Abstract
Study of the effects of pressure on macromolecular structure improves our understanding of the forces governing structure, provides details on the relevance of cavities and packing in structure, increases our understanding of hydration and provides a basis to understand the biology of high-pressure organisms. A study of DNA, in particular, helps us to understand how pressure can affect gene activity. Here we present the first high-resolution experimental study of B-DNA structure at high pressure, using NMR data acquired at pressures up to 200 MPa (2 kbar). The structure of DNA compresses very little, but is distorted so as to widen the minor groove, and to compress hydrogen bonds, with AT pairs compressing more than GC pairs. The minor groove changes are suggested to lead to a compression of the hydration water in the minor groove.
INTRODUCTION
Studying the effect of pressure upon DNA is important not only to deepen our understanding of expression and regulation of transcription under pressure, but also because pressure affects molecules via the partial molar volume of the system, and therefore provides information on hydration that is hard to obtain in other ways. For example, the binding of proteins and drugs to double-stranded B-DNA is weaker at high hydrostatic pressure (1), although the overall conformation of DNA is thought to alter very little with pressure (2). Moreover, the activity and selectivity of enzymes that act upon DNA, such as RNA polymerase, depend upon the hydrostatic pressure (1,3,4), such that for example restriction endonucleases become less active but more specific at high pressure, in that they have reduced non-specific ‘star’ activity (5,6). Are these effects due to changes to DNA, changes to the proteins or both? Because protein–DNA recognition often relies on deformation of the DNA, it is also important to understand how DNA deforms under pressure (7). This observation has been given increased importance by the discovery of deep-sea organisms living at pressures of up to 50 MPa (1 bar = 105 Pa ≈ 1 atm), some of which are strict piezophiles, i.e. they grow only at elevated pressure, while others grow equally well at atmospheric pressure (8). Pleiotropic effects on gene transcription have been observed in such organisms at elevated pressure, and an understanding of these effects will help us understand transcription regulation more generally (9).
Macromolecules in aqueous solution respond to pressure by reducing the partial molar volume of the system. The partial molar volume is the sum of the intrinsic or molecular volume and the ‘interaction volume’ (10), which is the (negative) effect of the macromolecule on the volume of the water that solvates the macromolecule: effectively, the presence of the macromolecule leads to a compression of the hydration layer around it, particularly in the vicinity of charges. The effect of pressure on solutions of macromolecules therefore provides information on hydration. It also gives information on volume fluctuations within the macromolecule since there is a thermodynamic relationship between volume fluctuation and compressibility (11).
In order to better understand such observations, and to increase our understanding of DNA hydration, we have investigated the conformational changes in a DNA hairpin on increasing the pressure from ambient to 200 MPa (2 kbar). The hairpin used was d(CTAGAGGATCCTUTTGGATCCT), in which the underlined residues form a helical stem, which has a standard B-DNA structure capped by a tetraloop (12). The results demonstrate that the DNA changes its structure very little. There is a slight reduction in the Watson–Crick hydrogen-bond distance, this reduction being greater for the AT than the GC base pairs, but otherwise there is almost no compression of the DNA, as expected from its high packing density and lack of internal cavities. The major structural change is a widening of the minor groove, most likely to allow the hydration water to occupy a smaller volume. This implies that changes in transcriptional activity with pressure are much more likely to be due to the protein component than to the DNA.
MATERIALS AND METHODS
The hairpin DNA was chemically synthesized by Ransom Hill Bioscience, Inc. (Ramona, CA, USA), gel purified and desalted (12), and dissolved in 50 mM tris, 0.5 mM EDTA, pH 7.0. The assignment of the homonuclear NMR spectrum has been described (12). NMR spectra were acquired at a range of pressures from ambient (using 3 MPa rather than 0.1 MPa, to avoid small air bubbles in the solution which degrade the NMR spectra) up to 200 MPa, in steps of 50 MPa. NMR measurements were carried out on a standard Bruker DMX-750 spectrometer into which was fitted a high-pressure cell constructed from quartz glass of outer diameter 3 mm, which could be pressurized using a hand pump connected to the high-pressure cell by stainless steel tubing (13). Chemical shift changes were linear with pressure. The data were therefore fitted to a linear dependence, from which the change in shift between ambient pressure and 200 MPa was obtained.
