Abstract
Botulinum neurotoxin (BoNT; serotypes A-G) and tetanus neurotoxin elicit flaccid and spastic paralysis, respectively. These neurotoxins are zinc proteases that cleave SNARE proteins to inhibit synaptic vesicle fusion to the plasma membrane. Although BoNT/B and tetanus neurotoxin (TeNT) cleave VAMP-2 at the same scissile bond, their mechanism(s) of VAMP-2 recognition is not clear. Mapping experiments showed that residues 60-87 of VAMP-2 were sufficient for efficient cleavage by BoNT/B and that residues 40-87 of VAMP-2 were sufficient for efficient TeNT cleavage. Alanine-scanning mutagenesis and kinetic analysis identified three regions within VAMP-2 that were recognized by BoNT/B and TeNT: residues adjacent to the site of scissile bond cleavage (cleavage region) and residues located within N-terminal and C-terminal regions relative to the cleavage region. Analysis of residues within the cleavage region showed that mutations at the P7, P4, P2, and P1′ residues of VAMP-2 had the greatest inhibition of LC/B cleavage (≥32- fold), whereas mutations at P7, P4, P1′, and P2′ residues of VAMP-2 had the greatest inhibition of LC/TeNT cleavage (≥64-fold). Residues within the cleavage region influenced catalysis, whereas residues N-terminal and C-terminal to the cleavage region influenced binding affinity. Thus, BoNT/B and TeNT possess similar organization but have unique residues to recognize and cleave VAMP-2. These studies provide new insights into how the clostridial neurotoxins recognize their substrates.
The botulinum neurotoxins (BoNTs)2 are the most potent protein toxins for humans (1). BoNTs have been used for malicious purposes (2) but also relieve numerous human neurological afflictions, such as blepharospasm, in addition to being used for cosmetic enhancement (3-5). Recent studies provide a more detailed understanding of BoNT action, which should provide insight for developing novel products and therapies to treat human suffering.
BoNTs are zinc proteases that cleave SNARE proteins to block synaptic vesicle fusion and neurotransmitter release to elicit flaccid paralysis. BoNTs are 150-kDa dichain proteins with AB structure-function properties; the N terminus encodes the zinc protease domain (light chain (LC)), and the C terminus encodes a translocation domain and a receptor binding domain (6, 7). The seven serotypes of BoNTs, termed A-G, are based upon antisera neutralization properties (8). Each BoNT serotype cleaves one of three neuronal SNARE proteins: SNAP-25, VAMP-2, or syntaxin 1a, except for BoNT/C, which cleaves both SNAP-25 and syntaxin 1a. BoNT/A and BoNT/E cleave SNAP-25 at different sites, whereas BoNT/B, BoNT/D, BoNT/F, and BoNT/G cleave VAMP-2 at distinct sites (9-12). In addition, tetanus neurotoxin (TeNT) shares structural homology with the BoNTs and cleaves VAMP-2 at the same site as BoNT/B. Despite this similarity, TeNT elicits spastic paralysis due to the ability to retrograde traffic within the peripheral neurons to target interneurons (6, 13-15).
Unlike other zinc proteases, BoNTs and TeNT recognize an extended region of the SNARE proteins for substrate cleavage (16-18). Recent studies (19) showed that SNAP-25 wraps around the LC/A with exosites that are N-terminal and C-terminal of the cleavage region. Biochemical studies identified a region N-terminal to the cleavage region of SNAP-25 that is involved in substrate recognition of LC/A and LC/E (20). Recognition within the cleavage region of SNAP-25 involves four pockets within LC/A: the S5, S3, S1′, and S4′ pockets (21), whereas recognition of the cleavage region of SNAP-25 involves three pockets within LC/E: the S3, S2, and S1′ pockets (22). Thus, although sharing a common mechanism, substrate recognition by LC/A and LC/E follow unique steps to enhance cleavage efficiency.
