Abstract
α-catenin has often been considered to be a non-regulatory intercellular adhesion protein, in contrast to β-catenin, which has well-documented dual roles in cell–cell adhesion and signal transduction. Recently, however, α-catenin has been found to be important not only in connecting the E-cadherin–β-catenin complex to the actin cytoskeleton, but also in coordinating actin dynamics and inversely correlating cell adhesion with proliferation. As the number of α-catenin-interacting partners increases, intriguing new connections imply even more complex regulatory functions for this protein.
In multicellular organisms, cell–cell contacts that are mediated by classic cadherins are essential in many fundamental processes, including morphogenesis, maintenance of tissue integrity, wound healing and cell polarity. The prototype classic cadherin is the transmembrane protein E-cadherin. This protein uses its extracellular domain to bind Ca2+ and interact with E-cadherin on adjacent cells, thereby forming adherens junctions. To establish efficient cell–cell junctions, E-cadherin uses its cytoplasmic domain to couple to catenins and the actin cytoskeleton. This association sets the classic cadherins apart form desmosomal cadherins, which form complexes with the intracellular proteins plakoglobin and desmoplakin to form a more robust Ca2+-induced adhesive interaction — the desmosome — that links the intermediate-filament cytoskeleton (see also the article by Lynne Chang and Robert Goldman in this issue). Many cells have both adherens junctions and desmosomes, which function coordinately in intercellular adhesion (FIG. 1).
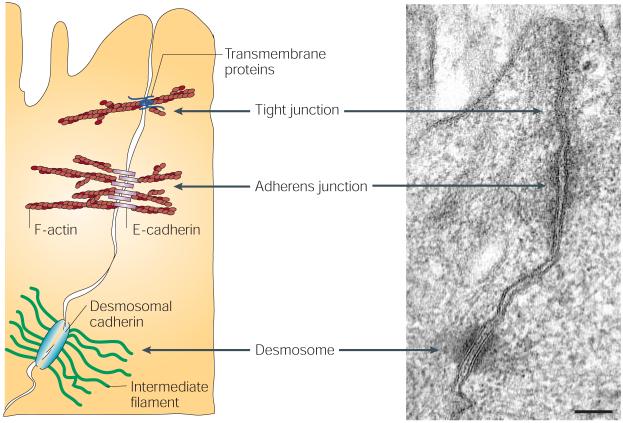
Figure 1. Intercellular junctions in skin.
The electron micrograph and corresponding schematic depict the main types of intercellular junction in epithelial cells. Tight junctions are composed of transmembrane proteins that link to the actin cytoskeleton and prevent the leakage of small molecules through intercellular spaces. Adherens junctions are formed by homophilic interactions between E-cadherin molecules, and are connected to the actin network through β- and α-catenins. They function to coordinate the actin cytoskeleton across an epithelial sheet. Desmosomes are composed of desmosomal cadherins that are linked to intermediate filaments and integrate the intermediate-filament network across the epithelial sheet. Electron micrograph courtesy of H. Amalia Pasolli, Rockefeller University, New York, USA.
- CLASSIC CADHERINS
Cadherins are transmembrane molecules that mediate Ca2+-dependent cell–cell adhesion. Classic cadherins are typified by an extracellular segment that consists of five distinct Ca2+-binding domains and a conserved cytoplasmic domain, which binds β-catenin. The extracellular part interacts homotypically with cadherins on the surface of neighbouring cells to form adherens junctions. The cytoplasmic tail links the actin cytoskeleton to adherens junctions.
- ADHERENS JUNCTION
A specialized intercellular junction of the plasma membrane, in which the cadherin molecules of adjacent cells interact in a Ca2+-dependent manner. Actin filaments are linked to this structure through catenins that are located underneath the junction.
- DESMOSOMES
Specialized junctional structures that form a tight connection between epithelial cells or cardiac myocytes. They consist of several transmembrane adhesive glycoproteins (desmogleins and desmocollins) and cytoplasmic plaque proteins (desmoplakins) that link to intermediate filaments.
- INTERMEDIATE FILAMENTS
Proteins that acquired their name from the diameter of their polymeric structure, which is midway between the diameters of thin actin microfilaments and thick microtubules. Their ability to form very stable filaments enables them to confer mechanical strength on the cytoskeleton.
Desmosomes use their attachments to intermediate filaments to provide mechanical strength to intercellular connections1-3. They are particularly important in tissues that are subjected to substantial physical stress, such as muscle and epidermis. By contrast, adherens junctions use their connections to the actomyosin network to remodel cell–cell interactions and provide flexible dynamic adhesion during wound repair of adult tissuesand embryonic development4-6. Whereas desmosomes and their associated intermediate filaments function as molecular clamps to reinforce intercellular junctions, actin dynamics and adherens-junction formation have been implicated in the initial steps of bringing membranes together and in the final steps of sealing membranes into epithelial sheets.
- ACTOMYOSIN NETWORK
A complex of myosin and actin filaments that is responsible for a range of cellular movements in eukaryotic cells. Myosins can translocate vesicles or other cargo on actin filaments.
Dysregulation of cadherin-mediated junctions can lead to severe developmental defects. Mutations in desmosomal genes often result in degenerative disorders, and several excellent reviews have recently described the composition and function of these robust adhesive structures1-3. Curiously, however, although mutations in adherens-junction proteins can sometimes cause tissue degeneration, in other cases they can contribute to carcinogenesis and metastasis4-6. With an increasing knowledge of the composition of adherens junctions, their assembly, and their relationship to other membrane junctions and receptors, new insights into some of these processes are beginning to unfold.
At the heart of the story lie the catenins, which associate with the cytoplasmic domain of cadherins to assemble a protein complex that can associate with the actin cytoskeleton, coordinate stable intercellular adhesion, and regulate indirect associations with desmosomes, tight junctions, growth-factor receptors and microtubules4. In addition, in response to WNT signalling, excess β-catenin that is not used in cell–cell junctions can accumulate and adopt a second function as a nuclear transcriptional co-activator for the lymphoid enhancer-binding factor-1 (LEF1)/T-cell-specific factor (TCF) family of DNA-binding proteins7-9. As sustained stabilization of β-catenin has been associated with a range of human cancers, this regulatory function has been the main focus of the cancer–cadherin link. The reader is referred to several elegant reviews that have covered, in depth, the role of β-catenin in signalling and cancer7-9. However, mutations in E-cadherin and α-catenin can also contribute to cancers10-17, and gene-targeting studies indicate that the proliferation that is sometimes associated with such mutations does not always result from the activity of nuclear β-catenin18,19.
- TIGHT JUNCTIONS
The most apical intercellular junctions, which function as selective (semi-permeable) diffusion barriers between individual cells. They are identified as a belt-like region in which two lipid-apposing membranes lie close together.
In this review, we focus on α-catenin, which is the protein that connects E-cadherin–β-catenin complexes with the actin cytoskeleton20,21. Whereas all other catenins (β-catenin, plakoglobin and p120 catenin) share considerable sequence similarity and belong to the Armadillo family of proteins, α-catenin differs notably in both sequence and structural organization. Although it was previously considered to be solely a structural protein, new roles have begun to emerge for α-catenin in both assembling the cytoskeleton and regulating its dynamics at cell–cell junctions. In addition, recent studies imply that the interactions of α-catenin with E-cadherin–β-catenin complexes might control the accessibility of these complexes to other cellular proteins. In the past few years, various binding partners for α-catenin have surfaced, which have shed new light on the functions of α-catenin and its underlying mechanisms of action (FIG. 2).
Figure 2. A multiprotein complex at adherens junctions.
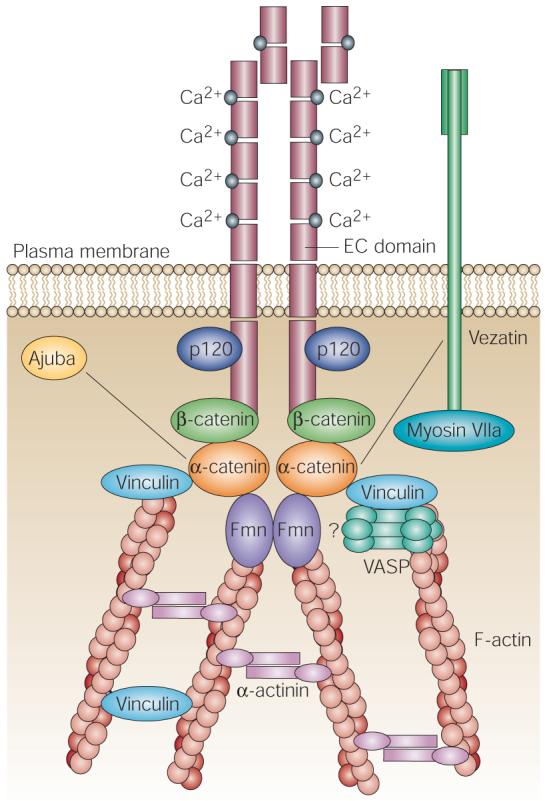
The extracellular region of E-cadherin, which contains extracellular cadherin (EC) domains, undergoes a Ca2+-dependent conformational change that allows it to homodimerize at the membrane. Through extracellular interactions with E-cadherins on a neighbouring cell, opposing cadherin dimers can integrate the actin cytoskeletons. Stabilization of intercellular adhesion requires the cytoplasmic domain of E-cadherin, which binds to β-catenin. β-catenin, in turn, binds α-catenin, which is central in recruiting a number of cytoskeletal proteins, including the filamentous (F)-actin-nucleating formin proteins (Fmn), and the actin-binding proteins vinculin, Ajuba, myosin VIIa, vezatin, α-actinin and members of the vasodilator-stimulated phosphoprotein (VASP) family of F-actin-elongating proteins. At least some of these interactions are essential in polymerizing and organizing actin into cables that help to seal membranes and integrate the actin cytoskeleton across the epithelial sheet. By contrast, p120 catenin (p120), which is related to β-catenin, binds to E-cadherin through a juxtamembrane domain and seems to function in cadherin turnover, perhaps by regulating cadherin trafficking. An emerging intercellular adhesion system (not shown), consisting of nectin and afadin, also has roles in the organization of a range of intercellular junctions. Nectin is a Ca2+-independent, immunoglobulin-like, intercellular adhesion molecule, and afadin is a nectin- and actin-filament-binding protein that connects nectin to the actin cytoskeleton78,79.