The methodology used for calculation of the high-pressure structure is based upon our previous use of NMR chemical shift changes to characterize structural changes in proteins with pressure (14–16), modified for calculation of DNA. In outline, we have earlier derived equations that relate structure to chemical shift (17,18). Using these equations, and starting from a known structure, restrained molecular dynamics (MD) can be applied to use the pressure-dependent change in chemical shift to calculate a change in structure. This methodology yields much more accurate structural changes than the more traditional nuclear overhauser effects and coupling constants, since chemical shifts are sensitive to structural changes on the order of hundredths of an ångström. The MD is used as an efficient way of reaching a minimally altered structure that matches the observed chemical shift changes, and hence the lowest possible temperature is used in the calculation to avoid perturbing the structure more than is necessary. For DNA, we used ring current shifts (19) and electric field effects only. Bond magnetic anisotropies were not used, because the overall effects of bond magnetic anisotropies in the bases are calculated reasonably well by ring current effects except at short distances; and because in nucleic acids (unlike proteins), bond magnetic anisotropies from double bonds do not have C2v symmetry, which makes them much more troublesome to use as restraints (20). As was done for the earlier protein calculations, restraints on hydrogen-bonded protons were calculated using electric field effects and ring currents only (i.e. no bond magnetic anisotropies), except that the partial atomic charges used were those appropriate for imino protons. This method was found to be the most accurate for proteins (18) and should be even better for nucleic acids because of the presence of smaller bond anisotropies.
The structure was refined in the standard full XPLOR force field, i.e. containing van der Waals forces (a Lennard–Jones potential out to 4 Å, decaying smoothly to zero at 5 Å) and coulombic forces with a 1/r-dependent dielectric constant (i.e. a distance-dependent dielectric of 4r) (21), switched off between 4 and 5 Å, using standard charges for DNA as listed in the XPLOR file topnshle.dna. All masses were set to 100 throughout the calculation. In addition to this, it was found necessary to add restraints on the sugar dihedral angles, base-pair planarity and base-pair hydrogen-bond lengths, to stop the structure distorting unreasonably during equilibration. For the planarity restraints, all atoms in a given base (except methyl hydrogens) were restrained to be in the same plane. Each pair of paired bases was also restrained to be in the same plane—i.e. the C2, C5 and N9 of adenine or guanine and the N1, N3 and C5 of the paired thymine or cytosine were restrained to the same plane. Planarity restraints were based on those used in the XPLOR example file brestraints.inp and used a weight of 400. For the hydrogen-bond restraints, base-pair hydrogen bonds were restrained to their initial distances ±0.3 Å. Sugar dihedral restraints were used to keep the ring puckered.
The first four unpaired bases of the experimental sequence were not used in the structure calculation. The starting NMR structure 1dgo of this hairpin from the Protein Data Bank (22) was refined iteratively until convergence, i.e. until further rounds of calculations produced no significant structural change. This was carried out in two stages. First, the NMR structure was subjected to 50 rounds of MD at 100 K, 10 times in parallel, to give 10 ‘multi-refined’ structures MREF1-10, which were averaged. Then, starting from the averaged MREF structure, further cycles of MD were conducted using the same procedure (except that only 25 rounds of MD were used rather than 50), to produce a ‘continued refined’ structure CREF1. This was repeated four more times to generate CREF2 and so on up to CREF5. For each round of calculation, the starting structure was followed for 2000 steps of 0.003 ps followed by 2000 steps of 0.002 ps; and this trajectory calculation was repeated nine more times and an average was calculated. The average is used as the starting structure for the next round. In practice, the MREF stage reached stability after approximately 20 rounds, and all five CREF structures are virtually identical, implying convergence has been reached already by the first structure. Therefore, CREF1 was in fact used as the starting structure for calculation of low- and high-pressure structures.
