Abstract
Superimposed on the activation of embryonic genome in preimplantation mouse embryos is the formation of a chromatin-mediated transcriptionally repressive state that arises in the late two-cell embryo and becomes more pronounced with development. In this study, we investigated expression and function of Class I histone deacetylases (HDAC) HDAC1, HDAC2, and HDAC3 during preimplantation development. HDAC1 is likely a major deacetylase in preimplantation embryos and its expression inversely correlates with changes in the acetylation state of histone H4K5 during preimplantation development. RNAi-mediated reduction of HDAC1 leads to hyperacetylation of histone H4 and a developmental delay even though expression of HDAC2 and HDAC3 is significantly induced in Hdac1-suppresssed embryos; increased expression of p21Cip1/Waf may contribute to the observed developmental delay. RNAi-mediated reduction of HDAC2 has no noticeable effect on preimplantation development, suggesting that individual HDACs have distinct functions during preimplantation development. Although RNAi-mediated targeting of Hdac3 mRNA was very efficient, maternal HDAC3 protein was stable during preimplantation development, thereby preventing an examination of its role. HDAC1 knockdown does not increase the rate of global transcription in late 2-cell embryos, but does result in elevated levels of expression of a subset of genes; this increased expression correlates with hyperacetylation of histone H4. Results of these experiments suggest that HDAC1 is involved in the development of a transcriptionally repressive state that initiates in 2-cell embryos.
Keywords: histone deacetylase, gene expression, preimplantation embryo development, histone acetylation, RNAi
Introduction
A fundamental problem in early mouse development is transforming the highly differentiated oocyte into totipotent blastomeres by the 2-cell stage. This transition, which is called the maternal-to-zygotic transition, first entails degradation of maternal mRNAs and then activation of the embryonic genome (Schultz, 2002). Genome activation results not only in replacing transcripts common to the oocyte and embryo, e.g., actin, but also in generating new transcripts and is essential for further development; mouse embryos that fail to undergo genome activation arrest at the 2-cell stage. Superimposed on genome activation, which results in a dramatic reprogramming of gene expression, is the development of a chromatin-mediated transcriptionally repressive state that is likely critical for generating the correct pattern of gene expression required for continued development (Schultz, 2002).
Changes in histone acetylation likely underlie development of the transcriptionally repressive state, because inducing histone hyperacetylation by using histone deacetylase (HDAC) inhibitors relieves the enhancer requirement for efficient expression of plasmid-borne reporter genes in 2-cell embryos (Wiekowski et al., 1993; Henery et al., 1995). Inducing histone hyperacetylation also relieves the repression for endogenous genes, e.g., Eif1a (Davis et al., 1996) and results in a further increase in global transcription as assessed by BrUTP incorporation (Aoki et al., 1997). Of note is that the strength of the transcriptionally repressive state progressively increases with development (Christians et al., 1994; Henery et al., 1995).
Histone acetylation is a particularly important modification of histone amino-termini, because, in general, increased levels of histone acetylation (hyperacetylation) are associated with transcriptionally-permissive chromatin, whereas decreased levels of acetylation (hypoacetylation) are associated with repression of gene expression (Marks et al., 2003). These changes in transcriptional activity promoted by histone acetylation may be linked to changes in chromatin structure (Thiagalingam et al., 2003; Peterson and Laniel, 2004; Shogren-Knaak et al., 2006). The steady-state level of histone acetylation is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs). In addition, HDACs control gene transcription by regulating acetylation of DNA sequence-specific transcription factors (Gu and Roeder, 1997; Wilson et al., 2006). Through these mechanisms, HDACs are emerging as critical regulators of gene expression.
Eighteen mammalian HDACs have been identified to date (Verdin et al., 2003). Based on their homology with yeast HDACs, the HDACs are grouped into four classes (Bolden et al., 2006). Class I HDACs (HDAC1, 2, 3 and 8) show homology to the yeast protein RPD3, are usually detected in the nucleus, and show ubiquitous expression in various mammalian cell lines and tissues. Class II HDACs (4,5,6,7,9 and 10) have a high degree of homology to the Hda1 protein and can shuttle between the nucleus and the cytoplasm. Class III HDACs are homologous to the yeast Sir2 HDAC and HDAC11 is the sole member of the class IV HDACs. HDACs are implicated in the development of cancer, regulation of cell proliferation, apoptosis and cell cycle (Mehnert and Kelly, 2007). The role of individual HDACs in these processes, however, has not been unambigously resolved, largely in part because the physiological role of HDACs during these processes was deduced using deacetylase inhibitors that block the majority of class I and class II enzymes.
Transcript profiling suggests that oocytes and preimplantation embryos express most Class I and II HDACs (e.g., 1,2,3,4,5,6,9, and SIR2, and possibly HDAC7 and 11; no convincing signal is detected for HDAC8)(Zeng et al, 2004, Zeng and Schultz, 2005; Pan et al., 2005). Recent studies suggest that Class II HDACs (e.g., 4 and 6) are not linked to transcription repression following genome activation (Verdel et al., 2003) (Kageyama et al., 2006). Thus, Class I HDACs may be involved in development of the transcriptionally repressive state. Consistent with this proposal is that microarray data reveal that only HDAC1 is sensitive to α-amanitin among the HDACs expressed in 2-cell embryos and Ingenuity Pathway Analysis placed HDAC1 at the hub of numerous interactions in a gene network that could contribute to development of the transcriptionally repressive state (Zeng and Schultz, 2005).
We report here that HDAC1 is likely to be critical for the overall state of hyperacetylated histones in preimplantation mouse embryos based on an inverse correlation between HDAC1 (and not HDAC2 and 3) expression and acetylation state of K5 of histone H4 (H4K5). RNAi-mediated reduction of HDAC1 in 2-cell embryos induces hyperacetylation of histone H4K5 despite up-regulation of HDAC2 and 3. In addition, expression of a subset of genes analyzed in HDAC1-depleted embryos is enhanced, including genes that normally become repressed. Development of HDAC1-depleted embryos is retarded, perhaps due to over-expression of p21Cip1/Waf. In contrast, depleting HDAC2 by RNAi has no effect on HDAC1 or HDAC3 protein levels, acetylation status of H4K5, or development to the blastocyst stage.
Materials and Methods
Oocyte and embryo collection, culture and microinjection
Cumulus cell-free germinal vesicle (GV)-intact oocytes were obtained from PMSG-primed CF-1 females as previously described (Schultz et al., 1983). The collection medium for oocytes was bicarbonate-free minimal essential medium (Earle’s salts) containing, 25 mM Hepes, pH 7.3, 3 mg/ml polyvinylpyrollidone (PVP) and 2.5 μM milrinone to prevent germinal vesicle (GV) breakdown. Oocytes were matured in vitro in Whitten’s medium (Whitten, 1971) containing 0.01% polyvinyl alcohol (PVA) (Whittens/PVA).
One-cell embryos were collected from eCG- and hCG-primed CF-1 female mice mated with B6D2F1/J males (Jackson Laboratory) as previously described (Temeles et al., 1994). Embryos were cultured in KSOM containing amino acids (Ho et al., 1995) for up to four days in 5% CO2 in air at 37°C. One-cell, 2-cell, 4-cell, 8-cell, morula, and blastocysts that developed in vivo were flushed from either oviducts or uteri at 20–21, 41–44, 60–61, 68, 75–77, and 92–96 h post-eCG, respectively. All animal experiments were approved by the Institutional Animal Use and Care Committee and were consistent with National Institutes of Health guidelines.
Microinjection of 1-cell embryos was performed essentially as previously described (Kurasawa et al., 1989). Prior to pronucleus formation, the embryos were injected with 10 pl of dsRNA using Picoliter Injector Microinjection System (Harvard Apparatus, Holliston, MA); the culture medium was bicarbonate-free Whitten’s medium containing 0.01% PVA and 25 mM Hepes, pH 7.3. Following microinjection, the embryos were cultured in KSOM containing amino acids medium as described above.