α-catenins in development and differentiation
α-catenin is conserved across the eukaryotic kingdom, where it functions broadly in intercellular adhesion during development and differentiation. In Drosophila melanogaster, cell adhesion is disrupted when α-catenin contains a mutation in the binding site for Armadillo, which is the D. melanogastor homologue of β-catenin22. Adherens junctions are also present in the nematode Caenorhabditis elegans, which expresses the homologues HMR-1 (cadherin), HMP-1 (α-catenin) and HMP-2 (β-catenin)23.
In mice, there are three α-catenins and one close relative, which all share substantial amino-acid sequence identity (TABLE 1): αE-catenin is most prevalent in epithelial tissues24; αN-catenin is restricted to neural tissues25,26; αT-catenin is expressed primarily in heart tissue27; and α-catulin, which is an α-catenin-like protein, is ubiquitously expressed28,29. A more distant relative is vinculin, which is ubiquitously expressed and localizes to both focal adhesions and adherens junctions30 (TABLE 2).
Table 1.
Amino-acid sequence identities (and similarities) between mouse α-catenins/vinculin*
| αE-catenin | αN-catenin | αT-catenin | α-catulin | Vinculin | |
|---|---|---|---|---|---|
| αE-catenin | 100% | 76.5% (83.1%) | 56.1% (73.7%) | 24.8% (33.9%) | 22.5% (31.0%) |
| αN-catenin | – | 100% | 58.5% (69.6%) | 25.6% (32.8%) | 22.3% (30.0%) |
| αT-catenin | – | – | 100% | 19.7% (32.1%) | 17.3% (30.3%) |
| α-catulin | – | – | – | 100% | 18.6% (23.7%) |
| Vinculin | – | – | – | – | 100% |
Data are from REF. 7.
Table 2.
Members of the α-catenin/vinculin family and their functions
| Family member | Chromosome localization (human/mouse) |
Cellular localization |
Phenotype |
|---|---|---|---|
| αE-catenin | 5q31/18q11.0 | Adherens junctions | Lethal at the blastocyst stage |
| αN-catenin | 2p12–p11.1/6-B3D | Adherens junctions | Cerebellar deficient folia |
| αT-catenin | 10q21–q23 | Adherens junctions | ND |
| α-catulin | 9q22–q32/4-C1 | ND | ND |
| Vinculin | 10q21–q23/14-A2 | Focal contacts and adherens junctions |
Lethal by embryonic day 10 |
The α-catenin/vinculin-family members are crucial for the formation of proper adherens junctions during development, as judged by the phenotypes of the knockout animals. αE-catenin is required at the blastocyst stage and without this protein, the outer epithelial covering (trophectoderm) of the developing embryo lacks integrity. Loss of αN-catenin results in the cerebellar deficient folia (cdf) phenotype with ataxia, cerebellar hypoplasia and abnormal cerebellar lobulation. Vinculin knockout results in heart and brain defects during embryonic development. Vinculin-mutant embryos are also 30–40% smaller, growth of their somites and limbs are retarded, and their ectodermal tissues are sparse and fragile112. ND, not determined.
- FOCAL ADHESIONS
A cell-to-substrate adhesion structure that anchors the ends of actin microfilaments (stress fibres) and mediates strong attachment to substrates.
αE-catenin
Formerly known as cadherin-associated protein (CAP)-102, αE-catenin is the founding member of the α-catenin family31,32. Localized on chromosome 18q11.0 (ref. 33), the murine gene encodes the 102-kDa αE-catenin32,34. The human αE-catenin gene (CTNNA1) localizes to chromosome 5q31 and consists of 16 coding exons (906 amino acids; 102 kDa) and at least one 5′ non-coding exon35. Human and mouse αE-catenin proteins are exceptionally highly conserved, sharing 99.2% identity. This evolutionary conservation might reflect the diversity of the interacting protein partners and functions of αE-catenin36.
αE-catenin was initially identified on the basis of its ability to associate with E-cadherin24,34. It also interacts with other classic cadherins, such as neuronal (N) and placental (P) cadherin24. When kidney epithelial cells are cultured in medium that contains Ca2+ at levels that induce the formation of adherens junctions, E-cadherin and catenins shift from the soluble to the cytoskeletal fraction (if the cells are lysed and fractionated) concomitant with the formation of stable adherens junctions37,38. Gene-targeting studies in mice indicate that αE-catenin is required to mediate the formation of adherens junctions in epithelial cells18,34,39. In addition, cancer cell lines that lack αE-catenin do not show proper cell–cell adhesion10 unless the wild-type gene is reintroduced and adherens-junction formation is restored14,25,40.
A gene-trap-induced mouse mutation of the αE-catenin gene, which specifically deletes the region that encodes the carboxy-terminal third of the protein, results in an embryonic-lethal phenotype that is similar to that of the E-cadherin-null-mutant mouse41. In both cases, the proteins are required at the blastocyst stage and, without αE-catenin, the outer epithelial covering (trophectoderm) of the developing embryo lacks integrity41,42.
- GENE TRAP
A methodology that is used to characterize new genes and analyse their importance in biological phenomena. The technique involves the use of mouse embryonic stem cells and reporter vectors that are designed to randomly integrate into the genome, tag an insertion site and generate a mutation.
- BLASTOCYST STAGE
The stage during embryonic development that is characterized by the formation of two cell types: the embryoblast (the inner cell mass on the inside of the blastocoel) and the trophoblast (the cells on the outside of the blastocoel).
To examine the functional importance of αE-catenin in other epithelial tissues, Vasioukhin and colleagues engineered a conditional αE-catenin-mutant mouse18. When αE-catenin expression was ablated from the developing skin around embryonic day 15, hair-follicle development was impaired and the architecture of the epidermal tissue was markedly altered. In the epidermis, the formation of adherens junctions seemed to be compromised, even though E-cadherin and β-catenin complexes still localized to cell–cell borders. Desmosomes loosely held the skin epithelium together, but partial defects in epithelial polarity arose, including suprabasal mitoses (mitosis is usually observed only in the basal layer of the epidermis). Perhaps most intriguing was the presence of epidermal hyperproliferation and multinucleate cells, which were accompanied by epithelial invaginations that resembled the precancerous lesions that are known as squamous-cell carcinomas in situ.
Although null mutations in αE-catenin have been associated with epithelial cancers10-14, it has usually been assumed that perturbations in cell–cell adhesion are late, rather than early, steps in carcinogenesis, and that they are preceded by mutations in cell-cycle-regulated genes that lead to uncontrolled growth. In the mice that lacked αE-catenin, however, the skin epithelium uniformly showed epidermal hyperproliferation shortly after αE-catenin-gene ablation, which prompted researchers to investigate alternative molecular explanations. In vitro and in vivo studies uncovered a sustained activation of the Ras–ERK/MAPK (extracellular signal-regulated kinase/mitogen-activated protein kinase) pathway, which seemed to involve the insulin–insulin-like-growth-factor signal-transduction pathway18. Curiously, these perturbations seemed to occur independently of the effects on intercellular adhesion and of β-catenin–LEF1/TCF-mediated gene transcription. Although the precise mechanism remains elusive, these findings indicate that when αE-catenin is absent, E-cadherin–β-catenin complexes might interact in new ways with components of signal-transduction pathways that are involved in cell-cycle regulation. Such a mechanism could be important for understanding the inverse link between αE-catenin mutations and tumorigenesis. In a ‘natural’ setting, the mechanism might be relevant to processes such as wound healing, in which proliferation is transiently enhanced and cell–cell adhesion is reduced during the epithelial regeneration. In this model, although αE-catenin would not be mutated, it might be transiently inactivated, perhaps through post-translational modification or associations with other proteins.
αN-catenin
αN-catenin (encoded by CTNNA2) or cadherin-associated-protein related (CAPR) was first characterized by Claverie and colleagues as a human cDNA encoding a protein that was 80% identical to αE-catenin (CAP102) but that contained a 48-residue insert26. The CTNNA2 gene maps to human chromosome 2p12–p11.1, which is homologous to the B3-D region of mouse chromosome 6 (34.2).
αN-catenin localizes to adherens junctions that border active zones in developing and mature synapses throughout the developing and postnatal brain43,44. The expression of specific cadherin subtypes delineates specific neuronal circuits, which indicates that cadherin–catenin complexes mediate adhesion between pre- and postsynaptic membranes45-47. Interestingly, the classic mouse mutation cerebellar deficient folia (cdf) results from a deletion in the αN-catenin gene, and cdf mice show ataxia, cerebellar hypoplasia and abnormal cerebellar lobulation48. Approximately 40% of the purkinje cells in these animals are located ectopically in the white matter and inner granule-cell layer (FIG. 3). Transgenic re-expression of the αN-catenin transgene in these mice restored normal cerebellar morphology. Taken together, these findings underline a role for αN-catenin in stabilizing N-cadherin-mediated synapse formation during the development of the central nervous system48.
Figure 3. Abnormal cerebellar development in αN-catenin-mutant mice.