To calculate the low-pressure structure, an MD trajectory was calculated using the same force field, with the addition of a chemical shift restraint such that all protons to which chemical shift restraints were applied in the high-pressure calculation were restrained to a chemical shift change of 0 p.p.m. The chemical shift restraints were added using a strong force constant of 4000 kcal mol−1 p.p.m.−2, as in our earlier studies (14,15). The purpose of this calculation is to act as a reference for comparison to the high-pressure calculation. To calculate the high-pressure structure, CREF1 was again used as the starting structure, but this time pressure-dependent chemical shift changes (listed in Supplementary Material) were applied as restraints. The restraints were applied to 96 protons (37 base protons and 59 sugar protons). Shifts from nucleotides in the tetraloop were not used as restraints.
DNA structural parameters were calculated using the program 3DNA (23). Molecular volumes were calculated using the program VOIDOO (24), which in our experience is much more reproducible and reliable for calculating volumes (of cavities in particular) than other methods such as Voronoi volumes and Swiss-PdbViewer.
RESULTS
Chemical shifts were measured for the B-DNA hairpin d(CTAGAGGATCCTUTTGGATCCT) using homonuclear 2D spectra at pressures from 3 MPa to 200 MPa. The observed chemical shift changes are linear with pressure. This has two important implications: first, that the compressibility is essentially independent of pressure; and second, that pressure does not favour the presence of different structures (as it does, for example with both lysozyme and protein G, where the presence of some curved pressure dependences demonstrated an increased population of alternative structures with water molecules inserted into cavities in the proteins) (14,15,25,26). The fact that compressibility is independent of pressure further implies that volume fluctuations of the DNA are not affected by an increase in pressure, because of the thermodynamic relationship mentioned above (11).
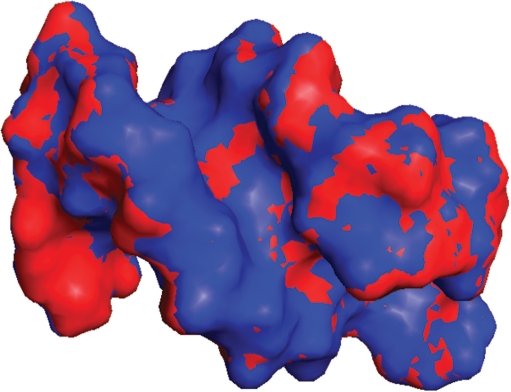
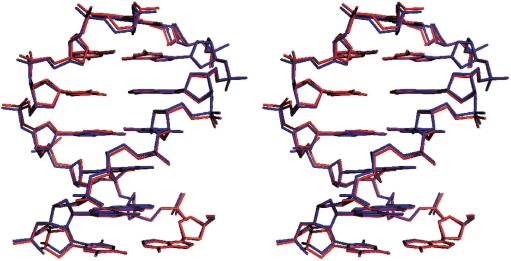
The changes in chemical shift from low to high pressure were used as restraints to calculate the change in structure with increase in pressure. Structural changes were calculated for the B-DNA stem only. The NMR structure 1dgo was energy minimized to produce a ‘starting structure’ CREF1. This structure was then subjected to three different low-temperature MD calculations: one with no chemical shift restraints, one with chemical shift restraints designed to keep the structure unchanged and one with shift restraints matching those observed on going from low to high pressure. The calculations with no or unchanged restraints had very little effect on the structure, as expected, but the high-pressure chemical shifts caused a change in structure of 0.17 Å (Table 1). The chemical shift restraints were satisfied in the resultant high-pressure structures, giving an root mean square (RMS) difference between calculated and observed shift in the high-pressure structure of 0.007 p.p.m., which is within the experimental error. The structural changes are described in Table 2, and the high- and low-pressure structures have been deposited with the Research Collaboratory of Structural Bioinformatics Protein Data Bank with code numbers 2vai and 2vah, respectively. The low- and high-pressure structures are shown as a surface representation in Figure 1 and in a stereo stick representation in Figure 2.
Table 1.
