional interchangeability. The present data demonstrate this effect specifically for the case of stabilizing small DNA repression loops closed by lac repressor tetramer. We have shown that complementation of the ΔHU defect in E. coli DNA looping can be provided by expression of a two-box rat HMGB protein (14) or by the yeast Nhp6A HMGB protein (this work), but not by an isolated HMG box A domain from human HMGB2, despite the fact that the latter protein shows strong DNA-kinking properties in vitro (16,17). It is unknown why the human HMGB2 derivative, though highly expressed in E. coli, did not functionally complement the ΔHU looping defect in the present study. It is possible that some subtle feature of this human protein domain distinguishes it from the corresponding rat and yeast proteins tested here, making it incompatible with function in the E. coli nucleoid.
In the cases where we have shown HMGB complementation of the ΔHU looping defect, removal of HMGB cationic tails does not block complementation. Complete removal of the Nhp6A cationic leader was not compatible with protein expression in E. coli, leaving open the possibility that three crucial cationic amino acids positioned in the compressed major groove of bent DNA are essential for function, as has been suggested by the additional DNA bending provided by such residues in vitro (44).
Despite the observation that isolated HMGB proteins can enhance the apparent flexibility of DNA molecules in vitro and in E. coli, the natural function of HMGB proteins remains mysterious in vivo (18,70–74). The mechanistic redundancy of HMGB proteins has limited the ability to interpret phenotypes of organisms with individual HMGB gene disruptions (75). Saccharomyces cerevisiae contains at least ten HMGB proteins (76). The mechanism of even the relatively well-studied yeast Nhp6A/B proteins remains obscure. Nhp6A/B are weakly bound as part of the FACT complex that remodels nucleosomes and facilitates transcription elongation (70,71), but it is unclear if this is their dominant role (72). It has recently been shown that loss of a different yeast HMGB protein, Spt2, increases the functional range of transcription activation (77). It is tempting to speculate that increased apparent DNA stiffness would produce such an effect, but it is not clear why the Spt2 defect would create such a clear phenotype in the presence of other yeast sequence-nonspecific HMGB proteins. Just as there is evidence that the E. coli HU protein may have a histone-like role in DNA compaction (66,78), in addition to its ability to facilitate DNA looping, it is important to consider that mammalian HMGB proteins may also be multifunctional.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Mayo Foundation and by NIH Grants GM054411 and GM075965 to L.J.M. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM075965.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hagerman PJ. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 2.Williams LD, Maher LJ., III Electrostatic mechanisms of DNA deformation. Annu. Rev. Biophys. Biomol. Struct. 2000;29:497–521. doi: 10.1146/annurev.biophys.29.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Crothers DM. Architectural elements in nucleoprotein complexes. Curr. Biol. 1993;3:675–676. doi: 10.1016/0960-9822(93)90065-v. [DOI] [PubMed] [Google Scholar]
- 4.Garcia HG, Grayson P, Han L, Inamdar M, Kondev J, Nelson PC, Phillips R, Widom J, Wiggins PA. Biological consequences of tightly bent DNA: the other life of a macromolecular celebrity. Biopolymers. 2007;85:115–130. doi: 10.1002/bip.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 6.Travers AA, Ner SS, Churchill MEA. DNA chaperones: a solution to a persistence problem. Cell. 1994;77:167–169. doi: 10.1016/0092-8674(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 7.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 9.Luijsterburg MS, Noom MC, Wuite GJL, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Harmer T, Wu M, Schleif R. The role of rigidity in DNA looping-unlooping by AraC. Proc. Natl Acad. Sci. USA. 2001;98:427–431. doi: 10.1073/pnas.98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lobell RB, Schleif RF. DNA looping and unlooping by AraC protein. Science. 1990;250:528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- 12.Matthews KS. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilar JM, Saiz L. DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr. Opin. Genet. Dev. 2005;15:136–144. doi: 10.1016/j.gde.