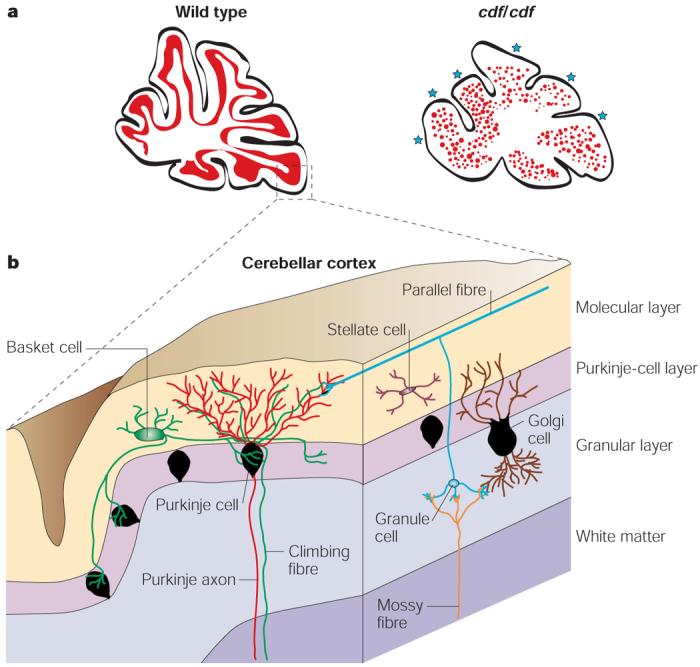
Mutations in the αN-catenin gene underlie the genetic defect in the ataxic cerebellar deficient folia (cdf)-mutant mice. The cerebella of cdf/cdf-mutant mice show hypoplasia and abnormal lobulation. a | A schematic representation of the midline sagittal sections from wild-type and cdf/cdf-mutant mouse brain cerebella shows scattered Purkinje cells in the more central white matter and abnormal lobulation (blue stars). The magnified region (b) shows the structure of the cerebellar cortex (the grey matter), which includes the cell bodies and dendrites of the Purkinje cells; the axons of the granule cells; and the cell bodies, dendrites and axons of the basket cells.
- ACTIVE ZONES
The sites along nerve terminals where synaptic vesicles dock and undergo Ca2+-dependent exocytosis during synaptic transmission.
- PURKINJE CELLS
Large neurons with extensive dendritic projections that form a layer near to the surface of the cerebellum.
αT-catenin
αT-catenin (encoded by CTNNA3) is predominantly expressed in the heart and testis27. The human and mouse CTNNA3 genes contain 18 exons, the boundaries of which are identical to those in the human CTNNA2 gene49. αT-catenin seems to be functionally equivalent to αE-catenin on the basis of its ability to restore cell–cell adhesion in a colon-cancer cell line in which the CTNNA1 gene is mutated27. Because of its chromosomal position (10q21) and its high level of expression in the heart, CTNNA3 has been deemed to be a candidate for a form of dilated cardiomyopathy that has been linked to human chromosome 10q21–q23 (CMD1C). However, mutation screening of all 18 exons of the CTNNA3 gene in one family that is affected by this disease failed to uncover any mutations49.
- DILATED CARDIOMYOPATHY
Also known as ‘congestive cardiomyopathy’, this is the most common form of myocardial disease, which causes decreased systolic function and increased ventricular volume.
α-catulin
Catenin α-like-1 — or α-catulin (encoded by CTNNAL1) — was first characterized as a 2.45-kb transcript that was downregulated in human pancreatic cancer cells28. In vitro transcription and translation of the cloned transcript yielded an 82-kDa protein that shares 24.8% identity with αE-catenin. The human CTNNAL1 gene maps to chromosome 9q22–q32 (mouse chromosome 4 band C1)28. α-catulin mRNA is expressed ubiquitously, although the levels are lower in neural tissues. At 734 amino acids, the predicted α-catulin polypeptide is smaller than αN- and αE-catenin; it lacks the central 110-residue region and has a shorter carboxyl terminus. It is not yet known whether α-catulin localizes to cell–cell borders or is a component of adherens junctions. However, the amino-terminal amphipathic helices of α-catenin that are thought to ensure an interaction with β-catenin are also present in α-catulin. Furthermore, amphipathic helices in the carboxy-terminal part of vinculin and α-catenin, which allow these proteins to bind to the actin cytoskeleton, are also present in α-catulin29. So, sequence homology between α-catulin and α-catenin or vinculin implies that α-catulin might have the potential to bind to β-catenin and the actin cytoskeleton.
- AMPHIPATHIC HELICES
Helical structures that consist of hydrophobic non-polar residues on one side of the helical cylinder, and hydrophilic and polar residues on the other side.
yeast two-hybrid analyses have uncovered the lymphoid blast crisis oncogene (LBC) Rho guanine nucleotide-exchange factor (LBC Rho-GEF) as a partner for α-catulin but not for αE-catenin, which indicates that α-catulin might have a new role in modulating signalling by the Rho pathway29. It will be interesting to see how the functions and associations of α-catulin differ from those of the ‘classic’ α-catenins.
- YEAST TWO-HYBRID ANALYSIS
A technique that is used to study protein–protein interactions in vivo in yeast cells.
- GUANINE NUCLEOTIDE-EXCHANGE FACTOR
A protein that facilitates the exchange of guanine diphosphate (GDP) for guanine triphosphate (GTP) in the nucleotide-binding pocket of a GTP-binding protein.
αE-catenin structure
Sequence similarities and structural relationships
Because of three extended regions of sequence similarity that are shared by αE-catenin and its distant cousin vinculin (known as the vinculin-homology (VH) domains VH1–VH3), αE-catenin has been described as a relative of vinculin24,34,50,51 (figs 4,5). Although the degree of identity is not high, these homologies are regarded as biologically meaningful, not only because both molecules anchor actin filaments to the plasma membrane, but also because some ligand-binding sites map to the regions that are more highly conserved between the proteins. Actin and α-actinin are well-characterized binding partners of both proteins52-56, and there is substantial evidence of a direct interaction between vinculin and β-catenin in vivo57.
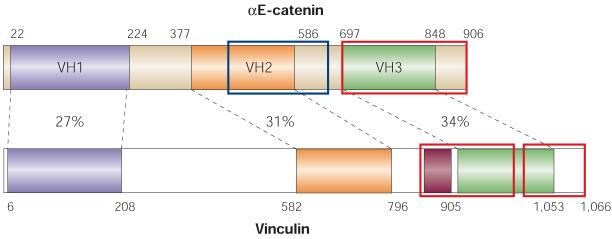
Figure 4. Sequence similarities between vinculin and αE-catenin.
The coloured areas represent regions of similarity between the two proteins. The degrees of amino-acid identity are indicated as percentages. The magenta region in vinculin represents the proline-rich hinge segment. Actin-binding domains are marked as the red open boxes (amino acids 697–906 for αE-catenin and 893–985/1016–1066 for vinculin). The intercellular adhesion modulation (M) domain (residues 509–643) of αE-catenin is marked as the blue open box. The numbers correspond to amino acids of the protein sequence. VH, vinculin-homology domain.
Figure 5. Comparison of the crystal structures of the αE-catenin M-fragment and vinculin tail.
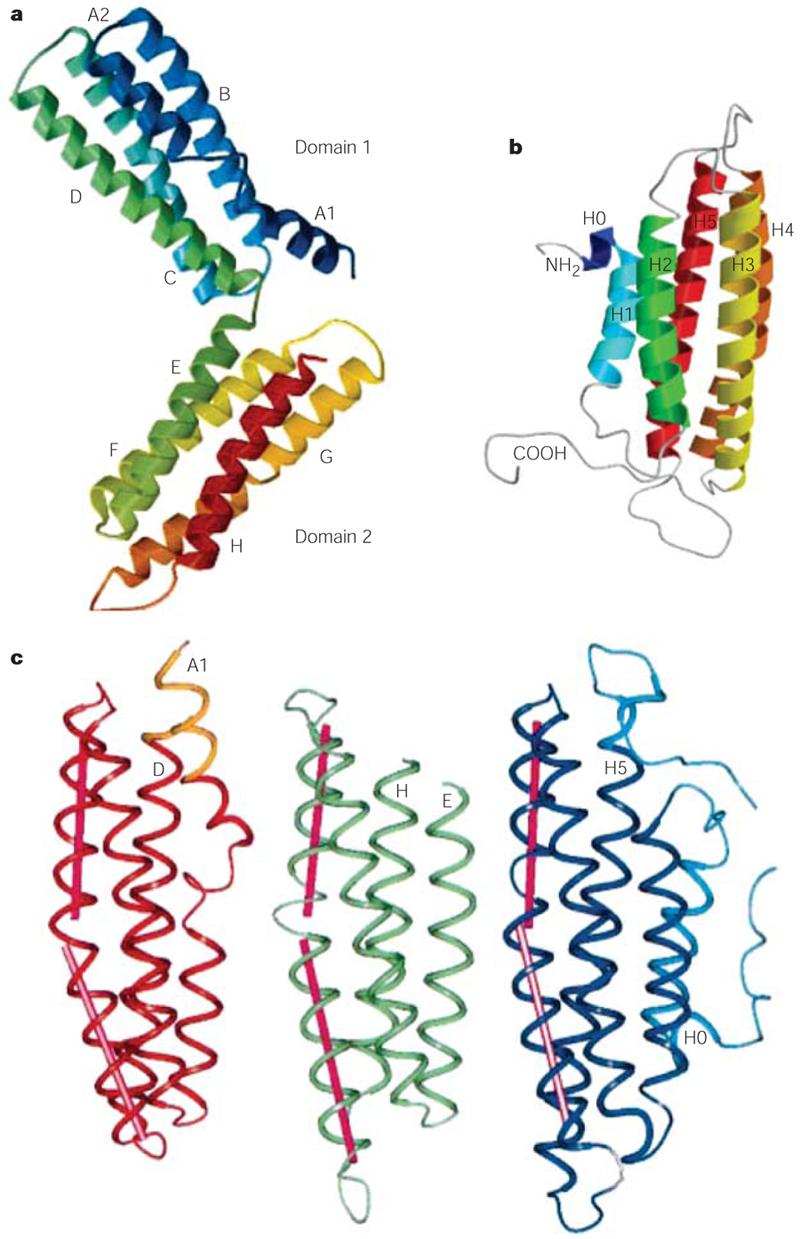
The crucial functions of αE-catenin and vinculin are to mediate protein–protein and protein–phospholipid interactions. Proteins composed of tandemly repeated α-helical bundles provide a structural framework for the assembly of multiprotein complexes. a | View of the αE-catenin M-fragment structure with two four-helix domains (domains 1 and 2) coloured from the amino terminus (blue) to the carboxyl terminus (red). Letter labels for each of the helices are shown. b | The crystal structure of the carboxy-terminal part of vinculin comprises five helices (H0–H4). c | A comparison of the αE-catenin M-fragment domain 1 (red/orange; left) and domain 2 (green; middle) with the vinculin-tail domain (dark/light blue; right). Parts a and c courtesy of D. Barford and reproduced with permission from ref. 67 © (2001) Macmillan Magazines Ltd; part b courtesy of D. R. Critchley, Biochemistry Department, University of Leicester, UK and reproduced with permission from ref. 113 © (1999) Elsevier.