Double-stranded RNA (dsRNA) preparation
Double-stranded RNA was prepared by annealing two complementary RNAs transcribed by T7 or SP6 polymerase in vitro. cDNA fragments were initially subcloned into the PCRII vector. Hdac1 dsRNA was a 517 bp fragment prepared using primers 5′-ATCCCTAATGAGCTGCCCTACA-3′ and 5′-ATGGAGAAGATGGGGCTGCAGA-3′. Hdac2 dsRNA was a 560 bp-fragment prepared with 5′-TGTTGCCCGATGTTGGACATAT-3′ and 5′-ATCTTATCCCAGAACGTGTCTCAC-3′. Hdac3 dsRNA was a 506 bp-fragment prepared with 5′-AATACTTCGAGTACTTTGCCCC-3′ and 5′-CCCTGAGAGGGACAATCATC-3′.
After in vitro transcription using T7 and SP6 RNA polymerase (Ambion), DNA templates were removed by DNase I treatment. The RNA products were extracted with phenol: chloroform and then precipitated with ethanol. To anneal sense and antisense RNAs, equimolar quantities of sense and anti-sense RNA were mixed in annealing buffer (10 mM Tris, pH 7.4, 0.1 mM EDTA) to a final concentration of 2 μM each, heated for 2 min at 94 °C, and then incubated at room temperature for at least 16 h. To remove unhybridized RNA, the mixture was treated with 2 μg/ml RNaseT1 (Calbiochem, San Diego, CA) and 1 μg/ml RNaseA (Sigma) for 30 min at 37°C. The dsRNA products were extracted with phenol: chloroform and ethanol precipitated, then dissolved in water. The quality of dsRNA was confirmed by electrophoresis in an agarose gel. Gfp dsRNA was prepared as previously described (Stein et al., 2003). The dsRNA samples were diluted to a final concentration of 1–2 mg/ml and stored at −80°C until used.
In vitro transcription assay
BrUTP incorporation assays were performed as previously described (Aoki et al., 1997). Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope. The intensity of fluorescence was quantified using NIH Image J software (National institutes of Health) as previously described (Aoki et al., 1997).
Immunostaining of oocytes/eggs/embryos and quantification of fluorescence intensity
Oocytes or embryos were fixed in 2% paraformaldehyde in PBS for 20 min at room temperature, and then permeabilized with 0.2% Triton X-100 in PBS for 10 min. Immunocytochemical staining was performed by incubating the fixed samples with primary antibodies for 60 min, followed by secondary antibodies conjugated with Cy5 or FITC for 60 min. Polyclonal antibodies against HDAC1 (Upstate Biotechnology), hyperacetylated histone H4 (Upstate Biotechnology) and histone H4 acetylated on K5 (Abcam) and monoclonal antibodies against HDAC2 and HDAC3 (Upstate Biotechnology, Lake Placid, NY) were diluted 1:200. The DNA was stained with 1 μM SYTOX Green (Molecular Probes, Eugene, OR). The cells were then washed and mounted under a coverslip with gentle compression in VectaShield antibleaching solution (Vector Labs). Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope.
For each HDAC (or H4K5) immunostaining, all samples, i.e., oocytes, eggs, and embryos, were processed simultaneously. For each HDAC (or H4K5) the laser power was adjusted so that the signal intensity was below saturation for the developmental stage that displayed the highest intensity and all images were then scanned at that laser power. Because all images in a developmental series were taken at the same laser power one can compare signal intensity changes for a given HDAC (or H4K5) with respect to developmental stage. One cannot, however, compare the signal intensity of different HDACs at the same developmental stage.
The intensity of fluorescence was quantified using NIH Image J software. Briefly, nuclear signal was outlined and mean fluorescence intensity was measured. This same encircled region was dragged to the cytoplasm of the same cell, and background fluorescence was measured. The specific signal was calculated by dividing nuclear values by cytoplasmic values.
TUNEL labeling assay
TUNEL (TdT-mediated dUTP nick end labeling) assays were carried out with In Situ Cell Death Detection Kit (Roche Diagnostics, Basel, Switzerland) according to the manufacture’s instructions.
RNA extraction and real time RT-PCR
Total RNA from 5 to 50 embryos was extracted using the Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA). The reverse transcription reaction, primed with random hexamers, was performed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Total RNA isolated was reverse transcribed in a 20 μl reaction volume. The resulting cDNA was quantified by real time PCR (qRT-PCR). qRT-PCR analysis was performed with the ABI Taqman Assay-on-demand probe/primer sets for Hdac1, Hdac2, Hdac3, p21Cip1/Waf, Eif1a and other genes as described previously (Zeng et al., 2004) (Table S1). One embryo equivalent of cDNA was used for each real-time PCR reaction with a minimum of three replicates as well as a minus RT and minus template controls for each gene. Unless otherwise stated, quantification was normalized to Ubf and histone H2A mRNA
Immunoblot analysis
Protein samples from embryos were solubilized in Laemmli sample buffer (Laemmli, 1970), resolved by SDS-PAGE (10% gel), and transferred to a nitrocellulose membrane. The membrane was blocked by soaking in Blotto (Tris-buffered saline with 0.1% Tween-20 and 5% non-fat dried milk) for 1.5 h and incubated overnight with the primary antibody in blocking solution. The membrane was then washed three times with TBST (Tris-buffered saline with 0.1% Tween-20), incubated with a secondary antibody conjugated with horseradish peroxidase for 45 min and washed five times with TBST. The signal was detected with the ECL Advance western blotting detection reagents (Amersham, Piscataway, NJ) following the manufacturer’s recommendations. The primary antibodies (HDAC1, HDAC2, HDAC3, β-tubulin and p21Cip1/Waf) were diluted 1:1000–1:10000 and secondary antibodies were diluted 1:20000. The antibody towards p21Cip1/Waf was purchased from BD Biosciences (San Jose, CA)
Statistical Analysis
Student t-test was used to evaluate the difference between groups, and differences of p<0.05 were considered significant.
Results
Temporal and spatial pattern of HDAC1, 2, and 3 expression during oocyte maturation and preimplantation development
Prior to conducting studies to address the role of HDAC1 in the chromatin-mediated repression of transcription that develops during the 2-cell stage, we first characterized the temporal and spatial expression pattern of HDAC1, 2, and 3. Each HDAC was concentrated in the nucleus of the fully-grown, GV-intact oocyte. At metaphase I (MI) and II (MII), however, only HDAC1 was associated with chromosomes congressed on the metaphase plate (Fig. 1A) and this localization correlated with loss of H4K5 acetylation. A decrease in histone acetylation occurs during oocyte maturation (Kim et al., 2003; Sarmento et al., 2004) and association of HDAC1 with chromosomes implicates HDAC1 as the responsible HDAC. Following fertilization, HDACs 1–3 again appear in the pronucleus/nucleus. The intensity of HDAC1 nuclear staining increased until the morula stage but the morula-blastocyst transition was accompanied by a decrease in staining intensity. The intensity of HDAC2 nuclear staining displayed a progressive decrease commencing at the 2-cell stage, whereas the intensity of HDAC3 nuclear staining displayed a progressive decrease during preimplantation development. Of note is that whereas both HDAC1 and 2 were localized throughout the nucleoplasm in oocytes and preimplantation embryos, HDAC3 was enriched around the nucleolus, which in turn is surrounded by a ring of heterchromatin (Fig. 2); a similar nuclear distribution of HDAC1–3 was also observed in preimplantation embryos (data not shown). HDAC3 could therefore be critical for heterochromatin formation and/or maintenance.
Figure 1.