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Becker NA, Kahn JD, Maher LJ., III Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005;349:716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 15.Becker NA, Kahn JD, Maher LJ., III Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007;35:3988–4000. doi: 10.1093/nar/gkm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCauley M, Hardwidge PR, Maher LJ, III, Williams MC. Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys. J. 2005;89:353–364. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCauley MJ, Zimmerman J, Maher LJ, III, Williams MC. HMGB binding to DNA: single and double box motifs. J. Mol. Biol. 2007;374:993–1004. doi: 10.1016/j.jmb.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paull TT, Johnson RC. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. J. Biol. Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 19.Ross ED, Hardwidge PR, Maher LJ., III HMG proteins and DNA flexibility in transcription activation. Mol. Cell. Biol. 2001;21:6598–6605. doi: 10.1128/MCB.21.19.6598-6605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross ED, Keating AM, Maher LJ., III DNA constraints on transcription activation in vitro. J. Mol. Biol. 2000;297:321–334. doi: 10.1006/jmbi.2000.3562. [DOI] [PubMed] [Google Scholar]
- 21.Stros M. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains – effect of flanking sequences. J. Biol. Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 22.Bonnefoy E, Takahashi M, Yaniv JR. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J. Mol. Biol. 1994;242:116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- 23.Hodges-Garcia Y, Hagerman PJ, Pettijohn DE. DNA ring closure mediated by protein HU. J. Biol. Chem. 1989;264:14621–14623. [PubMed] [Google Scholar]
- 24.Lewis DE, Geanacopoulos M, Adhya S. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol. Microbiol. 1999;31:451–461. doi: 10.1046/j.1365-2958.1999.01186.x. [DOI] [PubMed] [Google Scholar]
- 25.Pontiggia A, Negri A, Beltrame M, Bianchi ME. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 26.Skoko D, Wong B, Johnson RC, Marko JF. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 27.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lia G, Bensimon D, Croquette V, Allemand JF, Dunlap D, Lewis DE, Adhya S, Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. Histone-like protein HU and bacterial DNA topology: suppression of an HU deficiency by gyrase mutations. J. Mol. Biol. 1996;256:66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 33.Rouviere-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 34.Bianchi ME. Prokaryotic HU and eukaryotic HMG1: a kinked relationship. Mol. Microbiol. 1994;14:1–5. doi: 10.1111/j.1365-2958.1994.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 35.Allain FH, Yen YM, Masse JE, Schultze P, Dieckmann T, Johnson RC, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardman CH, Broadhurst RW, Raine AR, Grasser KD, Thomas JO, Laue ED. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1995;34:16596–16607. doi: 10.1021/bi00051a007. [DOI] [PubMed] [Google Scholar]
- 37.He Q, Ohndorf UM, Lippard SJ. Intercalating residues determine the mode of HMG1 domains A and B binding to cisplatin-modified DNA. Biochemistry. 2000;39:14426–14435. doi: 10.1021/bi001700j. [DOI] [PubMed] [Google Scholar]
- 38.Jamieson ER, Jacobson MP, Barnes CM, Chow CS, Lippard SJ. Structural and kinetic studies of a cisplatin-modified DNA icosamer binding to HMG1 domain B. J. Biol. Chem. 1999;274:12346–12354. doi: 10.1074/jbc.274.18.12346. [DOI] [PubMed] [Google Scholar]
- 39.Jung Y, Lippard SJ. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry. 2003;42:2664–2671. doi: 10.1021/bi026972w. [DOI] [PubMed] [Google Scholar]
- 40.Masse JE, Wong B, Yen YM, Allain FH, Johnson RC, Feigon J. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 41.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 42.Stott K, Tang GS, Lee KB, Thomas JO. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 2006;360:90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 43.Weir HM, Kraulis PJ, Hill CS, Raine ARC, Laue ED, Thomas JO. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill ME, Crane-Robinson C, Privalov PL. DNA binding and bending by HMG boxes: energetic determinants of specificity. J. Mol. Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Lorenz M, Hillisch A, Payet D, Buttinelli M, Travers A, Diekmann S. DNA bending induced by high mobility group proteins studied by fluorescence resonance energy transfer. Biochemistry. 1999;38:12150–12158. doi: 10.1021/bi990459+. [DOI] [PubMed] [Google Scholar]