BoNT serotypes and TeNT that cleave VAMP-2 also recognize extended regions of the substrate for efficient cleavage (16, 18, 23-26). In the current study, BoNT/B and TeNT, which share ∼51% primary amino acid identity and ∼68% similarity and cleave VAMP-2 at the same site, were used to study VAMP-2 recognition and cleavage. The results show that LC/B and LC/TeNT have similar organization but recognize unique residues for the efficient cleavage of VAMP-2.
MATERIALS AND METHODS
Plasmid Construction for Protein Expression—Plasmids for the expression of BoNT LC/B-(1-430) and LC/TeNT-(1-436) were constructed by amplifying DNA encoding the LCs from Clostridium botulinum serotype B Okra strain (27, 28) and Clostridium tetani, respectively. The products were subcloned into pET-15b and transformed into Escherichia coli BL21(DE3)-RIL (Stratagene). Protein expression and purification was achieved as previously described (20). Human VAMP-2-(1-97) was constructed by PCR amplifying a cDNA clone purchased from ATCC and subcloning into pGEX-2T. VAMP-2-(1-97) is a soluble form of VAMP-2 that lacks the transmembrane spanning region of full-length VAMP-2. pGEX-VAMP-2-(1-97) was transformed into E. coli BL21 (DE3). For VAMP-2 truncation derivatives, the GST fusion protein may cause some hindrance effect on some of the N-terminal truncation derivative as previously observed with GST-SNAP-25 truncations (20). Residues 1-21 of SNAP-25, which do not interact with LC/B and LC/TeNT, based on co-pull-down studies (data not shown), were used as an extension to reduce the hindrance effect of GST. VAMP-2 deletions were constructed by PCR amplification of the indicated sequence (Fig. 2), which was subcloned into pGEX-2T. VAMP-2 expression was achieved by culturing E. coli at 30 °C for 2 h with shaking (250 rpm), 0.75 mm IPTG was added, and the incubation was continued for an additional 2 h. GST-VAMP-2 was purified by glutathione-Sepharose 4B affinity chromatography (Amersham Biosciences) (20).
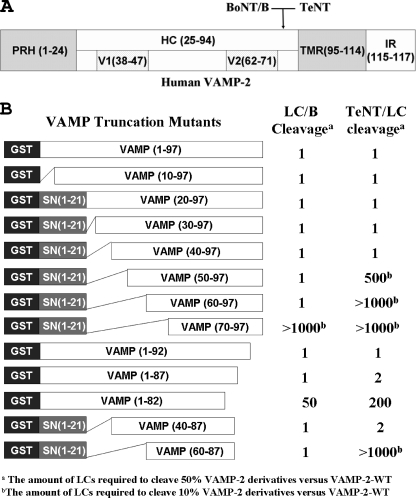
FIGURE 2.
Cleavage efficiency of VAMP-2 deletion peptides by LC/B and LC/TeNT. A, schematic of human VAMP-2 (117 residues) that contains four domains, proline-rich head (PRH), hydrophilic center (HC), transmembrane region (TMR), and several amino acids that represent the intracellular region (IR). There are two SNARE motifs, V1 and V2, in the HC domain. B, several VAMP-2 deletion derivatives were constructed as GST fusion proteins with an internal SNAP-25, SN-(1-21), extension to prevent GST hindrance effects on VAMP-2 cleavage. In the linear velocity assay, 6 nm LC/B cleaved 50% of the VAMP-2, whereas 120 nm LC/TeNT cleaved 50% cleavage of VAMP-2. A, values represent the amount of LC/B or LC/TeNT required for 50% cleavage of the indicated VAMP-2 derivative relative to 50% cleavage of VAMP-2-WT. B, VAMP-2 derivative that was an inefficient substrate (50% cleavage was not achieved); the values represent the amount of LC/B or LC/TeNT required for 10% cleavage of the indicated VAMP-2 derivative relative to 10% cleavage of VAMP-2-WT, where the limit of detection was ∼1000-fold.