The actin-binding domains in both vinculin (which contains a bipartite actin-binding site) and αE-catenin are localized in their carboxy-terminal segments (residues 893–985/1016–1066 and 697–906, respectively; FIG. 4)58-60. Vinculin is interesting in that its head and tail segments can interact with each other to provide a conformational state that precludes its ability to associate with focal-complex proteins and with the actin cytoskeleton61,62. On signal-transduction-mediated activation of focal-complex assembly, however, activated phospholipids can change the conformation of vinculin associated with its partners talin (an actin-binding protein that binds directly to integrins) and actin (reviewed in refs 63,64). A priori, it might be tempting to speculate that the similarity of αE-catenin to vinculin means that it too can undergo autoregulation. However, the head-to-tail interactions of vinculin have been strongly detected by yeast two-hybrid analyses, whereas corresponding αE-catenin interactions have not been found (ref. 50 and A.K. et al., unpublished data). One possibility is that the lack of a proline-rich hinge region in αE-catenin, which is present in vinculin, yields a less flexible structure, which might prevent the head-to-tail interaction65.
Despite sequence and structural differences that result in different modes of regulation and only partially overlapping binding partners, both vinculin and αE-catenin participate in coordinating actin dynamics at their respective adhesive junctions. Vinculin seems to be more important at integrin-mediated junctions, whereas αE-catenin seems to be exclusive for adherens junctions. Despite these differences, the association of αE-catenin with E-cadherin–β-catenin complexes and actin must be intricately regulated in a manner that seems conceptually, if not mechanistically, analogous to the coordinated association of vinculin with talin and actin. Whether αE-catenin changes its ligand-binding ability through post-translational modification, localized activation of phospholipids or dynamic interactions with other proteins remains an important question for the future.
Three-dimensional structure of the M-fragment of αE-catenin
To define the segments of αE-catenin that are necessary for mediating intercellular adhesion, sequences that encode the extracellular and transmembrane domains of E-cadherin were fused to various segments of αE-catenin cDNA, and the trans-genes were tested for the ability of their encoded fusion proteins to induce the rapid aggregation of mouse l cells66. This led to the identification of the intercellular adhesion modulation (M) domain (residues 509–643, which partially includes the VH2 domain) of αE-catenin. In this assay, weak intercellular adhesion was facilitated by an E-cadherin–M-domain fusion protein, whereas a fusion protein containing E-cadherin and the full-length αE-catenin conferred considerably stronger adhesion. Although other explanations are possible, the underlying difference is likely to reside, at least in part, in the need for not only the M-domain but also the domain that is involved in actin-based cytoskeletal interactions, which are known to be required for stable intercellular adhesion37,66.
- L CELLS
Cells of a mouse fibroblast line that is derived from connective tissue and does not express classic cadherin molecules.
Crystallographic analysis of the so-called M-fragment (residues 377–633), which includes the M-domain, detected a tandem repeat of a four-α-helix bundle. This is structurally related to the vinculin tail67 (FIG. 5). Overall, these findings indicate that αE-catenin probably comprises a series of repeating antiparallel α-helical domains.
In solution, the M-fragment can form relatively low-affinity dimers67. On the basis of the findings of Imamura and colleagues66, it seems likely that, through weak dimerization of the M-domain of αE-catenin, the lateral dimerization of E-cadherin molecules in the plasma membrane might be enhanced and/or stabilized, thereby promoting and/or strengthening cell–cell adhesion68,69.
Dimerization and β-catenin-binding domains of αE-catenin
In vitro binding assays indicate that classic cadherins, β-catenin and αE-catenin assemble into a complex with a 1:1:1 stoichiometry70. αE-catenin forms a homodimer in solution at micromolar concentrations56,62; however, in the presence of β-catenin or plakoglobin, the homodimer dissociates and a 1:1 heterodimer of αE-catenin and β-catenin/plakoglobin is observed62,71. Indeed, the two processes seem to be mutually exclusive, as judged by the fact that the site on αE-catenin for β-catenin binding (residues 54–148)62,71-74 overlaps with that for homodimerization (residues 82–279)62,71. The crystal structure of this domain indicates that αE-catenin dimerizes through the formation of a four-helix bundle in which two antiparallel helices are contributed by each protomer. The precise role of homodimerization in adherens-junction assembly remains unclear. One attractive possibility is that the transition between homo- and heterodimer occurs at the cell membrane, which enables a β-catenin-induced conformational change to influence the association of αE-catenin with other partners — for example, actin filaments and/or their polymerizing machinery. It is not clear how this relates to the potential dimerization of αE-catenin through the M-domain or the possible enhancement of E-cadherin lateral dimerization.
αE-catenin and the actin cytoskeleton
Proteins that link αE-catenin to actin
Studies of keratinocytes in which the αE-catenin gene (CTNNAI) has been conditionally inactivated have uncovered a role for αE-catenin in the formation of the radial actin cables that are necessary for stabilizing adherens junctions, sealing membranes and assembling epithelial sheets18,39. During the initial steps of epithelial-sheet formation, linear radial actin cables assemble at nascent intercellular adherens junctions; this process requires αE-catenin and actin polymerization.
- KERATINOCYTES
Differentiated epithelial cells of the skin.
αE-catenin was postulated to interact directly with filamentous (F)-actin75, and a direct interaction was confirmed by Rimm and co-workers56. Biochemical and genetic approaches have uncovered domains in αE-catenin that mediate further interactions with actin-binding proteins (FIG. 6), which have provided new insights into the importance of αE-catenin in actin dynamics (reviewed in refs 50,76). α-actinin and vinculin were among the first binding partners of αE-catenin to be described60,66,73,77; both interact with similar residues in αE-catenin and can bind actin directly. The carboxy-terminal 280 residues of αE-catenin interact with the zonula occludens-1 (ZO1) protein, which is also able to bind actin directly66.
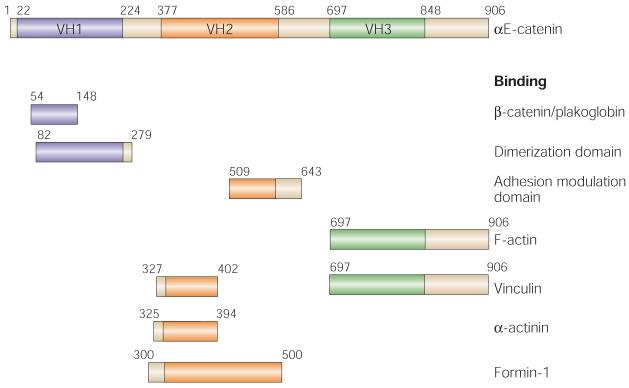
Figure 6. Functionally important regions of αE-catenin.
This schematic of αE-catenin illustrates its three vinculin homology (VH) domains (VH1–VH3). The direct binding partners for αE-catenin are listed on the right, and the corresponding domain diagrams denote the regions and encompassing amino-acid residues of αE-catenin that have been identified as biochemically essential for the interaction. αE-catenin interacts with E-cadherin–β-catenin complexes through its amino-terminal β-catenin-binding domain. In solution, αE-catenin forms a homodimer through its dimerization domain. When αE-catenin associates with E-cadherin–β-catenin complexes, however, the αE-catenin homodimer dissociates, and heterodimers of α- and β-catenin are formed. The intercellular adhesion modulation (M) domain, which partially includes the VH2 domain of αE-catenin, defines the segments of αE-catenin that are necessary for mediating intercellular adhesion. The carboxy-terminal region, which includes the VH3 domain of αE-catenin, overlaps with the filamentous (F)-actin-binding domain. The vinculin-, α-actinin- and formin-1-binding domains on αE-catenin facilitate organization of the F-actin cytoskeleton and the regulation of actin dynamics during the formation of stable intercellular adhesions.
Another binding partner of αE-catenin is afadin, which is known to bind to F-actin and might recruit it to sites of nascent adherens junctions. Afadin also associates with the protein nectin, which forms the transmembrane core of another intercellular adhesion system78,79. Other associates of αE-catenin include vasodilator-stimulated phosphoprotein (VASP) and Enabled (Ena), which localize to adherens junctions in an αE-catenin-dependent fashion39. Whether these interactions are direct or indirect is not yet clear, as αE-catenin also associates directly with members of the zyxin family, which can also bind actin and/or recruit members of the Ena/VASP families52. Irrespective of how VASP gets to the adherens junctions, both Ena and VASP can bind to the actin-monomer-binding protein profilin, which promotes the addition of ATP–actin monomers to actin filaments. VASP has recently been shown to compete with capping protein for the barbed end of pre-existing actin filaments, which provides a mechanism for how VASP promotes the extension of actin polymers (reviewed in ref. 4).