RMS distances between structures
| RMS to minimized 1DGO |
RMS to CREF1 |
|||||||
|---|---|---|---|---|---|---|---|---|
| CREF1 | No Shiftsa | Low P | High P | No shiftsa | Low P | High P | RMS to low P | |
| Aligned on base pairs | 1.544 | 1.545 | 1.556 | 1.575 | 0.034 | 0.092 | 0.170 | 0.171 |
| Aligned on P and C1′ | 2.386 | 2.389 | 2.378 | 2.495 | 0.048 | 0.069 | 0.248 | 0.247 |
aStructure computed the same way as the low- and high-pressure structures except that no proton shift term is included in the restraints.
Table 2.
Structural changes in the DNA double-stranded stem between 3 and 200 MPa (30 and 2000 bar)
| Low P | High P | % change | |
|---|---|---|---|
| Vol, 0 Å probe (Å3 × 103) | 3.330 | 3.328 | −0.06 |
| Vol, 1.4 Å probe (Å3 × 103) | 7.174 | 7.171 | −0.042 |
| Izza | 5630 | 5724 | +1.7 |
| Ixxa | 947 | 918 | −3.1 |
| Iyya | 704 | 676 | −4.0 |
| Stem length (Å)b | 18.84 | 19.06 | +1.2 |
| Mean rise (Å) | 2.85 | 2.87 | +0.55 |
aMoments of inertia. The z axis runs along the DNA helical axis.
bCalculated as the distance between the two terminal base pairs (calculating mean coordinates for the two pairs).
Figure 1.
Surface representation of the structure of the DNA hairpin, viewed from the minor groove. Low-pressure structure is blue and high-pressure is red.
Figure 2.
Stereo superposition of the low-pressure (blue) and high-pressure (red) DNA structures.
The change in structure of 0.17 Å is at the lower end of the range of structural changes seen in earlier calculations on proteins (14–16). The volume of the high-pressure structure is only 0.042% smaller than that of the low-pressure structure (based on the non-terminal stem bases only), corresponding to an intrinsic compressibility of ca. 0.6 × 10−4 ml mol−1 bar−1 per nucleotide. This is small compared to typical adiabatic molar compressibilities measured for DNA solutions (30–70 × 10−4 ml mol−1 bar−1) (27), suggesting that the DNA molecule itself is almost incompressible, and that almost all of the compressibility of the DNA solutions comes from the hydration layer, as noted by a number of authors (27–30). In approximate agreement with this result, the compressibility of the DNA molecule alone has been estimated to be much more similar to the change reported here, namely 5 × 10−4 ml mol−1 bar−1, based upon the very high packing coefficient of B-DNA of 0.87 and the small void volume of the structure (31).
These results allow us to estimate the compressibility of the hydration layer. Compressibilities can be expressed either as the partial specific compressibility β, which is defined as 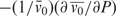 , where
, where 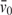 is the partial specific volume of the solute, or the partial compressibility φK, which is just
is the partial specific volume of the solute, or the partial compressibility φK, which is just 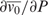 . Here we use partial compressibilities, since they are additive (extrinsic). The following is written in terms of adiabatic compressibility φKS, since this is what can be measured most easily. Ultrasound measurements of DNA solutions provide values for the apparent molar adiabatic compressibility (i.e. the compressibility that would be attributed to one mole of solute if it is assumed that the solvent contributes the same compressibility as it has in the pure state), which is given by ref. (27)
. Here we use partial compressibilities, since they are additive (extrinsic). The following is written in terms of adiabatic compressibility φKS, since this is what can be measured most easily. Ultrasound measurements of DNA solutions provide values for the apparent molar adiabatic compressibility (i.e. the compressibility that would be attributed to one mole of solute if it is assumed that the solvent contributes the same compressibility as it has in the pure state), which is given by ref. (27)
where KM is the intrinsic molar adiabatic compressibility of the solute, nh is the number of water molecules hydrating each nucleotide, and KSh and KS0 are the partial molar adiabatic compressibilities of water in the hydration shell and in the bulk solvent, respectively. φKS depends on the proportion of AT base pairs (27) and for our sequence is estimated as −45.3 × 10−4 ml mol−1 bar−1 (negative because the solute and the hydration layer are both less compressible than bulk water). KM is what we have measured here, nh has been estimated as 24 (27) and KS0 is 8.08 × 10−4 ml mol−1 bar−1 (27,32). This allows us to estimate KSh, the partial molar adiabatic compressibility of water in the hydration shell, as 6.2 ml mol−1 bar−1. This value is 75% of the compressibility of bulk water and is thus towards the upper end of previous estimates (27), largely because of the very small compressibility of DNA calculated here. In other words, we conclude that hydration water is slightly more similar to bulk water, in compressibility and by implication also in density, than previously estimated.