Alanine-scanning Mutagenesis of VAMP-2-(1-97)—Alanine-scanning mutagenesis was performed on residues 40-91 of VAMP-2, using QuikChange (Stratagene) protocols as previously described (20) with pGEX-VAMP-2-(1-97) as a template. Plasmids were sequenced to confirm the mutation and that additional mutations were not present within the open reading frame of VAMP-2. Mutated proteins were produced and purified as described above for VAMP-2-(1-97).
Linear Velocity and Kinetic Constant Determinations for LC/B and LC/TeNT Cleavage—Linear velocity reactions contained in 10 μl:5 μm VAMP-2 or the indicated VAMP-2 derivatives incubated with varying concentrations of LC/B or LC/TeNT in 10 mm Tris-HCl (pH 7.6) with 20 mm NaCl at 37 °C for 10 min. Reactions were stopped by adding SDS-PAGE buffer, and the substrate and cleaved product were resolved by SDS-PAGE. The amount of VAMP-2 cleaved was determined by densitometry. Km and kcat determinations were performed with the same assay, where VAMP-2 concentrations were adjusted to between 1 and 300 μm to achieve <10% cleavage by LC/B and LC/TeNT. Reaction velocity versus substrate concentration was fit to the Michaelis-Menten equation, and kinetic constants were derived using the GraphPad Program (San Diego, CA).
Trypsin Digestion of VAMP-2-WT and VAMP-2 Derivatives—120 μm GST-VAMP-2-(1-97) or the indicated VAMP-2 derivative was incubated with 20 nm trypsin in a 20-μl reaction volume for 20 min at 37 °C. The reactions were stopped and resolved in SDS-polyacrylamide gels.
Molecular Modeling—The complex structures of LC/B-VAMP-2 and LC/TeNT-VAMP-2 were modeled and analyzed using SWISS-MODEL and refined by PyMol (available on the World Wide Web) as described previously (22). Protein Data Bank coordinates used in this analysis were 1f82 for LC/B, 1z7h for LC/TeNT and 1xtg for LC/A-SNAP-25.
RESULTS
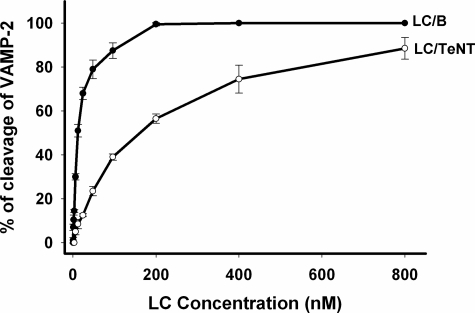
Although LC/B and LC/TeNT cleave VAMP-2 at the same site, LC/B is ∼20-fold more active than LC/TeNT (18) (Fig. 1), where in a linear velocity reaction, 6 nm LC/B and 120 nm LC/TeNT cleaved 50% of VAMP-2. LC/B had a Km of 1.6 μm for VAMP-2 and kcat of 65 min-1, whereas LC/TeNT had a Km of 4.0 μm for VAMP-2 and a kcat of 9.4 min-1 (see Table 2). Thus, recombinant LC/B used in this study has a higher affinity and faster catalytic rate for VAMP-2 cleavage than recombinant LC/TeNT.
FIGURE 1.
Cleavage of VAMP-2 by LC/B and LC/TeNT. The linear velocity for LC/B and LC/TeNT was performed in 10-μl reactions including 5 μm VAMP-2 and varying concentrations of LC/B or LC/TeNT in 10 mm Tris-HCl (pH 7.6) with 20 mm NaCl at 37 °C for 10 min. Reactions were stopped by adding SDS-PAGE buffer, and the substrate and cleaved product were resolved by SDS-PAGE. The amount of VAMP-2 cleaved was determined by densitometry and is the average of three independent determinations.
TABLE 2.