- BARBED END
The fast-polymerizing end of an actin filament, which is defined by the arrowhead-shaped decoration of actin filaments with myosin fragments.
Although the presence of Ena/VASP proteins is sufficient to explain the addition of actin subunits to preexisting actin filaments, it might not be sufficient to account for de novo actin polymerization. Recently, however, we identified isoforms of formin-1 (FMN1), the founding member of the formin family, as binding partners for αE-catenin and as candidates for generating the radial actin cables that assemble at nascent adherens junctions80. Using yeast two-hybrid and in vitro analyses, a region in the amino-terminal segment of several isoforms of FMN1 was shown to interact specifically with residues ∼300–500 of αE-catenin. The conserved formin homology (FH) domains in the carboxy-terminal portion of FMN1 contain sites for the nucleation of actin polymerization (reviewed in ref. 81). The carboxy-terminal segment of FMN1 can nucleate the polymerization of unbranched actin filaments in vitro and function in transfected cells in an αE-catenin-dependent fashion to promote the polymerization of radial actin filaments80. When transiently expressed, the carboxy-terminal portion of another formin, diaphanous protein homologue-1 (Dia1) also localizes to adherens junctions (reviewed in ref. 81). Because the FMN1 domain that associates with αE-catenin does not seem to be conserved in Dia1, it remains unclear how this carboxy-terminal segment of Dia1 is recruited to junctions. However, recent in vitro studies indicate that Dia1 might also nucleate actin polymerization82, and then associate with, and move processively along, the growing end of actin filaments in cells83.
- FORMIN FAMILY
A family of multidomain scaffold proteins that are involved in actin-dependent morphogenetic events. They are conserved from fungi to humans and are characterized by the presence of two conserved carboxy-terminal regions:the formin homology (FH) domains FH1 and FH2.
Although FMN1 and Dia1 have surfaced as good candidates to initiate the formation of linear actin cables at sites of nascent adherens junctions, it remains to be determined whether these, or any other formins in their native state, are functional at these sites, and, if so, how they are regulated to control actin polymerization. Given the complexity and possible redundancy within the mammalian formin family, resolving formin functions might be best addressed in lower eukaryotic systems. Irrespective of mechanism, once they are assembled at nascent junctions, actin filaments seem to bundle into linear cables. α-actinin is a candidate for this bundling, as it is known to form antiparallel dumbbell-like structures and thereby place an F-actin-binding site at each pole of an α-actinin dimer. Radial actin cables at junctions seem to function with myosins and generate tension across a growing epithelial sheet84. In searching for a binding partner for myosin VIIA, Kussel-Andermann and colleagues85 have identified another protein that links adherens junctions and the actomyosin-based contractile system37. Known as vezatin, this protein interacts with myosin VIIA and the carboxy-terminal region of αE-catenin. This interaction has prompted speculation that myosin VIIA, when anchored by vezatin to the adherens junctions, might use the actin cytoskeleton to generate tension, thereby strengthening cell–cell adhesion.
Finally, αE-catenin interacts directly with spectrin, which is an important component of the skeleton that underlies and reinforces the plasma membrane. The binding of spectrin to adherens-junction complexes might stabilize intercellular adhesion by integrating the complexes into this macromolecular membrane structure86.
Why so many connections?
Given the nature of the many associates of αE-catenin, it seems likely that this protein is a central player in nucleating the assembly of a multiprotein complex that links E-cadherin–β-catenin complexes to F-actin. Certainly, the CTNNA1-null keratinocytes described above illustrate the importance of this process not only for stabilizing intercellular junctions, but also for coordinating actin dynamics at these sites18,39 (FIG. 7). But why are there so many different ways to connect αE-catenin with actin dynamics?
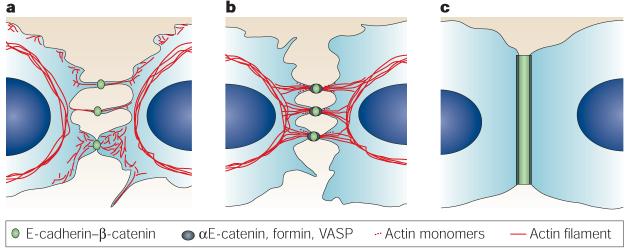
Figure 7. Model of adherens-junction formation in keratinocytes.
a | Initial cell–cell contacts are formed by opposing E-cadherin–β-catenin complexes at the tips of filopodia or lamellipodia. These initial contacts are nascent adherens junctions and are termed puncta. b | In the presence of αE-catenin, formin and other proteins such as VASP (vasodilator-stimulated phosphoprotein) that bind to αE-catenin and/or the actin cytoskeleton, bundles of radial actin cables assemble on each side of the puncta and form anchors to the underlying cortical actin ring. c | As a consequence of actin reorganization, mature adherens junctions are formed.
One factor that might help to explain the diversity in potential binding surfaces for actin at adherens junctions is the range of cytoskeletal changes that must occur in epithelial cells for them to establish stable cell–cell contacts and assemble into epithelial sheets. Some insights into these dynamics stem from video-microscopy studies of cells that express actin or cadherin bound to green fluorescent protein (GFP) while Ca2+ levels are raised and epithelial sheets form37,84,87.
During the initial steps of establishing cell–cell contacts, activated Cdc42 and Rac, which are members of the Rho family of small GTPases, stimulate the extension of filopodia and lamellipodia, respectively5,37,39,84,88. The arp2/3 protein complex has been implicated in nucleating actin polymerization and producing the extensive branched cortical meshwork of F-actin that develops beneath the membrane at Rac-stimulated lamellipodia (reviewed in ref. 89). Lamellipodial extensions generate large dynamic contacts between the surfaces of apposing cells. By contrast, filopodia-like extensions involve the polymerization of linear actin filaments and seem to be much more rapidly formed. Filopodia have been implicated in the generation of homophilic interactions between two opposing E-cadherin–β-catenin complexes at the tips of the cell–cell contact sites.
- FILOPODIA
Thin, transient actin protrusions that extend out from the cell surface and are formed by the elongation of bundled actin filaments in its core.
- LAMELLIPODIA
Cellular protrusions that contain extensively branched arrays of actin filaments, which are orientated with their plus (barbed) ends toward the plasma membrane.
- Arp2/3 PROTEIN COMPLEX
A complex that consists of two actin-related proteins, Arp2 and Arp3, along with five smaller proteins. When activated, the Arp2/3 complex binds to the side of an existing actin filament and nucleates the assembly of a new actin filament. The resulting branch structure is Y-shaped.
Once nascent adherens junctions (known as puncta) have assembled from clusters of transmembrane E-cadherin molecules and their cytoplasmic associates (β-catenin and αE-catenin), the contacts are stabilized by a second step that involves the attachment and/or de novo assembly of a linear radial actin cable at the tip of each punctum38,39,90. Proteins of the VASP and mammalian orthologue of Ena (MENA) families localize to these sites, and studies using a dominant-negative form of VASP imply that VASP might have a role in mediating intercellular adhesion in both cultured cells and transgenic mice39. As outlined above, functional studies support a role for these proteins in the elongation of pre-existing actin filaments89, and there are numerous ways in which developing adherens-junction complexes can bind to actin and potentially help to recruit F-actin.
The findings that imply a role for formins at adherens junctions are also intriguing, particularly given the broad role of formin-family members in stimulating de novo polymerization of linear radial actin cables (for reviews, see refs 81,91). As outlined above, if native formins localize to cell–cell junctions and participate in regulating the formation of linear actin cables, then an interesting issue to address will be whether different formin-family members have redundant or distinct roles in intercellular adhesion. Equally important will be whether Ena/VASP and formin-family members work together to assemble radial actin cables, or whether these proteins function independently, perhaps in response to different cellular cues.
αE-catenin binding to E-cadherin–β-catenin
Many events in organized epithelia, such as cytokinesis and cell migration, require dynamic remodelling of adhesive contacts. Once again, αE-catenin seems to be intricately involved in these processes. Whereas newly synthesized E-cadherin associates with β-catenin in the endoplasmic reticulum, αE-catenin seems to join the E-cadherin–β-catenin complex only after it has reached the plasma membrane92. Conversely, detachment of αE-catenin and actin from the E-cadherin–β-catenin complex has been observed during the disassembly of adherens junctions93.
Although it is not yet clear precisely how the association of αE-catenin with E-cadherin–β-catenin complexes is regulated, the Armadillo subfamily of catenins (β-catenin, plakoglobin and p120 catenin) seems to be involved. On signalling from tyrosine-kinase growth-factor receptors, or in response to overexpression of non-receptor tyrosine kinases, β-catenin is phosphorylated — an event that is accompanied by a decrease in adhesion4,94 and a loss of cytoskeletal anchorage to cell–cell junctions95. Moreover, in cells that are treated with pervanadate, which is an inhibitor of phosphotyrosine phosphatases, tyrosine phosphorylation of β-catenin and plakoglobin correlates with the dissociation of αE-catenin from the E-cadherin–β-catenin complex96.
Rho-family GTPases also participate in regulating cadherin-mediated cell adhesion (reviewed in ref. 97). Although several mechanisms are possible and, in fact, are likely to account for this, it is known that IQGAP1, which is a target of activated Cdc42 and Rac1 GTPases, binds to β-catenin and induces the dissociation of αE-catenin from the E-cadherin–β-catenin complex98. On the basis of these data, the association of αE-catenin with the E-cadherin–β-catenin complex seems a probable target for the functional modulation of this complex.