DISCUSSION
By Le Chatelier's principle, high hydrostatic pressure leads to structural changes that reduce the overall molar volume of the system. Because the volume of the DNA itself is almost unchanged, this implies that the changes seen in DNA in our calculations are such as to reduce the volume of the hydration layer (1,29). We have therefore analysed changes to the overall shape of DNA. The most obvious change is an increase in the width of the minor groove (Table 2), seen clearly in Figure 1. Crystal structures of B-DNA indicate a line of water molecules hydrating the minor groove. Because this spine of hydration generally has all of the four potential hydrogen bonding positions of water used (although with a non-ideal geometry), it has relatively large volume. The widening of the minor groove allows water to adopt different structures with lower partial molar volume. This explanation is consistent with previous studies of hydration in B-DNA (27,30), which show that water in the major groove is of low compressibility, because charged groups in the major groove already act to reduce the volume and compressibility of water, by the process known as electrostriction (27).
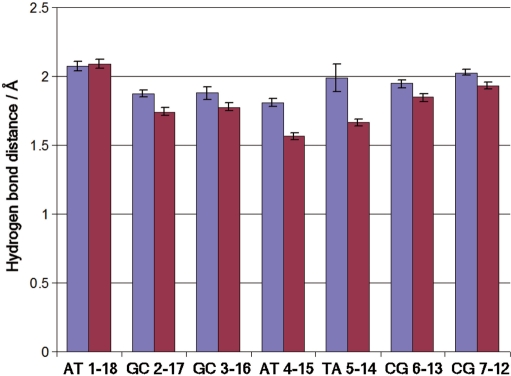
Hydrogen-bond lengths in the Watson–Crick base pairs are reduced in the high-pressure structure, by significantly more than the 0.02 Å seen in earlier MD simulations (Figure 3) (33). The reduction in hydrogen-bond length is approximately 0.11 Å for the GC pairs in the stem and 0.29 Å for the AT pairs. Thus, the AT pairs compress more than twice as much as the GC pairs, as one might expect because they are linked by two hydrogen bonds rather than the three in GC pairs, making the GC hydrogen bonds stiffer.
Figure 3.
Hydrogen-bond distances in base pairs within the B-DNA stem, at low (blue) and high (magenta) pressure.
We note that the overall length of the stem is actually slightly increased at high pressure, by 1.2% (Table 2). The bases remain coplanar and stacked, with a slight increase in slide. This observation demonstrates that DNA bases are already in close van der Waals contact at ambient pressure, and the energy required to compress them further is so great that other structural changes are preferred. A MD simulation also saw essentially no change in the length of the DNA stem with pressure (33).