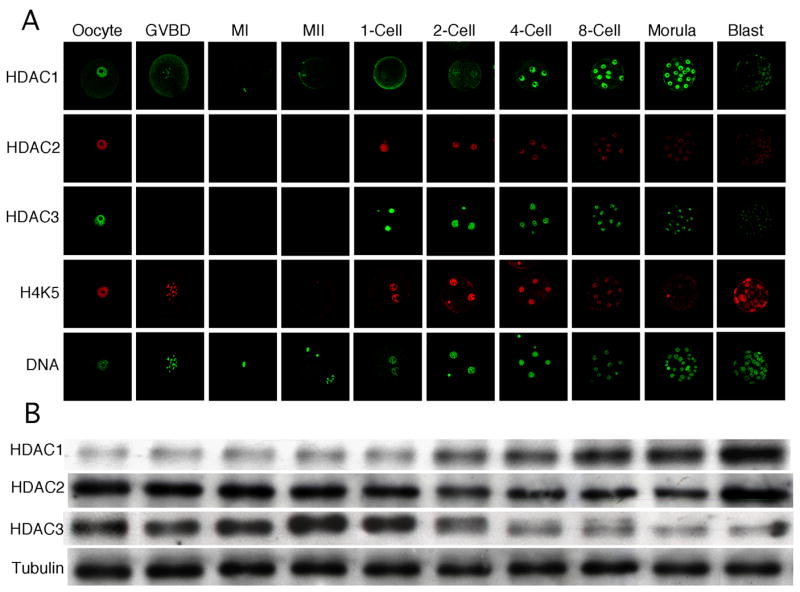
Temporal and spatial pattern of HDAC expression during oocyte maturation and preimplantation development. (A) Immunocytochemical analysis of HDAC1–3 expression and acetylation state of H4K5. All samples for a given HDAC were processed for immunocytochemistry together and all images were taken at the same laser power, thereby enabling direct comparison of signal intensities. When possible, oocytes/embryos were processed for more than one protein. The experiment was conducted 3 times and at least 25 oocytes/embryos were analyzed for each sample. Shown are representative examples. (B) Immunoblot analysis of HDAC1–3 expression. All samples for a given HDAC were processed for immunoblotting together thereby enabling direct comparison of signal intensities. One hundred oocytes/embryos were loaded per lane and the experiment was conducted twice and β-tubulin was used as a loading control. Shown are representative examples. GVBD, germinal vesicle breakdown; MI, metaphase I; MII, metaphase II.
Figure 2.
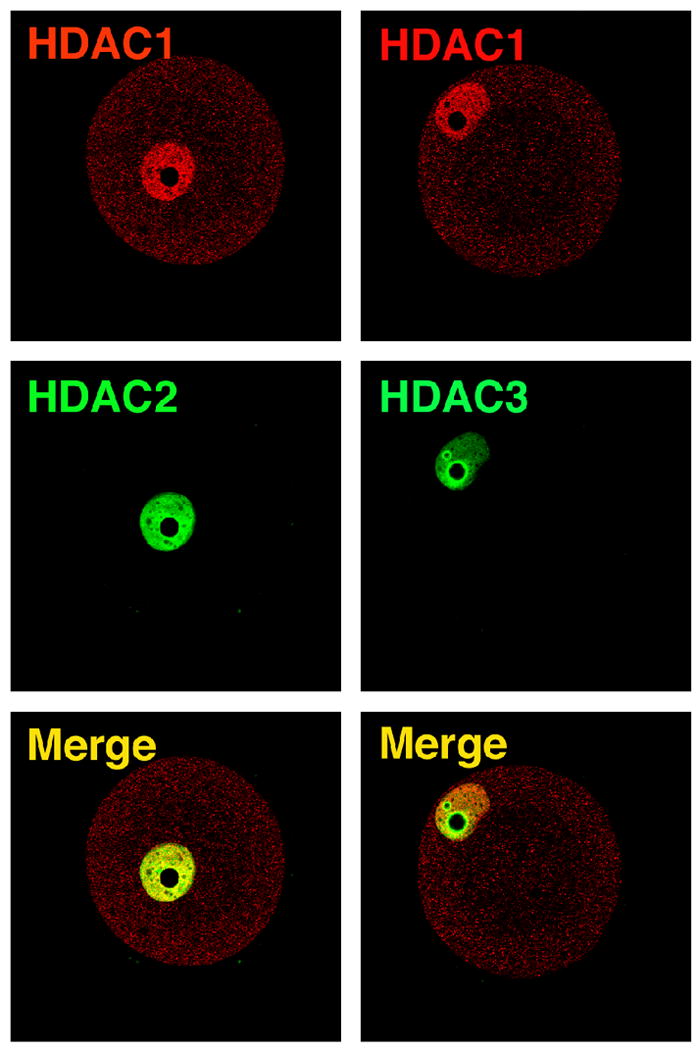
Localization of HDAC1, 2, and 3 in oocytes. HDAC1 and 2 and HDAC1 and 3 were detected by immunocytochemistry in the same oocyte; the HDAC1 antibody was raised in rabbits whereas mouse monoclonal antibodies were used to detect HDAC2 and HDAC3. Note that whereas HDAC1 and 2 co-localize mainly throughout the nucleoplasm, HDAC3 displays a pronounced enrichment around the nucleolus.
We also assessed changes in the acetylation state of H4K5 to evaluate developmental changes in chromatin structure. In somatic cells, acetylation of histone H4 occurs initially on K16, and then on K8 or K12, and ultimately on K5 (Ren et al., 2005; Turner and Fellows, 1989), an order that is also observed in mouse embryonic stem cells (Keohane et al., 1996). Thus, acetylation of H4K5 reflects hyperacetylated histone H4, which is strongly correlated with transcriptionally permissive chromatin (Urnov and Wolffe, 2001). Interestingly, the staining intensity of H4K5 inversely correlated with the intensity of HDAC1 nuclear staining (Fig. 1A). This correlation suggests that HDAC1 may be the responsible HDAC for changes in acetylation of H4K5.
The temporal pattern of Hdac1–3 expression assessed by qRT-PCR revealed that Hdac1 is zygotically expressed, confirming our previous microarray data (Zeng and Schultz, 2005)(Fig. 3). The relative transcript abundance for Hdac1 and Hdac2 was mirrored by the amount of HDAC protein as determined by immunoblotting (Fig. 1.B); the decrease in the amount of HDAC3 implies that its rate of degradation exceeds its rate of synthesis during the morula to blastocyst transition. The decrease in nuclear staining intensity for all three HDACs may reflect either a decrease in antibody accessibility or HDAC translocation to the cytoplasm, where the large dilution would obscure detecting the signal by immunofluorescence.
Figure 3.
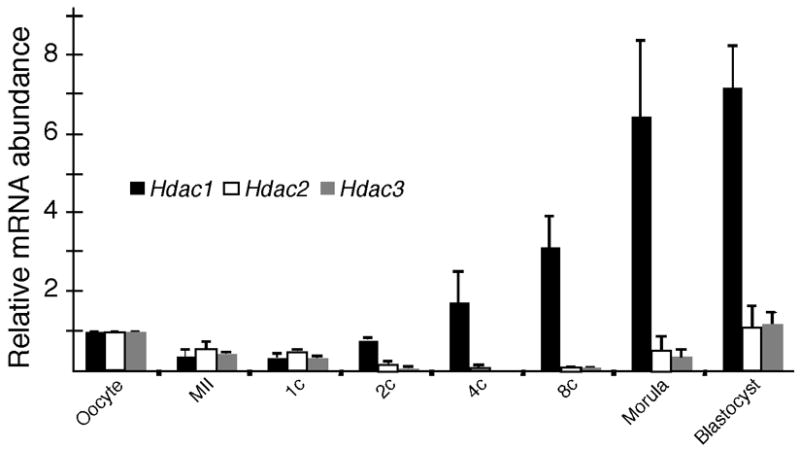
Temporal pattern of expression of Hdac1, 2, and 3. The relative abundance of Hdac1, 2, and 3 transcripts was determined by qRT-PCR. The experiment was conducted twice and the data are expressed as mean ± range and are expressed relative to the value obtained for oocytes. 1C, 1-cell embryo; 2C, 2-cell embryo; 4C, 4-cell embryo; 8C, 8-cell embryo.
RNAi-mediated ablation of HDAC1 results in acetylation of H4K5
The results described above implicate Hdac1 in development of the transcriptionally repressive state that initiates during the 2-cell stage (Wiekowski et al., 1993; Davis et al., 1996) and becomes more pronounced with further development (Christians et al., 1994; Henery et al., 1995). To address the role of HDAC1 in this process, 1-cell embryos were injected with Hdac1 dsRNA to ablate both maternal and zygotic Hdac1 mRNA; control embryos were injected with Gfp dsRNA. Results of these experiments demonstrated that Hdac1 mRNA was efficiently targeted—the relative abundance of Hdac1 mRNA was reduced by 80% for at least 72 h post-injection—and resulted in a decrease in HDAC1 nuclear staining and amount of HDAC1 protein as determined by immunoblotting (Fig. 4A, B). RNAi-mediated reduction of Hdac1 mRNA, however, resulted in a transient increase in the relative abundance of both Hdac2 (~50%) and Hdac3 (~150%) mRNA 24 h and 48 h post-injection, respectively. This transient increase in transcript abundance (Hdac2 at 24 h and Hdac3 at 48 h) translated into a increase in the amount of HDAC2 and 3 protein as detected by immunoblotting and immunocytochemistry (HDAC2 by 48 h and HDAC3 by 72 h) (Fig. 4B, C). Despite the increased amount of HDAC2 and 3 in HDAC1-depleted embryos, an increase in acetylated H4K5 was observed in these embryos (Fig. 4D, E). This finding provides further evidence that HDAC1 is the HDAC largely responsible for the acetylation status of H4K5 because increased expression of HDAC2 and 3 did not compensate for loss of HDAC1.