αE-catenin and tumour formation
Several studies imply that the disruption of cadherin-based adhesion is a main step in the progression of human epithelial cancers. Reduced levels of E-cadherin and αE-catenin proteins seem to be characteristic of many different human cancers, including malignant tumours of the breast, colon, stomach, oesophagus, bladder and liver13,15-17,99-101. The proportion of such tumours that fail to express one or both of these proteins has been reported to be as high as 80%, and the loss of E-cadherin or αE-catenin often correlates with the degree of tumour differentiation and metastasis13,102-104. The loss of αE-catenin seems to be the more common event, and also correlates better than the loss of E-cadherin with the level of invasiveness of a tumour and with its degree of differentiation13,102. αE-catenin is absent or markedly depleted in several human cancer cell lines, including the lung cancer cell line PC9 (ref. 10) and the prostate adenocarcinoma cell line PC3 (refs 11,12). If αE-catenin is artificially reintroduced into PC3 cells, E-cadherin-dependent adhesion is restored and the invasive phenotype is suppressed11. In this respect, αE-catenin seems to function as a tumour suppressor, similar to several of the classic cadherins.
The seminal work of Shimoyama and co-workers associated a loss of αE-catenin-gene function with human cancer by showing downregulation of the expression of αE-catenin in PC9 cells following homozygous deletion of part of the αE-catenin gene10. Bullions and colleagues, however, have taken the connection one step further by identifying a mutation in the αE-catenin gene in cells that are derived from an ovarian carcinoma14. The Ov2008 αE-catenin mutation results in an amino-terminally truncated form of the protein that localizes to the cytoplasm and does not bind β-catenin efficiently. Again, restoration of wild-type αE-catenin expression restricted the uncontrolled growth of the cells and restored a normal adhesive morphology.
As mentioned above, mutations in E-cadherin have also been found in human cancers and E-cadherin has been shown to have growth-suppressor activity. On the basis of cell-culture studies, some of the tumour-suppressor effects of E-cadherin might be adhesion independent. As E-cadherin binds to β-catenin, cells that have appreciable soluble pools of β-catenin might respond to elevated E-cadherin levels by suppressing the transcription of LEF1/TCF-regulated genes105. Conversely, a loss of E-cadherin expression might, in certain instances, resemble sustained activation of Wingless/WNT-related (WNT) signalling, which has a well-established link to some types of human cancer.
The ability of E-cadherin to compete with transcription factors for β-catenin raises the question of whether the loss of αE-catenin might also affect the status of WNT signalling — perhaps by negatively affecting the stability of E-cadherin–β-catenin complexes and increasing the soluble pool of β-catenin. Although this is theoretically possible, the gene-targeting studies that have been carried out so far do not support this idea; for example, when mice in which αE-catenin was conditionally deleted in the developing skin were mated with WNT-reporter-expressing mice, no increase in the activity of β-catenin–transcription-factor complexes was detected, despite the increase in epidermal-cell proliferation18. So, although αE-catenin expression clearly restores the normal phenotype to αE-catenin-mutant cancer cells, the mechanisms by which αE-catenin loss influences tumorigenesis might be distinct from those caused by either E-cadherin loss or β-catenin overactivation. The sustained activation of the Ras–ERK/MAPK pathway in skin epidermis that lacks αE-catenin expression provides one possible mechanism, which goes beyond a simple loss of cell–cell adhesion.
Potential nuclear partners for αE-catenin
The discovery that β-catenin can associate with and activate transcription factors in response to WNT signalling has been followed by more than a decade of research linking intercellular adhesion to transcriptional regulation and cancer7-9. Other interactions between cell-junction proteins and nuclear factors have since been detected106, and αE-catenin seems to be no exception.
Ajuba, which is a member of the zyxin family of proteins, has been shown to interact directly with αE-catenin107. Interestingly, whereas zyxin localizes to focal adhesions, which can be considered to be cell–substratum junctions, Ajuba localizes to cell–cell junctions. Through its dual association with the amino-terminal segment of αE-catenin and with F-actin, Ajuba might contribute to the coupling of adherens junctions to the actin cytoskeleton. Like zyxin, however, Ajuba has also been detected in the nucleus, and so has the intriguing potential to coordinate the regulation of transcription and cell adhesion108. Although further studies will be necessary to assess whether this hypothesis is correct, Ajuba joins β-catenin as a protein that localizes to both the nucleus and adherens junctions.
Another potentially interesting αE-catenin associate that is worth mentioning in this context is FMN1. Fmn1 was first identified as the gene that was mutated in mouse embryos that were homozygous for the limb deformity (ld) mutation109. FMN1 is the founding member of the formin family, and it has only more recently become apparent that a mutation in a formin gene leads to a disruption in the establishment of the Sonic hedgehog (SHH)–FGF4–BMP (where FGF stands for fibroblast growth factor and BMP is bone morphogenetic protein) feedback loop that is necessary for the formation of the apical ectodermal ridge (the layer of surface ectodermal cells at the apex of the embryonic limb bud). Interestingly, expression of Gremlin, which is a BMP-inhibitory protein, is lost in the ld-mutant mouse and grafting Gremlin-expressing cells into ld-mutant limb buds restores the feedback loop110. Recent studies imply that mutations in the Grem1 gene are sufficient to cause the ld phenotype without apparent consequence to Fmn1 expression111. As the Grem1 mutation fails to complement the ld mutation, it has been suggested that the ld phenotype might reflect defects in Grem1-gene transcription, rather than unveiling a role for FMN1 per se111. Although this reveals a functional role for Gremlin, it leaves open the functional importance of FMN1. Further studies, removing redundancies among formin-family members, will probably be required to address this intriguing issue.
Conclusions and perspectives
Although it has not been as extensively studied as its prominent partner β-catenin, αE-catenin has come into its own in recent years as an essential component of adherens junctions that can also integrate adhesion with other essential cellular events. It is well established that αE-catenin is a central protein not only in linking the actin cytoskeleton to E-cadherin–β-catenin complexes, but also in recruiting a range of other important proteins to developing intercellular junctions. Some of these proteins, such as members of the Ena/VASP and formin families, seem to be important in regulating actin polymerization at developing junctions. Others, such as vezatin and α-actinin, probably participate in generating force or tension at the interface between actin and adherens junctions. Others still, such as spectrin, might integrate the adherens junctions with the underlying cortical network, whereas Ajuba might generate crosstalk between junctions and the transcriptional state of the cell.
Studies of many of these connections are still in their infancy. In the future, it will be important to establish what conformational changes or protein modifications are involved in the selective recruitment of αE-catenin to E-cadherin–β-catenin complexes at the plasma membrane. What are the main molecular steps that are involved in regulating how αE-catenin selectively associates with and activates the actin-polymerizing/organizing machinery? Do formins have overlapping functions, particularly in epithelial-sheet formation? It is also possible that individual formins or formin isoforms might have distinct functions.
In addition, the precise roles of the Rho-family GTPases and the Ena/VASP-family members in regulating cadherin-mediated junction formation remain to be discovered. It is not known how broadly used these mechanisms are, or how they are involved in the functions of specific α-catenin-family members. Does αE-catenin strictly regulate actin-cytoskeletal dynamics at adherens junctions, or does it also have a role in polarizing the microtubule cytoskeleton? It is not clear whether Ajuba or other αE-catenin-associated proteins function as integrators of cell adhesion and transcriptional regulation, and, if so, under what circumstances this might occur. Finally, what mechanisms are involved in cadherin and αE-catenin dysregulation, which occurs during tumorigenesis and metastasis?
Finding the answers to these questions will keep investigators in the α-catenin and associated actin-dynamics fields occupied for the coming decade. However, as these answers unfold, we will be in a considerably better position to determine the extent to which α-catenin fits the role of the gatekeeper at the crossroads of adherens junctions, cytoskeletal dynamics and gene expression.
Acknowledgements
We are grateful to D. Barford and D. R. Critchley for providing us with images. E.F. is an investigator at the Howard Hughes Medical Institute, New York, USA. A.K. is a research associate supported by the US National Institutes of Health.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
The following terms in this article are linked online to:
Entrez Gene:
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
CTNNA1 | CTNNA2 | CTNNA3 | CTNNAL1 | Fmn1 | Grem1
Swiss-Prot: http://www.expasy.ch
Ajuba | Armadillo | β-catenin | α-catulin | desmoplakin | DIA1 | E-cadherin | αE-catenin | FGF4 | FMN1 | gremlin | HMP1 | HMP-2 | HMR-1 | IQGAP1 | LEF1 | myosin VIIA | αN-catenin | p120 catenin | plakoglobin | SHH | VASP | vezatin | vinculin | ZO1
Access to this interactive links box is free online.