To date, there are high-pressure crystal structures of several proteins [hen egg-white lysozyme (34), T4 lysozyme (35), metmyoglobin (36), urate oxidase (37) and cowpea mosaic virus (38)], together with an NMR structure of ubiquitin (39) and chemical shift-based NMR structures of lysozyme, BPTI and protein G (14–16). These studies are in agreement over the general features: proteins compress their molecular volume by approximately 0.05%/GPa (0.5%/kbar), with changes in coordinates of approximately 0.1 Å and shortening of amide hydrogen bonds by about 0.01 Å. Helices compress fairly uniformly, whereas sheets twist and distort. There is also a recent crystal structure of DNA, this being of short A-DNA helices surrounding a disordered B-DNA molecule (40). The crystal study reports very small changes of the DNA, which are linear up to approximately 1 GPa (10 kbar). For both the A-DNA and B-DNA, base planes move closer together by 0.15 Å/GPa (0.015 Å/kbar), and for A-DNA Watson–Crick hydrogen bonds shorten by 0.04 Å/GPa (0.004 Å/kbar) (this parameter could not be measured for B-DNA because of its disorder in the crystal). Thus, both X-ray and NMR studies of DNA at high pressure demonstrate that the structure of DNA is affected less than that of proteins by pressure, but they disagree over the details of the structural changes: the present study shows substantial shortening of the Watson–Crick hydrogen bonds and an increase in interbase stacking distance, whereas the crystal structure shows little change of hydrogen-bond lengths and a significant reduction in stacking distance. The origin of this disagreement is as yet unclear (R Fourme, personal communication), and awaits further experimental investigation. The crystal structure also demonstrates a large increase in the number of ordered hydration water seen in contact with the DNA at high pressure, again implying a significant change in hydration with pressure.
In summary, we have shown that structural change of the DNA itself is very small, with most of the changes being in the hydration layer. In proteins, pressure produces a relatively much larger but simple compression within helices, but a twisting of sheets with relatively little compression (15). Increased pressure also leads to a marked reduction in protein–protein association (41). Most common DNA-binding motifs use helices for DNA recognition. It is therefore likely that the altered DNA binding of proteins and transcriptional profile seen at high pressure are not due to changes in the DNA structure but can be ascribed mainly to two factors: a reduction in protein–protein association at high pressure, and alterations in the hydration layers of both DNA and protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Prof. Umesh Varshney (IISc Bangalore) for the DNA and the referees for their comments on this article. This work was supported by Biotechnology and Biological Sciences Research Council (BB/D521230/1); Department of Science and Technology, India; Department of Biotechnology, India; Council of Scientific and Industrial Research; Tata Institute of Fundamental Research, Mumbai. Funding to pay the Open Access publication charges for this article was provided by BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Macgregor RB. The interactions of nucleic acids at elevated hydrostatic pressure. Biochim. Biophys. Acta. 2002;1595:266–276. doi: 10.1016/s0167-4838(01)00349-1. [DOI] [PubMed] [Google Scholar]
- 2.Macgregor RB. Effect of hydrostatic pressure on nucleic acids. Biopolymers. 1998;48:253–263. doi: 10.1002/(sici)1097-0282(1998)48:4<253::aid-bip5>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Kawano H, Nakasone K, Matsumoto M, Yoshida Y, Usami R, Kato C, Abe F. Differential pressure resistance in the activity of RNA polymerase isolated from Shewanella violacea and Escherichia coli. Extremophiles. 2004;8:367–375. doi: 10.1007/s00792-004-0397-0. [DOI] [PubMed] [Google Scholar]