Figure 4.
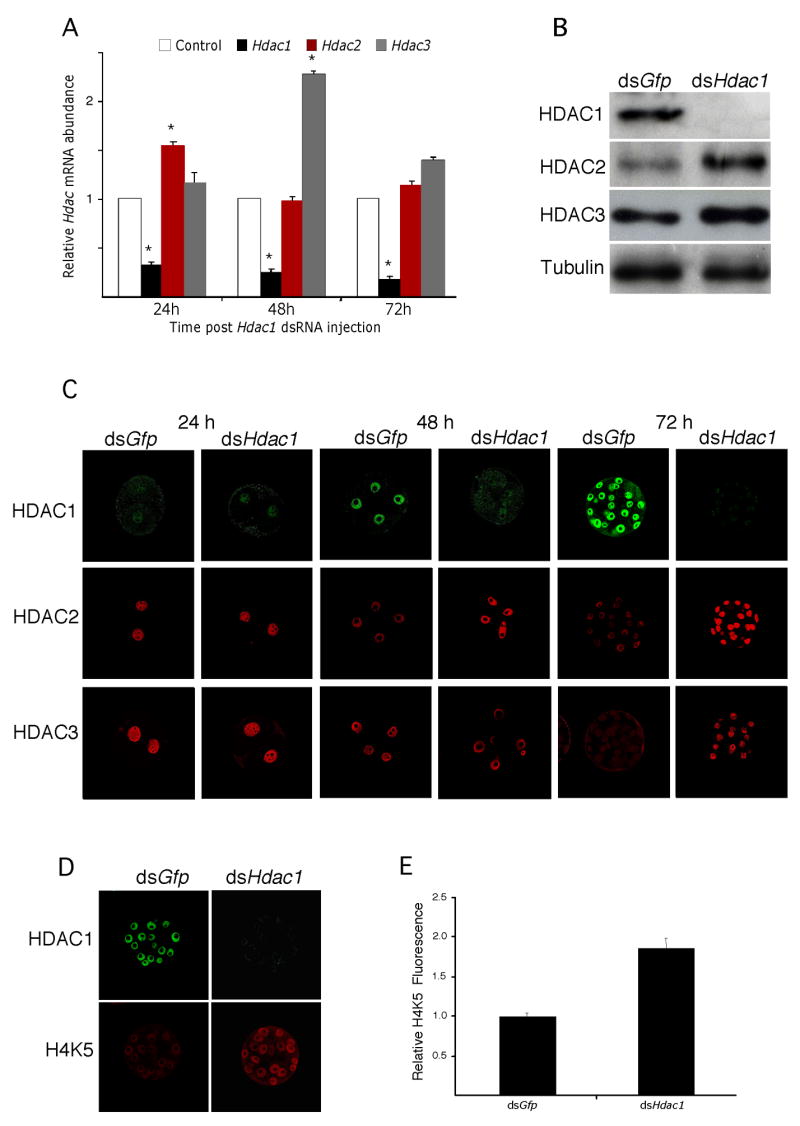
RNAi-mediated knockdown of Hdac1 mRNA. (A) Early1-cell embryos prior to pronucleus (PN) formation were injected with either Gfp dsRNA (control) or Hdac1 dsRNA and then cultured for 24 h, 48 h or 72 h, at which time the relative abundance of Hdac1, 2 and 3 transcripts was assayed by qRT-PCR and expressed relative to controls. The experiment was performed 4 times and the data expressed as mean ± SEM. *, p<0.05. (B) The experiment was performed as described in A and the relative amount of HDAC1, 2, or 3 was determined by immunoblot analysis after 72 h of culture. The experiment was performed twice and shown is an immunoblot. (C) The experiment was performed as described in A and the samples processed for immunocytochemical detection of HDAC1, 2, or 3 at the indicated times. (D) The samples in C were also processed for simultaneous detection of HDAC1 and acetylated H4K5 after 72 h of culture. (E) Quantification of the fluorescent signal in D. The data are expressed as mean ± SEM, p<0.05).
HDAC1 depletion leads to delayed preimplantation development and increased p21Cip1/Waf expression
Although depleting HDAC1 by RNAi had little or no effect on development to the compacted 8-cell/morula stage, a significantly smaller fraction of embryos developed to the fully-expanded blastocyst stage (Fig. 5A, B). The developmental delay was not due to an increase in apoptosis because there was no increase in the incidence of apoptotic cells as detected by TUNEL in Hdac1 dsRNA-injected embryos when compared to control Gfp dsRNA-injected embryos (Fig. S1); the number of TUNEL positive cells was 2.8±1.4 and 2.7±1.6 (mean ±SEM) in control and experimental embryos, respectively. Consistent with this finding is that depleting HDAC1 protein did not increase expression of either the pro-apoptotic Bax or Bcl2 transcripts (Fig. S1).
Figure 5.
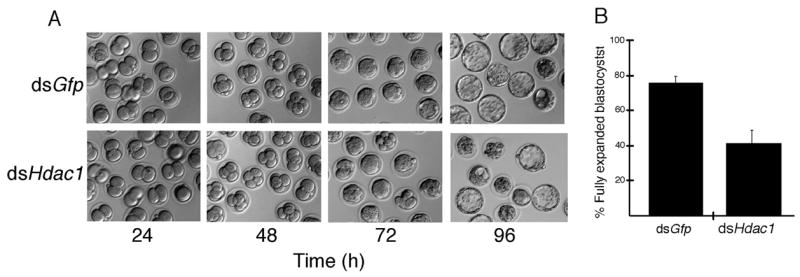
Effect of RNAi-mediated reduction of HDAC1 on development. (A) Early1-cell embryos prior to pronucleus (PN) formation were injected with either Gfp dsRNA (control) or Hdac1 dsRNA and then cultured for 24 h, 48 h, 72 h or 96 h and scored for development. Development to the compacted 8-cell stage was relatively unaffected, whereas development to the fully expanded blastocyst stage was inhibited. The experiment was performed 5 times and at least 50 embryos were examined for each sample. (B) Quantification of developmental delay following reduction of HDAC1. Data are expressed as mean ± SEM, p<0.05.
To gain a better understanding of the basis for the developmental delay of the HDAC1-depleted embryos, we assayed by qRT-PCR expression of p21cip1/WAF in these embryos. The rationale for doing so is that HDAC inhibitors induce expression of p21Cip1/Waf (as well as other CDK inhibitors) in many cells and tumors, leading to cell cycle arrest (Marks et al., 2003; Zhu et al., 2004). In addition, induction of p21Cip1/Waf was observed in Hdac1-deficient embryonic stem cells (Lagger et al., 2002). We noted a 2.5-fold increase in p21Cip1/Waf mRNA level 72 h post-injection (Fig. 6A), as well as a similar increase in p21CIP1/WAF protein 96 h post-injection (Fig. 6B).
Figure 6.