Kinetic constants of VAMP-2-WT and VAMP-2 derivative cleavage by LC/B and LC/TeNT
| Light chain/VAMP-2-(1-97) | Region | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μM | min−1 | min−1/μM | ||
| LC/B | ||||
| WT | 1.6 | 65 ± 5 | 40 | |
| L61A | 1.5 | 62 ± 4.8 | 41 | |
| A69S | 1.6 | 60 ± 8.2 | 38 | |
| L63A | NTa | 13 | 14 ± 1.3 | 1.1 |
| A67S | NT | 14 | 31 ± 3.6 | 2.2 |
| D68A | NT | 12 | 43 ± 6.8 | 3.6 |
| L70A | CR-P7 | 6.5 | 1.1 ± 0.5 | 0.2 |
| G73A | CR-P4 | 3.5 | 4.5 ± 1 | 1.2 |
| S75A | CR-P2 | 1.6 | 1.4 ± 0.5 | 0.8 |
| Q76A | CR-P1 | 1.9 | 9.4 ± 1.4 | 5.0 |
| F77A | CR-P1' | 1.4 | 0.2 ± 0.1 | 0.1 |
| E78A | CR-P2' | 2.6 | 13 ± 2.4 | 5.0 |
| S80A | CR-P4' | 2.1 | 16 ± 2.8 | 7.9 |
| K83A | CT | 13 | 54 ± 5.6 | 3.6 |
|
K85A |
CT |
13 |
56 ± 2.3 |
4.2 |
| LC/TeNT | ||||
| WT | 4.0 | 9.4 ± 1.3 | 2.4 | |
| L61A | 4.2 | 9.0 ± 1.8 | 2.1 | |
| A69S | 4.1 | 9.7 ± 1.2 | 2.4 | |
| L70A | CR-P7 | 17 | 0.1 ± 0.03 | 0.01 |
| G73A | CR-P4 | 8.2 | 0.2 ± 0.1 | 0.03 |
| A74S | CR-P3 | 4.0 | 0.5 ± 0.1 | 0.1 |
| Q76A | CR-P1 | 4.0 | 1.3 ± 0.2 | 0.3 |
| F77A | CR-P1' | 3.8 | 0.01 | 0.002 |
| E78A | CR-P2' | 4.3 | 0.05 | 0.01 |
| S80A | CR-P4' | 4.8 | 1.3 ± 0.6 | 0.3 |
| L84A | CT | 53 | 8.8 ± 0.8 | 0.2 |
| K85A | CT | 19 | 9.2 ± 1.4 | 0.4 |
| R86A | CT | 17 | 9.6 ± 1.8 | 0.6 |
NT, N-terminal region; CR, cleavage region; CT, C-terminal region.
Mapping Regions of VAMP-2 That LC/B and LC/TeNT Recognize for Efficient Cleavage—Earlier studies showed that efficient cleavage of VAMP-2 by the clostridial neurotoxins required an extended substrate (18, 23, 29); however, a complete mapping of the recognition domain of VAMP-2 was not determined. A series of VAMP-2 derivatives with C-terminal and N-terminal truncations (Fig. 2) were used to determine the minimal VAMP-2 substrate for efficient cleavage by LC/B and LC/TeNT.
Deletion mapping showed that removal of the N-terminal 60 residues or C-terminal residues 87-97 of VAMP-2 did not reduce the efficiency of LC/B cleavage of VAMP-2. Correspondingly, LC/B cleaved VAMP-2-(60-87) with a similar efficiency as VAMP-2-WT (Fig. 2). This indicated that LC/B recognized a relatively short domain within VAMP-2, residues 60-87, as a minimal efficient substrate and that the SNARE motif V1 (residues 38-47) did not contribute to LC/B cleavage of VAMP-2.
For LC/TeNT, truncation of the N-terminal 40 amino acids of VAMP-2 did not affect LC/TeNT cleavage, but truncation of the N-terminal 50 amino acids reduced the efficiency of LC/TeNT cleavage by ∼500-fold relative to VAMP-2-WT (Fig. 2). The deletion of the C-terminal 87-97 residues of VAMP-2 had ∼2-fold reduction upon LC/TeNT cleavage. LC/TeNT cleaved VAMP-2-(40-87) at about 50% of the efficiency of VAMP-2-WT, indicating that VAMP-2-(40-87) was the minimal efficient substrate for LC/TeNT.