- 4.Kawano H, Nakasone K, Abe F, Kato C, Yoshida Y, Usami R, Horikoshi K. Protein-DNA interactions under high-pressure conditions, studied by capillary narrow-tube electrophoresis. Biosci. Biotechnol. Biochem. 2005;69:1415–1417. doi: 10.1271/bbb.69.1415. [DOI] [PubMed] [Google Scholar]
- 5.Robinson CR, Sligar SG. Hydrostatic pressure reverses osmotic pressure effects on the specificity of E. coli DNA interactions. Biochemistry. 1994;33:3787–3793. doi: 10.1021/bi00179a001. [DOI] [PubMed] [Google Scholar]
- 6.Robinson CR, Sligar SG. Heterogeneity in molecular recognition by restriction endonucleases: osmotic and hydrostatic pressure effects on BamHI, PvuLI, and EcoRV specificity. Proc. Natl Acad. Sci. USA. 1995;92:3444–3448. doi: 10.1073/pnas.92.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon DR, Pruski AM, Dixon LRJ. The effects of hydrostatic pressure change on DNA integrity in the hydrothermal-vent mussel Bathymodiolus azoricus: implications for future deep-sea mutagenicity studies. Mutat. Res. 2004;552:235–246. doi: 10.1016/j.mrfmmm.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 9.Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles. 2005;9:65–73. doi: 10.1007/s00792-004-0414-3. [DOI] [PubMed] [Google Scholar]
- 10.Chalikian TV. Volumetric properties of proteins. Ann. Rev. Biophys. Biomol. Struct. 2003;32:207–235. doi: 10.1146/annurev.biophys.32.110601.141709. [DOI] [PubMed] [Google Scholar]
- 11.Cooper A. Thermodynamic fluctuations in protein molecules. Proc. Natl Acad. Sci. USA. 1976;73:2740–2741. doi: 10.1073/pnas.73.8.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh M, Kumar NV, Varshney U, Chary KVR. Structural basis for uracil DNA glycosylase interaction with uracil: NMR study. Nucleic Acids Res. 2000;28:1906–1912. doi: 10.1093/nar/28.9.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada H, Nishikawa K, Honda M, Shimura T, Akasaka K, Tabayashi K. Pressure-resisting cell for high-pressure, high-resolution nuclear magnetic resonance measurements at very high magnetic fields. Rev. Sci. Instrum. 2001;72:1463–1471. [Google Scholar]
- 14.Williamson MP, Akasaka K, Refaee M. The solution structure of bovine pancreatic trypsin inhibitor at high pressure. Protein Sci. 2003;12:1971–1979. doi: 10.1110/ps.0242103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Refaee M, Tezuka T, Akasaka K, Williamson MP. Pressure-dependent changes in the solution structure of hen egg-white lysozyme. J. Mol. Biol. 2003;327:857–865. doi: 10.1016/s0022-2836(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 16.Wilton DJ, Tunnicliffe RB, Kamatari YO, Akasaka K, Williamson MP. Pressure induced changes in the solution structure of the GB1 domain of protein G. Proteins. 2008;71:1432–1440. doi: 10.1002/prot.21832. [DOI] [PubMed] [Google Scholar]
- 17.Williamson MP, Asakura T. Empirical comparisons of models for chemical shift calculation in proteins. J. Magn. Reson. Ser. B. 1993;101:63–71. [Google Scholar]
- 18.Asakura T, Taoka K, Demura M, Williamson MP. The relationship between amide proton chemical shifts and secondary structure in proteins. J. Biomol. NMR. 1995;6:227–236. doi: 10.1007/BF00197804. [DOI] [PubMed] [Google Scholar]
- 19.Case DA. Calibration of ring-current effects in proteins and nucleic acids. J. Biomol. NMR. 1995;6:341–346. doi: 10.1007/BF00197633. [DOI] [PubMed] [Google Scholar]
- 20.Packer MJ, Zonta C, Hunter CA. Complexation-induced chemical shifts: ab initio parameterization of transferable bond anisotropies. J. Magn. Reson. 2003;162:102–112. doi: 10.1016/s1090-7807(03)00037-5. [DOI] [PubMed] [Google Scholar]
- 21.Wijmenga SS, Kruithof M, Hilbers CW. Analysis of 1H chemical shifts in DNA: assessment of the reliability of 1H chemical shift calculations for use in structure refinement. J. Biomol. NMR. 1997;10:337–350. doi: 10.1023/A:1018348123074. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. 1994;D50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 25.Williamson MP. Many residues in cytochrome c populate alternative states under equilibrium conditions. Proteins. 2003;53:731–739. doi: 10.1002/prot.10464. [DOI] [PubMed] [Google Scholar]