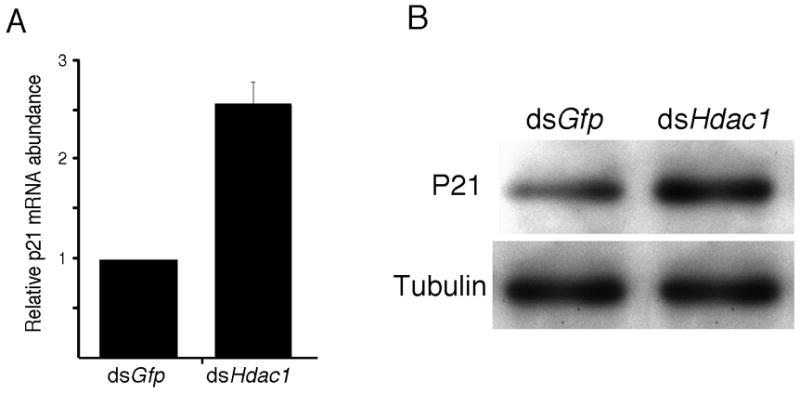
Effect of HDAC1 knockdown on p21Cip1/Waf expression. The relative amounts of p21Cip1/Waf mRNA and protein were determined 72 and 96 h following injection, respectively. (A) The experiment was performed 4 times and the data (mean ± SEM) are expressed relative to controls. The differences are significant, p<0.01. (B) The experiment was performed twice and shown is an immunoblot. β-Tubulin was used as a loading control.
Hdac1 is involved in development of the transcriptionally repressive state
The ability of HDAC inhibitors to relieve the requirement for an enhancer for efficient expression of plasmid-borne reporter genes (Wiekowski et al., 1993; Henery et al., 1995) and to maintain expression of endogenous genes that are transiently expressed during the 2-cell stage (Davis et al., 1996) led to the proposal that a chromatin-mediated transcriptionally repressive state develops during the late two-cell stage. To ascertain whether expression of Hdac1 is central to development of the transcriptionally repressive state, we next analyzed HDAC1-suppressed embryos at the late 2-cell stage in greater detail.
Zygotes injected with Hdac1 dsRNA were collected 35 h after injection, a time when the embryos were at the late 2-cell stage. We first examined expression of HDAC1, HDAC2 and HDAC3 using immunocytochemistry and observed that HDAC1 staining was reduced by 55% compared with control Gfp dsRNA-injected embryos, whereas there was no significant effect on either HDAC2 or HDAC3 (Fig. 7A, B). In addition, histone H4 became hyperacetylated in these injected embryos (Fig. 7A, B), providing further support that HDAC1 is a major regulator of histone acetylation in the preimplantation embryo.
Figure 7.
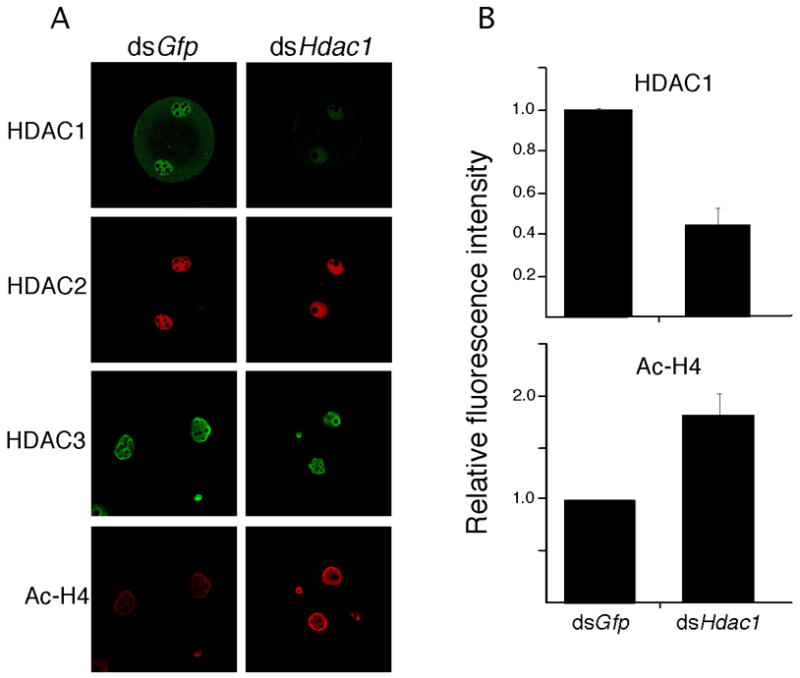
Effect of RNAi-mediated reduction of Hdac1 mRNA on expression of HDAC1, 2, and 3 and histone acetylation in late 2-cell embryos. (A) Immunocytochemical detection of HDAC1–3 and hyperacetylated histone H4. Note marked reduction in HDAC1 and enhanced staining for hyperacetylated histone H4 but little effect on HDAC2 and HDAC3. The experiment was performed 4 times and at least 40 embryos were analyzed for each sample. Shown are representative examples. (B) Quantification of the reduction in HDAC1 and increase in hyperacetylated H4. The data are expressed as mean ± SEM and relative to controls. In both cases the differences are significant, p<0.05.
We had previously noted that TSA treatment of 2-cell embryos results in a marked increase in acetylation of H4K5 and a global increase in transcription using a transcription run-on assay that monitors BrUTP incorporation (Aoki et al., 1997). The increase in acetylation of H4K5 is much greater than that observed in HDAC1-depleted embryos. Conducting similar experiments using HDAC1-depleted embryos, however, did not reveal any significant increase in BrUTP incorporation, i.e., there was no apparent increase in overall transcription (Fig. S2). Nevertheless, analysis of a battery of transcripts in these HDAC1-depleted late 2-cell embryos revealed that expression of about half were significantly enhanced (Fig. 8). Thus, HDAC1 appears to regulate expression of a subset of transcripts, which is consistent with its presence in a subset of transcription complexes (Yang and Seto, 2003, 2008).
Figure 8.
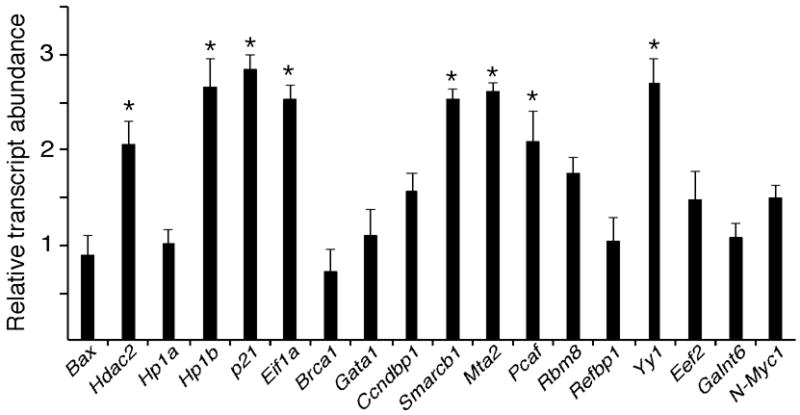
Effect of RNAi-mediated degradation of Hdac1 mRNA on expression of selected genes in late 2-cell embryos (i.e., 35 h post-injection). The data (mean ±SEM, n=3) are expressed relative to controls. That data were normalized to human GAPDH mRNA that added as an external control prior to RNA isolation. *, p<0.05
RNAi targeting of Hdac2 and Hdac3 transcripts
To confirm a specific role for HDAC1 in modulating the acetylation status of H4K5 and the development of a transcriptionally repressive state, we used RNAi to target Hdac2 and Hdac3 transcripts. Among mammalian class I HDACs (subtypes 1, 2, 3 and 8), HDAC1 and HDAC2 are highly similar, with an overall sequence identity of ~ 82%, and found in the ubiquitously expressed mSin3a, NURD/Mi2/NRD, and CoRest co-repressor complexes (Yang and Seto, 2003, 2008). In vivo, HDAC1 and HDAC2 display activity within co-repressor complexes, which is consistent with our observation that HDAC1 and HDAC2 co-localize in preimplantation mouse embryos (Fig. 2).
RNAi-mediated targeting of Hdac2 resulted in a marked reduction in Hdac2 mRNA for at least 72 h post-injection of Hdac2 dsRNA into zygotes (Fig. 9A). Targeting was specific in that no effect was observed on the relative abundance of Hdac1 mRNA. Interestingly, there was a transient increase in expression of Hdac3 mRNA (Fig. Fig. 9A). Immunoblotting and immunocytochemical analyses revealed that RNAi-mediated targeting of Hdac2 mRNA resulted in a dramatic reduction in the amount of HDAC2 protein (Fig. 9B, C). Similar analyses revealed no apparent effect on the amount of HDAC1 and HDAC3 protein. Thus, even though there was a transient increase in the relative amount of Hdac3 mRNA in response to targeting Hdac2 mRNA, this increase did not result in any observable increase in the amount of HDAC3 protein.