References
- 1.Huber O. Structure and function of desmosomal proteins and their role in development and disease. Cell Mol. Life Sci. 2003;60:1872–1890. doi: 10.1007/s00018-003-3050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii K. Greater diversity of desmosomal cadherins. J. Invest. Dermatol. 2003;120:IX–X. doi: 10.1046/j.1523-1747.2003.12110.x. [DOI] [PubMed] [Google Scholar]
- 3.Garrod DR, Merritt AJ, Nie Z. Desmosomal cadherins. Curr. Opin. Cell Biol. 2002;14:537–545. doi: 10.1016/s0955-0674(02)00366-6. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 5.Tepass U. Adherens junctions: new insight into assembly, modulation and function. Bioessays. 2002;24:690–695. doi: 10.1002/bies.10129. [DOI] [PubMed] [Google Scholar]
- 6.Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 7.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 8.Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J. Cancer Res. Clin. Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi A. Tumor formation by genetic mutations in the components of the Wnt signaling pathway. Cancer Sci. 2003;94:225–229. doi: 10.1111/j.1349-7006.2003.tb01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimoyama Y, et al. Cadherin dysfunction in a human cancer cell line: possible involvement of loss of α-catenin expression in reduced cell–cell adhesiveness. Cancer Res. 1992;52:5770–5774. [PubMed] [Google Scholar]
- 11.Ewing CM, et al. Chromosome-5 suppresses tumorigenicity of PC3 prostate-cancer cells: correlation with reexpression of α-catenin and restoration of E-cadherin function. Cancer Res. 1995;55:4813–4817. [PubMed] [Google Scholar]
- 12.Morton RA, Ewing CM, Nagafuchi A, Tsukita S, Isaacs WB. Reduction of E-cadherin levels and deletion of the α-catenin gene in human prostate cancer cells. Cancer Res. 1993;53:3585–3590. [PubMed] [Google Scholar]
- 13.Kadowaki T, et al. E-cadherin and α-catenin expression in human esophageal cancer. Cancer Res. 1994;54:291–296. [PubMed] [Google Scholar]
- 14.Bullions LC, Notterman DA, Chung LS, Levine AJ. Expression of wild-type α-catenin protein in cells with a mutant α-catenin gene restores both growth regulation and tumor suppressor activities. Mol. Cell. Biol. 1997;17:4501–4508. doi: 10.1128/mcb.17.8.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moll R, Mitze M, Frixen UH, Birchmeier W. Differential loss of E-cadherin expression in infiltrating ductal and lobular breast carcinomas. Am. J. Pathol. 1993;143:1731–1742. [PMC free article] [PubMed] [Google Scholar]
- 16.Rimm DL, Sinard JH, Morrow JS. Reduced α-catenin and E-cadherin expression in breast cancer. Lab. Invest. 1995;72:506–512. [PubMed] [Google Scholar]
- 17.Shimazui T, Giroldi LA, Bringuier PP, Oosterwijk E, Schalken JA. Complex cadherin expression in renal cell carcinoma. Cancer Res. 1996;56:3234–3237. [PubMed] [Google Scholar]
- 18.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. Conditional ablation of αE-catenin in developing skin causes defects in hair-follicle development and epidermal morphogenesis. Adherens-junction formation, intercellular adhesion and epithelial polarity are all affected. Although differentiation occurs, the epidermis shows hyperproliferation, suprabasal mitoses and multinucleate cells. [DOI] [PubMed] [Google Scholar]
- 19.Tinkle CL, Lechler T, Pasolli HA, Fuchs E. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc. Natl Acad. Sci. USA. 2004;13:552–557. doi: 10.1073/pnas.0307437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 21.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 22.Orsulic S, Peifer M. An in vivo structure–function study of Armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for Wingless signaling. J. Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simske JS, et al. The cell junction protein VAB-9 regulates adhesion and epidermal morphology in C. elegans. Nature Cell Biol. 2003;5:619–625. doi: 10.1038/ncb1002. [DOI] [PubMed] [Google Scholar]
- 24.Nagafuchi A, Takeichi M, Tsukita S. The 102 kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell. 1991;65:849–857. doi: 10.1016/0092-8674(91)90392-c. [DOI] [PubMed] [Google Scholar]
- 25.Hirano S, Kimoto N, Shimoyama Y, Hirohashi S, Takeichi M. Identification of a neural α-catenin as a key regulator of cadherin function and multicellular organization. Cell. 1992;70:293–301. doi: 10.1016/0092-8674(92)90103-j. [DOI] [PubMed] [Google Scholar]
- 26.Claverie JM, et al. Characterization and chromosomal assignment of a human cDNA encoding a protein related to the murine 102-kDa cadherin-associated protein (α-catenin) Genomics. 1993;15:13–20. doi: 10.1006/geno.1993.1004. [DOI] [PubMed] [Google Scholar]
- 27.Janssens B, et al. αT-catenin: a novel tissue-specific β-catenin-binding protein mediating strong cell–cell adhesion. J. Cell Sci. 2001;114:3177–3188. doi: 10.1242/jcs.114.17.3177. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JS, et al. Identification and chromosomal localization of CTNNAL1, a novel protein homologous to α-catenin. Genomics. 1998;54:149–154. doi: 10.1006/geno.1998.5458. [DOI] [PubMed] [Google Scholar]
- 29.Park B, et al. Association of Lbc Rho guanine nucleotide exchange factor with α-catenin-related protein, α-catulin/CTNNAL1, supports serum response factor activation. J. Biol. Chem. 2002;277:45361–45370. doi: 10.1074/jbc.M202447200. [DOI] [PubMed] [Google Scholar]
- 30.Burridge K, Fath K. Focal contacts: transmembrane links between the extracellular matrix and the cytoskeleton. Bioessays. 1989;10:104–108. doi: 10.1002/bies.950100403. [DOI] [PubMed] [Google Scholar]
- 31.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagafuchi A, Takeichi M. Transmembrane control of cadherin-mediated cell adhesion: a 94 kDa protein functionally associated with a specific region of the cytoplasmic domain of E-cadherin. Cell Regul. 1989;1:37–44. doi: 10.1091/mbc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guenet JL, Simon-Chazottes D, Ringwald M, Kemler R. The genes coding for α and β catenin (Catna1 and Catnb) and plakoglobin (Jup) map to mouse chromosomes 18, 9, and 11, respectively. Mamm. Genome. 1995;6:363–366. doi: 10.1007/BF00364802. [DOI] [PubMed] [Google Scholar]
- 34.Herrenknecht K, et al. The uvomorulin-anchorage protein α-catenin is a vinculin homologue. Proc. Natl Acad. Sci. USA. 1991;88:9156–9160. doi: 10.1073/pnas.88.20.9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oda T, et al. Cloning of the human α-catenin cDNA and its aberrant mRNA in a human cancer cell line. Biochem. Biophys. Res. Commun. 1993;193:897–904. doi: 10.1006/bbrc.1993.1710. [DOI] [PubMed] [Google Scholar]
- 36.Furukawa Y, et al. Structure, expression and chromosome assignment of the human catenin (cadherin-associated protein) α1 gene (CTNNA1) Cytogenet. Cell Genet. 1994;65:74–78. doi: 10.1159/000133603. [DOI] [PubMed] [Google Scholar]
- 37.Adams CL, Nelson WJ. Cytomechanics of cadherin-mediated cell–cell adhesion. Curr. Opin. Cell Biol. 1998;10:572–577. doi: 10.1016/s0955-0674(98)80031-8. [DOI] [PubMed] [Google Scholar]
- 38.Yonemura S, Itoh M, Nagafuchi A, Tsukita S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between nonpolarized fibroblasts and polarized epithelial cells. J. Cell Sci. 1995;108:127–142. doi: 10.1242/jcs.108.1.127. [DOI] [PubMed] [Google Scholar]
- 39.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. During the formation of cadherin-mediated intercellular adhesions, Ca2+ stimulates filopodia, which penetrate and embed into neighbouring cells. E-cadherin complexes cluster at the filopodia tips and generate a two-rowed zipper of embedded puncta. Apposing cell surfaces are clamped by desmosomes, whereas vinculin, zyxin, VASP and MENA are recruited to adhesion zippers by a mechanism that requires α-catenin. [DOI] [PubMed] [Google Scholar]
- 40.Watabe M, Nagafuchi A, Tsukita S, Takeichi M. Induction of polarized cell–cell association and retardation of growth by activation of the E-cadherin catenin adhesion system in a dispersed carcinoma line. J. Cell Biol. 1994;127:247–256. doi: 10.1083/jcb.127.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres M, et al. An α-E-catenin gene trap mutation defines its function in preimplantation development. Proc. Natl Acad. Sci. USA. 1997;94:901–906. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl Acad. Sci. USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki SC, Inoue T, Kimura Y, Tanaka T, Takeichi M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol. Cell. Neurosci. 1997;9:433–447. doi: 10.1006/mcne.1997.0626. [DOI] [PubMed] [Google Scholar]
- 46.Colman DR. Neurites, synapses, and cadherins reconciled. Mol. Cell. Neurosci. 1997;10:1–6. doi: 10.1006/mcne.1997.0648. [DOI] [PubMed] [Google Scholar]
- 47.Serafini T. An old friend in a new home: cadherins at the synapse. Trends Neurosci. 1997;20:322–323. doi: 10.1016/s0166-2236(97)01126-0. [DOI] [PubMed] [Google Scholar]
- 48.Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding αN-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nature Genet. 2002;31:279–284. doi: 10.1038/ng908. Shows that mice that are homozygous for the cerebellar deficient folia (cdf) mutation are ataxic, and have cerebellar hypoplasia and abnormal lobulation of the cerebellum. The deletion on chromosome 6 includes part of Catna2, which encodes the αN-catenin protein that links the classic cadherins to the neuronal cytoskeleton. [DOI] [PubMed] [Google Scholar]
- 49.Janssens B, et al. Assessment of the CTNNA3 gene encoding human αT-catenin regarding its involvement in dilated cardiomyopathy. Hum. Genet. 2003;112:227–236. doi: 10.1007/s00439-002-0857-5. [DOI] [PubMed] [Google Scholar]
- 50.Rudiger M. Vinculin and α-catenin: shared and unique functions in adherens junctions. Bioessays. 1998;20:733–740. doi: 10.1002/(SICI)1521-1878(199809)20:9<733::AID-BIES6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 51.Izard T, et al. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- 52.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 53.Menkel AR, et al. Characterization of an F-actin-binding domain in the cytoskeletal protein vinculin. J. Cell Biol. 1994;126:1231–1240. doi: 10.1083/jcb.126.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGregor A, Blanchard AD, Rowe AJ, Critchley DR. Identification of the vinculin-binding site in the cytoskeletal protein α-actinin. Biochem. J. 1994;301:225–233. doi: 10.1042/bj3010225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kroemker M, Rudiger AH, Jockusch BM, Rudiger M. Intramolecular interactions in vinculin control α-actinin binding to the vinculin head. FEBS Lett. 1994;355:259–262. doi: 10.1016/0014-5793(94)01216-4. [DOI] [PubMed] [Google Scholar]