- 26.Tunnicliffe RB, Waby JL, Williams RJ, Williamson MP. An experimental investigation of conformational fluctuations in proteins G and L. Structure. 2005;13:1677–1684. doi: 10.1016/j.str.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Chalikian TV, Sarvazyan AP, Plum GE, Breslauer KJ. Influence of base composition, base sequence, and duplex structure on DNA hydration: apparent molar volumes and apparent molar adiabatic compressibilities of synthetic and natural DNA duplexes at 25 °C. Biochemistry. 1994;33:2394–2401. doi: 10.1021/bi00175a007. [DOI] [PubMed] [Google Scholar]
- 28.Chalikian TV, Breslauer KJ. Volumetric properties of nucleic acids. Biopolymers. 1998;48:264–280. doi: 10.1002/(sici)1097-0282(1998)48:4<264::aid-bip6>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 29.Giel-Pietraszuk M, Barciszewski J. A nature of conformational changes of yeast tRNAPhe: high hydrostatic pressure effects. Int. J. Biol. Macromol. 2005;37:109–114. doi: 10.1016/j.ijbiomac.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Barciszewski J, Jurczak J, Porowski S, Specht T, Erdmann VA. The role of water structure in conformational changes of nucleic acids in ambient and high-pressure conditions. Eur. J. Biochem. 1999;260:293–307. doi: 10.1046/j.1432-1327.1999.00184.x. [DOI] [PubMed] [Google Scholar]
- 31.Buckin VA, Kankiya BI, Sarvazyan AP, Uedaira H. Acoustical investigation of poly(dA).poly(dT), poly[d(A-T)].Poly[d(A-T)], poly(A).poly(U) and DNA hydration in dilute aqueous solutions. Nucleic Acids Res. 1989;17:4189–4203. doi: 10.1093/nar/17.11.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalikian TV, Sarvazyan AP, Breslauer KJ. Hydration and partial compressibility of biological compounds. Biophys. Chem. 1994;51:89–109. doi: 10.1016/0301-4622(94)85007-0. [DOI] [PubMed] [Google Scholar]
- 33.Norberg J, Nilsson L. Constant pressure molecular dynamics simulations of the dodecamers: d(GCGCGCGCGCGC)2 and r(GCGCGCGCGCGC)2. J. Chem. Phys. 1996;104:6052–6057. [Google Scholar]
- 34.Kundrot CE, Richards FM. Crystal structure of hen egg-white lysozyme at a hydrostatic pressure of 1000 atmospheres. J. Mol. Biol. 1987;193:157–170. doi: 10.1016/0022-2836(87)90634-6. [DOI] [PubMed] [Google Scholar]
- 35.Collins MD, Quillin ML, Hummer G, Matthews BW, Gruner SM. Structural rigidity of a large cavity-containing protein revealed by high-pressure crystallography. J. Mol. Biol. 2007;367:752–763. doi: 10.1016/j.jmb.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urayama P, Phillips GN, Gruner SM. Probing substates in sperm whale myoglobin using high-pressure crystallography. Structure. 2002;10:51–60. doi: 10.1016/s0969-2126(01)00699-2. [DOI] [PubMed] [Google Scholar]
- 37.Colloc'h N, Girard E, Dhaussy AC, Kahn R, Ascone I, Mezouar M, Fourme R. High pressure macromolecular crystallography: the 140-MPa crystal structure at 2.3 Å resolution of urate oxidase, a 135-kDa tetrameric assembly. Biochim. Biophys. Acta. 2006;1764:391–397. doi: 10.1016/j.bbapap.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Girard E, Kahn R, Mezouar M, Dhaussy AC, Lin TW, Johnson JE, Fourme R. The first crystal structure of a macromolecular assembly under high pressure: CpMV at 330 MPa. Biophys. J. 2005;88:3562–3571. doi: 10.1529/biophysj.104.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: Ubiquitin at 30 bar-3 kbar. J. Mol. Biol. 2005;347:277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 40.Girard E, Prange T, Dhaussy AC, Migianu-Griffoni E, Lecouvey M, Chervin JC, Mezouar M, Kahn R, Fourme R. Adaptation of the base-paired double-helix molecular architecture to extreme pressure. Nucleic Acids Res. 2007;35:4800–4808. doi: 10.1093/nar/gkm511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva JL, Weber G. Pressure stability of proteins. Ann. Rev. Phys. Chem. 1993;44:89–113. doi: 10.1146/annurev.pc.44.100193.000513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.