Figure 9.
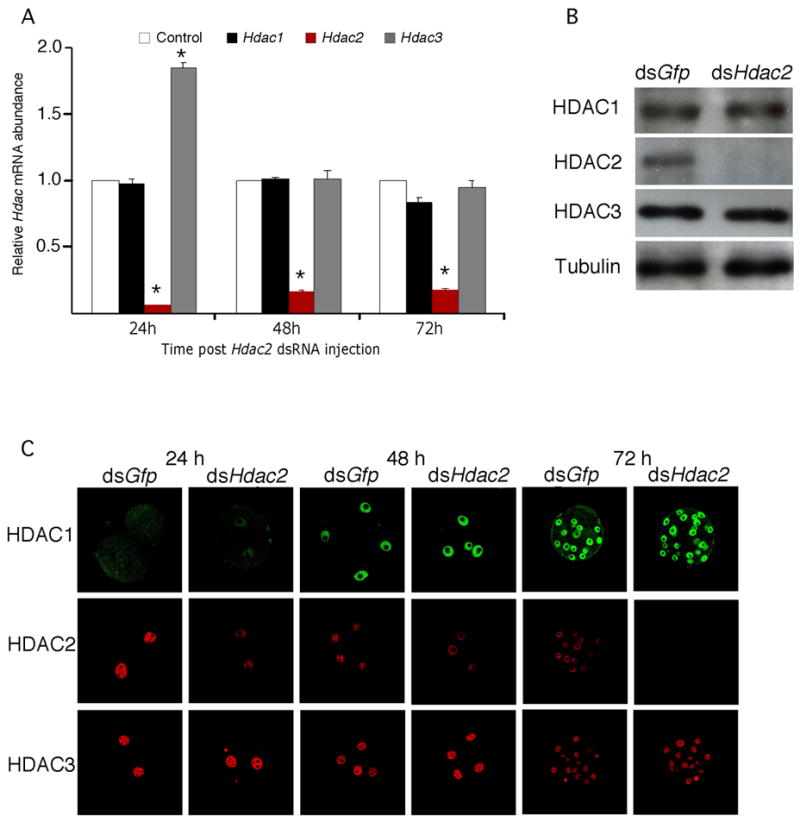
Effect of RNAi-mediated reduction of Hdac2 mRNA on Hdac expression. (A) Early1-cell embryos prior to pronucleus (PN) formation were injected with either Gfp dsRNA (control) or Hdac2 dsRNA and then cultured for 24 h, 48 h or 72 h, at which time the relative abundance of Hdac1, 2, and 3 transcripts was assayed by qRT-PCR and expressed relative to controls. The experiment was performed 4 times and the data expressed as mean ± SEM. *, p<0.05. (B) The experiment was performed as described in A and the relative amount of HDAC1, 2, or 3 was determined by immunoblot analysis after 72 h. The experiment was performed twice and shown is an immunoblot; β-tubulin was used as a loading control. (C) The experiment was performed as described in A and the samples processed for immunocytochemical detection of HDAC1, 2, or 3 at the indicated times. At least 40 embryos were analyzed for each sample and shown are representative images.
We next investigated the effects of targeting Hdac2 mRNA on preimplantation development. We routinely observed that ~80% of the Hdac2 dsRNA injected eggs reached the blastocyst stage during the four-day culture period. A comparable rate of development was also observed for Gfp dsRNA-injected eggs (Fig. 5A). In addition, the acetylation state of H4K5 was unaffected, and no increase in the relative abundance of either Eif1a or p21cip1/WAF mRNA was observed (Fig. S3); the relative abundance of each of these transcripts was increased following targeting of Hdac1 mRNA (Fig. 3A). These results strongly suggest that HDAC2 is not required for preimplantation development.
Although structurally related to HDAC1 and HDAC2, HDAC3 is component of the NCoR-SMRT co-repressor complex (Guenther et al., 2002), which is distinct from co-repressor complexes that typically contain HDAC1 and HDAC2 (Yang and Seto, 2003). We found that HDAC3 showed a distribution in preimplantation embryos similar to that observed in oocytes, i.e., enriched on heterochromatin surrounding the nucleolus (data not shown). This suggests that HDAC3 may possess unique roles during mouse early embryogenesis.
RNAi-mediated targeting of Hdac3 mRNA was very efficient but did not result in any significant difference in HDAC3 protein as assessed by immunocytochemistry (Fig. S4). This stability of HDAC3 precluded our ability to ascertain whether HDAC3 is critical for preimplantation development.
TSA treatment preferentially stimulates HDAC1 accumulation
An increase in histone H4 acetylation occurs during the 1-cell stage (Adenot et al., 1997). Histone acetylation, which persists through mitosis, could serve as molecular memory to mark genes for expression following entry into interphase. We previously demonstrated (Zeng and Schultz, 2005) and confirmed in this study that Hdac1 is zygotically expressed. To determine whether histone acetylation may contribute to the formation of a chromatin structure that underlies preferential expression of Hdac1, we assessed the effect of inducing histone hyperacetylation by a 6h TSA treatment of 2-cell embryos on Hdac1–3 expression. As expected, a 6-h TSA treatment led to a massive increase in histone acetylation (Fig. 10A). TSA treatment also led to a dramatic increase in Hdac1 expression as determined by the amount of HDAC1 protein, which was mainly localized to the cytoplasm. This increase in the amount of HDAC1 protein was preceded by an increase in the relative abundance of Hdac1 mRNA (Fig. 10B). In contrast, there was no apparent effect on either Hdac2 or Hdac3 expression after a 6-h TSA treatment. A 12-h TSA treatment also resulted in an increase in HDAC2, but not HDAC3 (Fig. 10A). Thus, histone acetylation may provide a molecular mark for Hdac1 expression in the 2-cell embryo.
Figure 10.
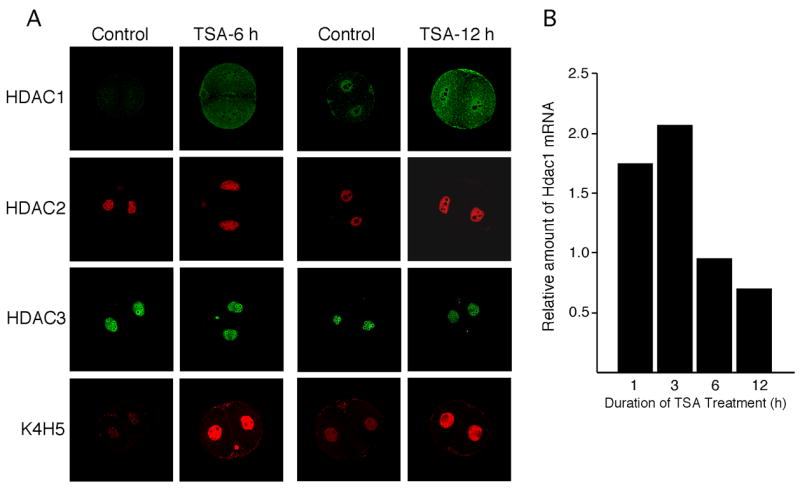
Effect of TSA treatment on Hdac expression and histone acetylation. (A) Two-cell embryos were incubated in the presence or absence of TSA for the indicated times at which time the embryos were processed for immunocytochemical detection of HDAC1, 2, and 3 and acetylated H4K5. The experiment was performed twice and a total of 24 embryos for each treatment group were examined. Shown are representative images. (B) Quantification of the effect of TSA treatment on the relative amount of Hdac1 mRNA with respect to time of TSA treatment; the data are expressed relative to untreated embryos that were cultured for the same length of time as those treated with TSA.
Discussion
Results of experiments reported here strongly implicate HDAC1 as the HDAC primarily responsible for establishing the steady-state level of histone acetylation in preimplantation mouse embryos, assuming that acetylation of H4K5 is a proxy for histone hyperacetylation. This conclusion is based on (1) qRT-PCR and microarray data indicating that Hdac1 mRNA is the major Class I Hdac transcript present in the preimplantation embryo, (2) the inverse relationship between HDAC1 nuclear localization and the acetylation status of H4K5, (3) RNAi-mediated reduction in HDAC1 resulting in an increase in acetylation of H4K5 despite a marked compensatory increase in HDAC2 and HDAC3, (4) expression of Hdac1, but not Hdac2 and Hdac3, accompanying genome activation, and (5) Hdac1 expression, which is stimulated by histone acetylation in other cell types (Hauser et al., 2002; Schuettengruber et al., 2003), being most sensitive to histone hyperacetylation induced by TSA.