- 46.Manning GS, Ebralidse KK, Mirzabekov AD, Rich A. An estimate of the extent of folding of nucleosomal DNA by laterally asymmetric neutralization of phosphate groups. J. Biomol. Struct. Dyn. 1989;6:877–889. doi: 10.1080/07391102.1989.10506519. [DOI] [PubMed] [Google Scholar]
- 47.Strauss JK, Maher LJ., III DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 48.Bellomy G, Mossing M, Record M. Physical properties of DNA in vivo as probed by the length dependence of the lac operator looping process. Biochemistry. 1988;27:3900–3906. doi: 10.1021/bi00411a002. [DOI] [PubMed] [Google Scholar]
- 49.Kramer H, Niemoller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Law SM, Bellomy GR, Schlax PJ, Record MT., Jr In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J. Mol. Biol. 1993;230:161–173. doi: 10.1006/jmbi.1993.1133. [DOI] [PubMed] [Google Scholar]
- 51.Mossing MC, Record MT., Jr Upstream operators enhance repression of the lac promoter. Science. 1986;233:889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- 52.Muller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 53.Oehler S, Eismann ER, Kramer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, McEwen AE, Crothers DM, Levene SD. Statistical-mechanical theory of DNA looping. Biophys. J. 2006;90:1903–1912. doi: 10.1529/biophysj.105.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, McEwen AE, Crothers DM, Levene SD. Analysis of in-vivo LacR-mediated gene repression based on the mechanics of DNA looping. PLoS ONE. 2006;1:e136. [Google Scholar]
- 56.Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 57.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol. Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 58.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc. Natl Acad. Sci. USA. 2005;102:5397–5402. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maher LJ., III DNA kinks available … if needed. Structure. 2006;14:1479–1480. doi: 10.1016/j.str.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Megraw TL, Chae CB. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 1993;268:12758–12763. [PubMed] [Google Scholar]
- 61.Whipple FW. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller J. New York: Cold Spring Harbor Laboratory Press; 1992. A Short Course in Bacterial Genetics. [Google Scholar]
- 64.O’Gorman RB, Rosenberg JM, Kallai OB, Dickerson RE, Itakura K, Riggs AD, Matthews KS. Equilibrium binding of inducer to lac repressor.operator DNA complex. J. Biol. Chem. 1980;255:10107–10114. [PubMed] [Google Scholar]
- 65.Kahn JD. Topological effects of the TATA box binding protein on minicircle DNA and a possible thermodynamic linkage to chromatin remodeling. Biochemistry. 2000;39:3520–3524. doi: 10.1021/bi992263f. [DOI] [PubMed] [Google Scholar]
- 66.Guo F, Adhya S. Spiral structure of Escherichia coli HUalphabeta provides foundation for DNA supercoiling. Proc. Natl Acad. Sci. USA. 2007;104:4309–4314. doi: 10.1073/pnas.0611686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald RJ, Kahn JD, Maher LJ., III DNA bending by bHLH charge variants. Nucleic Acids Res. 2006;34:4846–4856. doi: 10.1093/nar/gkl552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 69.Murphy FVT, Sweet RM, Churchill ME. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brewster NK, Johnston GC, Singer RA. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 2001;21:3491–3502. doi: 10.1128/MCB.21.10.3491-3502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome- binding factor SPN. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laser H, Bongards C, Schüller J, Heck S, Johnsson N, Lehming N. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl Acad. Sci. USA. 2000;97:13732–13737. doi: 10.1073/pnas.250400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreira JM, Holmberg S. Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J. 2000;19:6804–6813. doi: 10.1093/emboj/19.24.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paull TT, Carey M, Johnson RC. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 75.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein HMG1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Gen. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 76.Kamau E, Bauerle KT, Grove A. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J. Biol. Chem. 2004;279:55234–55240. doi: 10.1074/jbc.M409459200. [DOI] [PubMed] [Google Scholar]
- 77.Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kar S, Edgar R, Adhya S. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl Acad. Sci. USA. 2005;102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.