- 56.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. α1(E)-catenin is an actin-binding and-bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl Acad. Sci. USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. Shows αE-catenin to be a new actin-binding and -bundling protein, and supports a model in which αE-catenin is responsible for organizing and tethering actin filaments at the zones of E-cadherin-mediated cell–cell contact. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL. Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 1997;272:32448–32453. doi: 10.1074/jbc.272.51.32448. [DOI] [PubMed] [Google Scholar]
- 58.Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur. J. Biochem. 1997;247:1136–1142. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- 59.Tempel M, Goldmann WH, Isenberg G, Sackmann E. Interaction of the 47-kDa talin fragment and the 32-kDa vinculin fragment with acidic phospholipids: a computer analysis. Biophys. J. 1995;69:228–241. doi: 10.1016/S0006-3495(95)79894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss EE, Kroemker M, Rudiger AH, Jockusch BM, Rudiger M. Vinculin is part of the cadherin–catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Molony L, Burridge K. Molecular shape and self-association of vinculin and metavinculin. J. Cell. Biochem. 1985;29:31–36. doi: 10.1002/jcb.240290104. [DOI] [PubMed] [Google Scholar]
- 62.Koslov ER, Maupin P, Pradhan D, Morrow JS, Rimm DL. α-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with β-catenin. J. Biol. Chem. 1997;272:27301–27306. doi: 10.1074/jbc.272.43.27301. Reports that α-catenin exists as a homodimer in solution, whereas β-catenin exists as a monomer. When both are present, they form α–β-catenin heterodimers. The site of α-catenin dimerization localizes to the β-catenin-binding site. [DOI] [PubMed] [Google Scholar]
- 63.Craig SW, Johnson RP. Assembly of focal adhesions: progress, paradigms, and portents. Curr. Opin. Cell Biol. 1996;8:74–85. doi: 10.1016/s0955-0674(96)80051-2. [DOI] [PubMed] [Google Scholar]
- 64.Jockusch BM, et al. The molecular architecture of focal adhesions. Annu. Rev. Cell Dev. Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- 65.Winkler J, Lunsdorf H, Jockusch BM. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol. 1996;116:270–277. doi: 10.1006/jsbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 66.Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 1999;144:1311–1322. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. Describes the crystal structure of a region of αE-catenin termed the M-fragment. The region of αE-catenin previously defined as an adhesion M-domain corresponds to the carboxy-terminal four-helix bundle of the M-fragment and these domains exist as dimers in the crystal lattice, which might explain the biological activity of αE-catenin in promoting cell–cell adhesion by inducing lateral dimerization of the associated cadherin molecule. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yap AS, Brieher WM, Pruschy M, Gumbiner BM. Lateral clustering of the adhesive ectodomain: a fundamental determinant of cadherin function. Curr. Biol. 1997;7:308–315. doi: 10.1016/s0960-9822(06)00154-0. [DOI] [PubMed] [Google Scholar]
- 69.Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aberle H, et al. Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J. Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- 71.Pokutta S, Weis WI. Structure of the dimerization and β-catenin-binding region of α-catenin. Mol. Cell. 2000;5:533–543. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 72.Huber O, Krohn M, Kemler R. A specific domain in α-catenin mediates binding to β-catenin or plakoglobin. J. Cell Sci. 1997;110:1759–1765. doi: 10.1242/jcs.110.15.1759. [DOI] [PubMed] [Google Scholar]
- 73.Nieset JE, et al. Characterization of the interactions of α-catenin with α-actinin and β-catenin/plakoglobin. J. Cell Sci. 1997;110:1013–1022. doi: 10.1242/jcs.110.8.1013. [DOI] [PubMed] [Google Scholar]
- 74.Obama H, Ozawa M. Identification of the domain of α-catenin involved in its association with β-catenin and plakoglobin (γ-catenin) J. Biol. Chem. 1997;272:11017–11020. doi: 10.1074/jbc.272.17.11017. [DOI] [PubMed] [Google Scholar]
- 75.Ozawa M, Ringwald M, Kemler R. Uvomorulin–catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc. Natl Acad. Sci. USA. 1990;87:4246–4250. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Provost E, Rimm DL. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr. Opin. Cell Biol. 1999;11:567–572. doi: 10.1016/s0955-0674(99)00015-0. [DOI] [PubMed] [Google Scholar]
- 77.Watabe-Uchida M, et al. α-catenin–vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 1998;142:847–857. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ikeda W, et al. Afadin: a key molecule essential for structural organization of cell–cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 1999;146:1117–1132. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nature Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 82.Li F, Higgs HN. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 83.Higashida C, et al. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 84.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell. 2002;3:367–381. doi: 10.1016/s1534-5807(02)00259-9. Reports that during the formation of an epidermal sheet, a polarized network of nascent intercellular junctions and radial actin cables assemble in the apical plane of the monolayer. This polarized cytoskeleton is dependent on α-catenin, Rho and ROCK, and its regulation might be important for wound healing and/or stratification, in which coordinated tissue movements are involved. [DOI] [PubMed] [Google Scholar]
- 85.Kussel-Andermann P, et al. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin–catenins complex. EMBO J. 2000;19:6020–6029. doi: 10.1093/emboj/19.22.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pradhan D, Lombardo CR, Roe S, Rimm DL, Morrow JS. α-catenin binds directly to spectrin and facilitates spectrin-membrane assembly in vivo. J. Biol. Chem. 2001;276:4175–4181. doi: 10.1074/jbc.M009259200. [DOI] [PubMed] [Google Scholar]
- 87.Ehrlich JS, Hansen MD, Nelson WJ. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell–cell adhesion. Dev. Cell. 2002;3:259–270. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harden N. Signaling pathways directing the movement and fusion of epithelial sheets: lessons from dorsal closure in Drosophila. Differentiation. 2002;70:181–203. doi: 10.1046/j.1432-0436.2002.700408.x. [DOI] [PubMed] [Google Scholar]
- 89.Mullins RD. How WASP-family proteins and the Arp2/3 complex convert intracellular signals into cytoskeletal structures. Curr. Opin. Cell Biol. 2000;12:91–96. doi: 10.1016/s0955-0674(99)00061-7. [DOI] [PubMed] [Google Scholar]
- 90.Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- 92.Hinck L, Nathke IS, Papkoff J, Nelson WJ. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J. Cell Biol. 1994;125:1327–1340. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oda H, Tsukita S, Takeichi M. Dynamic behavior of the cadherin-based cell–cell adhesion system during Drosophila gastrulation. Dev. Biol. 1998;203:435–450. doi: 10.1006/dbio.1998.9047. [DOI] [PubMed] [Google Scholar]
- 94.Daniel JM, Reynolds AB. Tyrosine phosphorylation and cadherin/catenin function. Bioessays. 1997;19:883–891. doi: 10.1002/bies.950191008. [DOI] [PubMed] [Google Scholar]
- 95.Hoschuetzky H, Aberle H, Kemler R. β-catenin mediates the interaction of the cadherin–catenin complex with epidermal growth factor receptor. J. Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ozawa M, Kemler R. Altered cell adhesion activity by pervanadate due to the dissociation of α-catenin from the E-cadherin–catenin complex. J. Biol. Chem. 1998;273:6166–6170. doi: 10.1074/jbc.273.11.6166. [DOI] [PubMed] [Google Scholar]
- 97.Braga VM. Cell–cell adhesion and signalling. Curr. Opin. Cell Biol. 2002;14:546–556. doi: 10.1016/s0955-0674(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 98.Fukata M, et al. Cdc42 and Rac1 regulate the interaction of IQGAP1 with β-catenin. J. Biol. Chem. 1999;274:26044–26050. doi: 10.1074/jbc.274.37.26044. [DOI] [PubMed] [Google Scholar]
- 99.Kozyraki R, et al. Expression of cadherins and α-catenin in primary epithelial tumors of the liver. Gastroenterology. 1996;110:1137–1149. doi: 10.1053/gast.1996.v110.pm8613003. [DOI] [PubMed] [Google Scholar]
- 100.Ochiai A, et al. Frequent loss of α-catenin expression in scirrhous carcinomas with scattered cell growth. Jpn. J. Cancer Res. 1994;85:266–273. doi: 10.1111/j.1349-7006.1994.tb02092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shiozaki H, et al. Immunohistochemical detection of α-catenin expression in human cancers. Am. J. Pathol. 1994;144:667–674. [PMC free article] [PubMed] [Google Scholar]
- 102.Matsui S, et al. Immunohistochemical evaluation of α-catenin expression in human gastric cancer. Virchows Arch. 1994;424:375–381. doi: 10.1007/BF00190559. [DOI] [PubMed] [Google Scholar]
- 103.Schipper JH, et al. E-cadherin expression in squamous cell carcinomas of head and neck: inverse correlation with tumor dedifferentiation and lymph node metastasis. Cancer Res. 1991;51:6328–6337. [PubMed] [Google Scholar]
- 104.Vermeulen SJ, et al. Transition from the noninvasive to the invasive phenotype and loss of α-catenin in human colon cancer cells. Cancer Res. 1995;55:4722–4728. [PubMed] [Google Scholar]
- 105.Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting β-catenin signaling in an adhesion-independent manner. J. Cell Biol. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Daniel JM, Reynolds AB. The catenin p120 interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marie H, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with α-catenin. J. Biol. Chem. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- 108.Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeller R, Jackson-Grusby L, Leder P. The limb deformity gene is required for apical ectodermal ridge differentiation and anteroposterior limb pattern formation. Genes Dev. 1989;3:1481–1492. doi: 10.1101/gad.3.10.1481. [DOI] [PubMed] [Google Scholar]
- 110.Zuniga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 111.Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nature Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- 112.Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 113.Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]