Consistent with HDAC1 serving as the major HDAC in preimplantation mouse embryos are results from a previous study that found a marked reduction in total HDAC activity in HDAC1-deficient embryonic stem cells (Lagger et al., 2002). This decrease in total HDAC activity occurred despite a compensatory increase in HDAC2 and HDAC3 (Lagger et al., 2002), an increase similar to that we observed following RNAi-mediated reduction of HDAC1. We also find that RNAi-mediated reduction of HDAC2 has no effect on the acetylation state of H4K5, further supporting a role for HDAC1 as the major HDAC in preimplantation mouse embryos.
Oocyte maturation is accompanied by changes in histone modifications. Although global levels of the Me(K4)H3, Me(K9)H3 and Ph(S1)H4/H2A appear unchanged (Sarmento et al. 2004), a decrease in hyperacetylated H4, Me(R17)H3 and Me(R3)H4 occurs (Endo et al., 2005; Wang et al., 2006; Kim et al., 2003; Sarmento et al., 2004); histone deacetylation does not occur during mitosis (Kruhlak et al., 2001). The maturation-associated decrease in acetylation is due to HDAC activity and an apparent lack of HAT activity (Kim et al., 2003). TSA treatment prevents the maturation-associated decrease in histone acetylation, which results in a decrease in the extent of chromosome condensation. Reduced chromosome condensation is the likely basis for the observed increase the incidence of aneuploidy (Akiyama et al., 2006). Of interest is that histone deacetylation accompanying maturation of oocytes obtained from old mice is less pronounced (Akiyama et al., 2006) and could contribute to the age-associated increase in aneuploidy observed in females (Hassold, 1986; Hunt and Hassold, 2002). We also noted a maturation-associated decrease in H4K5 acetylation and our finding that HDAC1, but not HDAC2 or HDAC3, is associated with condensed chromosomes at MI and MII strongly suggests that HDAC1 mediates this decrease. Curiously, although porcine oocytes display a maturation-associated decrease in histone acetylation, HDAC1 is not associated with metaphase chromosomes in this species (Wang et al., 2006).
Targeted deletion of Hdac1, which results in embryo death post-implantation (Lagger et al., 2002), leads to increased expression of specific cyclin-dependent kinase inhibitors (p21Cip1Waf and p27Kip1) but no increase in apoptosis (Lagger et al., 2002). Enhanced expression of these genes likely contributes to the decrease in cell proliferation in Hdac1-deficient ES cells and post-implantation embryos; up-regulation of p21/Cip1Waf and p27Kip1 expression is likely due to increased histone acetylation of their promoters. In addition, CDK inhibitors are often induced in cells and tumors exposed to HDAC inhibitors with cell cycle arrest in the absence of apoptosis (Marks et al., 2003; Zhu et al., 2004). We find that reducing the amount of Hdac1 mRNA results in a developmental delay/arrest with the effect becoming most apparent during the 8-cell/morula to blastocyst transition; this developmental delay/arrest occurs in the absence of an increase in apoptosis. Up-regulation of p21Cip1Waf in Hdac1-deficient embryos may serve as a primary cause for this delay by increasing cell cycle time.
Our results indicate that HDAC1 is a major contributor to the development of the transcriptionally repressive state that is superimposed on genome activation. The presence of the transcriptionally repressive state is unmasked by the increased rate of transcription observed following inducing histone hyperacetylation (Aoki et al, 1997) and by the observation that an enhancer is no longer required for efficient expression of plasmid-born reporter genes (Wiekowski et al., 1993). We observe a progressive decrease in the steady-state amount of acetylated H4K5 between the 2-cell and morula stages of development. Given the positive correlation between histone acetylation and gene expression, the global decrease in histone acetylation, which correlates well with the decrease in synthesis of hyperacetylated H4 between the 1-cell and 8-cell stages (Wiekowski et al., 1997), may underlie the increasing strength of the transcriptionally repressive state that occurs between the 2-cell and morula stages.
N-terminal histone modifications (e.g., phosphorylation, methylation) are interdependent (Jenuwein and Allis, 2001; Strahl, 2000; Turner, 2002) and a decrease in H3K9 methylation occurs in HDAC1-deficient ES cells (Lagger et al., 2002). If H3K9 methylation is positively linked to Hdac1 expression in preimplantation embryos, an increase in H3K9 methylation, which is associated with repression of transcription via binding of heterochromatic protein 1 (HP1) (Lomberk et al., 2006), could contribute to the strength of the developmentally acquired transcriptionally repressive state. Interestingly, reducing HDAC1 resulted in an increase in Hp1b expression, as well as expression of Yy1, which recruits HDACs 1–3 to mediate its repressive function (Thomas and Seto, 1999; Yang et al., 1996). Increased expression of these genes, like that observed for Hdac2 and Hdac3 in HDAC1-deficient embryos, may represent another attempt mounted by the preimplantation embryo to initiate development of the transcriptionally repressive state in the absence of HDAC1. Whether other histone modifications that result in repression of transcription (e.g., methylation of H3K27 via its ability to recruit polycomb repressive complex 1 (PCR1) (Lyko et al., 2006)), occur as a consequence of depleting HDAC1 was not investigated.
We previously demonstrated that inducing histone hyperacetylation with HDAC inhibitors stimulates global transcription in late 2-cell stage embryos by 60% (Aoki et al., 1997). In contrast, we find no apparent increase in BrUTP incorporation following HDAC1 knockdown. A likely explanation is that the increase in histone acetylation observed following treatment with TSA/trapoxin is markedly greater than that observed following depletion of HDAC1. Consistent with this difference is that whereas HDAC1-depleted embryos readily develop to the 4-cell stage, 2-cell embryos treated with HDAC inhibitors fail to cleave (Ma et al., 2001). Because HDAC1 was knocked down by 55%, it is possible that total ablation of HDAC1, which would presumably result in a more pronounced increase in histone acetylation, would have resulted in an increase in BrUTP incorporation. This is unlikely to be the case, however, because by the morula stage when HDAC1 is virtually undetectable, the increase in acetylation of H4K5 still remains substantially lower than that observed following TSA treatment. Thus, it is unlikely that HDAC1 is a regulator of global transcription, but rather that HDAC1 affects expression of a subset of genes. This proposal is consistent with (1) the presence of HDAC1 in a subset of transcription factor complexes (Yang and Seto, 2003, 2008) (2) our finding that only a subset of transcripts are up-regulated following RNAi-mediated reduction of HDAC1 in late 2-cell embryos, and (3) microarray analysis revealing ~ 5% of genes up-regulated in Hdac1-deficient ES cells (Zupkovitz et al., 2006).
In summary, our results provide support for a seminal function for HDAC1 in preimplantation development by modulating gene expression. Future studies will address the impact of depleting HDAC1 on global patterns of gene expression during the course of genome activation to identify direct and indirect targets essential for development, as well as HDAC1’s role in oocyte development, which is characterized by formation of a transcriptionally quiescent state.
Supplementary Material
Figure S1. Lack of effect of depleting HDAC1 on apoptosis as determined by (A) TUNEL labeling and (B) expression of pro-apoptotic genes. The data are expressed as mean ± SEM and the differences between control and experimental groups are not significant.
Figure S2. Effect of RNAi-mediated reduction of Hdac1 mRNA on BrUTP incorporation. (A) Immunocytochemical detection of BrUTP incorporation and (B) quantification of the data. The experiment was performed 3 times and a total of 24 embryos examined. The difference between control and experimental embryos is not significant, p>0.05.
Figure S3. Lack of effect of RNAi-mediated reduction of Hdac2 mRNA on development, histone acetylation and gene expression. (A) Newly inseminated eggs were injected with either Gfp dsRNA (control) or Hdac2 dsRNA and cultured for 96 h at which time they were scored. (B) The injected embryos were processed for immunocytochemical detection of acetylated histone H4 and (C) the data quantified. Embryos harvested at the indicated times following injection were also processed for quantification of the relative amounts of Eif1a (D) and p21Cip1/Waf (E) mRNA. None of the differences between control and experimental embryos is significant, p>0.05.
Figure S4. RNAi-mediated degradation of Hdac3 mRNA (A) does not result in decrease in HDAC3 protein (B and C).
Acknowledgments
This research was supported by a grant from the NIH (HD22681) to RMS. The authors thank Paula Stein for thoughtful comments regarding the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci USA. 2006;103:7339–7344. doi: 10.1073/pnas.0510946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplanation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Christians E, Rao VH, Renard JP. Sequential acquisition of transcriptional control during early embryonic development in the rabbit. Dev Biol. 1994;164:160–172. doi: 10.1006/dbio.1994.1188. [DOI] [PubMed] [Google Scholar]
- Davis W, Jr, DeSousa PD, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: Identification by mRNA differential display and the role of DNA replication. Dev Biol. 1996;181:296–307. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- Endo T, Naito K, Aoki F, Kume S, Tojo H. Changes in histone modifications during in vitro maturation of porcine oocytes. Mol Reprod Dev. 2005;71:123–128. doi: 10.1002/mrd.20288. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16:3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T. Chromosome abnormalities in human reproduction. Trends in Genetics. 1986;2:105–110. [Google Scholar]
- Hauser C, Schuettengruber B, Bartl S, Lagger G, Seiser C. Activation of the mouse histone deacetylase 1 gene by cooperative histone phosphorylation and acetylation. Mol Cell Biol. 2002;22:7820–7830. doi: 10.1128/MCB.22.22.7820-7830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henery CC, Miranda M, Wiekowski M, Wilmut I, DePamphilis ML. Repression of gene expression at the beginning of mouse development. Dev Biol. 1995;169:448–460. doi: 10.1006/dbio.1995.1160. [DOI] [PubMed] [Google Scholar]
- Ho Y, Wigglesworth K, Eppig JE, Schultz RM. Preimplantation development of mouse embryos in KSOM: Augmentation by amino acids and analysis of gene expression. Mol Reprod Dev. 1995;41:232–238. doi: 10.1002/mrd.1080410214. [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kageyama S, Liu H, Nagata M, Aoki F. Stage specific expression of histone deacetylase 4 (HDAC4) during oogenesis and early preimplantation development in mice. J Reprod Dev. 2006;52:99–106. doi: 10.1262/jrd.17044. [DOI] [PubMed] [Google Scholar]
- Kim JM, Liu H, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol. 2003;162:37–46. doi: 10.1083/jcb.200303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett-Jones DP. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- Kurasawa S, Schultz RM, Kopf GS. Egg-induced modifications of the zona pellucida of mouse eggs: Effects of microinjected inositol 1,4,5-trisphosphate. Dev Biol. 1989;133:295–304. doi: 10.1016/0012-1606(89)90320-5. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Beisel C, Marhold J, Paro R. Epigenetic regulation in Drosophila. Curr Top Microbiol Immunol. 2006;310:23–44. doi: 10.1007/3-540-31181-5_3. [DOI] [PubMed] [Google Scholar]
- Ma J, Svoboda P, Schultz RM, Stein P. Regulation of zygotic gene activation in the preimplantation mouse embryo: global activation and repression of gene expression. Biol Reprod. 2001;64:1713–21. doi: 10.1095/biolreprod64.6.1713. [DOI] [PubMed] [Google Scholar]
- Marks PA, Miller T, Richon VM. Histone deacetylases. Curr Opin Pharmacol. 2003;3:344–351. doi: 10.1016/s1471-4892(03)00084-5. [DOI] [PubMed] [Google Scholar]
- Mehnert JM, Kelly WK. Histone deacetylase inhibitors: biology andmechanism of action. Cancer J. 2007;13:23–29. doi: 10.1097/PPO.0b013e31803c72ba. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Ren C, Zhang L, Freitas MA, Ghoshal K, Parthun MR, Jacob ST. Peptide mass mapping of acetylated isoforms of histone H4 from mouse lymphosarcoma cells treated with histone deacetylase (HDACs) inhibitors. J Am Soc Mass Spectrom. 2005;16:1641–1653. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128:301–311. doi: 10.1530/rep.1.00297. [DOI] [PubMed] [Google Scholar]
- Sarmento OF, Digilio LC, Wang Y, Perlin J, Herr JC, Allis CD, Coonrod SA. Dynamic alterations of specific histone modifications during early murine development. J Cell Sci. 2004;117:4449–4459. doi: 10.1242/jcs.01328. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Simboeck E, Khier H, Seiser C. Autoregulation of mouse histone deacetylase 1 expression. Mol Cell Biol. 2003;23:6993–7004. doi: 10.1128/MCB.23.19.6993-7004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Schultz RM, Montgomery RR, Belanoff JR. Regulation of mouse oocyte maturation: Implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol. 1983;97:264–273. doi: 10.1016/0012-1606(83)90085-4. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Stein P, Svoboda P, Anger M, Schultz RM. RNAi: Mammalian oocytes do it without RNA-dependent RNA polymerase. RNA. 2003;9:187–192. doi: 10.1261/rna.2860603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Temeles GL, Ram PT, Rothstein JL, Schultz RM. Expression patterns of novel genes during mouse preimplantation embryogenesis. Mol Reprod Dev. 1994;37:121–129. doi: 10.1002/mrd.1080370202. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Turner BM, Fellows G. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur J Biochem. 1989;179:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Wolffe AP. Chromatin remodeling and transcriptional activation: the cast (in order of appearance) Oncogene. 2001;20:2991–3006. doi: 10.1038/sj.onc.1204323. [DOI] [PubMed] [Google Scholar]
- Verdel A, Seigneurin-Berny D, Faure AK, Eddahbi M, Khochbin S, Nonchev S. HDAC6-induced premature chromatin compaction in mouse oocytes and fertilised eggs. Zygote. 2003;11:323–328. doi: 10.1017/s0967199403002387. [DOI] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- Wang Q, Yin S, Ai JS, Liang CG, Hou Y, Chen DY, Schatten H, Sun QY. Histone deacetylation is required for orderly meiosis. Cell Cycle. 2006;5:766–774. doi: 10.4161/cc.5.7.2627. [DOI] [PubMed] [Google Scholar]
- Whitten WK. Nutrient requirements for the culture of preimplantation mouse embryo in vitro. Adv Biosci. 1971;6:129–139. [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol. 1993;156:366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, Nothias JY, DePamphilis ML. Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. J Cell Sci. 1997;110:1147–1158. doi: 10.1242/jcs.110.10.1147. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283:40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Zhu P, Huber E, Kiefer F, Gottlicher M. Specific and redundant functions of histone deacetylases in regulation of cell cycle and apoptosis. Cell Cycle. 2004;3:1240–1242. doi: 10.4161/cc.3.10.1195. [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, Seiser C. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Lack of effect of depleting HDAC1 on apoptosis as determined by (A) TUNEL labeling and (B) expression of pro-apoptotic genes. The data are expressed as mean ± SEM and the differences between control and experimental groups are not significant.
Figure S2. Effect of RNAi-mediated reduction of Hdac1 mRNA on BrUTP incorporation. (A) Immunocytochemical detection of BrUTP incorporation and (B) quantification of the data. The experiment was performed 3 times and a total of 24 embryos examined. The difference between control and experimental embryos is not significant, p>0.05.
Figure S3. Lack of effect of RNAi-mediated reduction of Hdac2 mRNA on development, histone acetylation and gene expression. (A) Newly inseminated eggs were injected with either Gfp dsRNA (control) or Hdac2 dsRNA and cultured for 96 h at which time they were scored. (B) The injected embryos were processed for immunocytochemical detection of acetylated histone H4 and (C) the data quantified. Embryos harvested at the indicated times following injection were also processed for quantification of the relative amounts of Eif1a (D) and p21Cip1/Waf (E) mRNA. None of the differences between control and experimental embryos is significant, p>0.05.
Figure S4. RNAi-mediated degradation of Hdac3 mRNA (A) does not result in decrease in HDAC3 protein (B and C).