Deletion mapping also showed that C-terminal residues 82-87 of VAMP-2 affected the ability of LC/TeNT to cleave VAMP-2 to a greater extent than LC/B (Fig. 2). Relative to VAMP-2-WT, LC/TeNT was ∼200-fold less efficient in the cleavage of VAMP-2-(1-82), whereas LC/B was ∼50-fold less efficient (Fig. 2).
Contribution of Individual Residues of VAMP-2 on LC/B and TeNT/LC Cleavage—Alanine-scanning mutagenesis was performed to generate individual point mutations within VAMP-2 from residues 40 and 91. Residues adjacent to the scissile bond were designated as Px for residues N terminal to the cleavage site and P′x for residues C-terminal of the cleavage site, the numerical value of x increasing with the distance from the cleavage site.
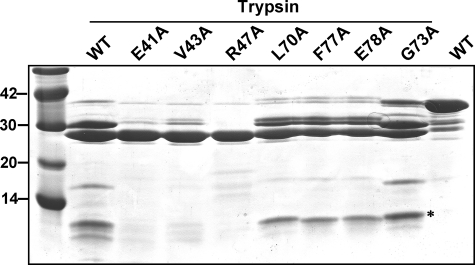
LC/B Cleavage of VAMP-2—Individual point mutations within residues 40-52 of VAMP-2 inhibited or enhanced substrate cleavage by LC/B (Table 1). This was unexpected, since deletion mapping indicated that deletion of the first 60 residues of VAMP-2 did not influence LC/B cleavage efficiency. A partial trypsin digestion of point mutations within residues 40-52 of VAMP-2 showed a different digestion profile relative to VAMP-2-WT, whereas point mutations within residues 63-78 of VAMP-2, which changed the efficiency of LC/B cleavage, did not affect trypsin cleavage patterns (Fig. 3). This indicated that mutations within residues 40-52 of VAMP-2 caused a structural change, which affected VAMP-2 recognition and cleavage distal to the minimal substrate, VAMP-2-(60-87), defined by deletion mapping. Note that undigested mutated VAMP-2 proteins had the same protein profile as undigested wild type VAMP, which is shown in Fig. 3. Thus, the affect of mutations within residues 40-52 were considered to yield global changes to the protein and therefore not subjected to further analysis.
TABLE 1.
Effect of VAMP-2 point mutations on the cleavage of VAMP-2 by LC/B and LC/TeNT
|
Point Mutation within VAMP-2 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
NT*-LC/TeNT;
V1(38-47) |
UR-LC/B |
|||||||||||||||||||||||||||||||||||||||||||||||||||
| Positions | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | ||||||||||||||||||||||||||
| VAMP-2 residue | D | E | V | V | D | I | M | R | V | N | V | D | K | V | L | E | R | D | Q | K | L | S | E | L | D | D | ||||||||||||||||||||||||||
| Mutation | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||
| LC/B Cleavage | 4a | 8 | 0.3 | 0.3 | 0.3 | 0.3 | 4 | 1 | 1 | 16 | 32 | 4 | 8 | 0.7 | 0.5 | 0.5 | 1 | 0.5 | 1 | 1 | 1 | 1 | 1 | 32 | 8 | 8 | ||||||||||||||||||||||||||
|
LC/TeNT Cleavage |
1b |
320 |
320 |
320 |
80 |
10 |
5 |
>320c |
2 |
10 |
10 |
10 |
1 |
2 |
2 |
1 |
2 |
2 |
2 |
6 |
1 |
1 |
1 |
1.5 |
1 |
2 |
||||||||||||||||||||||||||
|
Point mutation within VAMP-2 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
V2(62-71) |
CR*-LC/B
and LC/TeNT |
CT*-LC/B
and LC/TeNT |
||||||||||||||||||||||||||||||||||||||||||||||||||
| P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1' | P2' | P3' | P4' | ||||||||||||||||||||||||||||||||||||||||||
| Positions | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | ||||||||||||||||||||||||||
| VAMP-2 Sequence | R | A | D | A | L | Q | A | G | A | S | Q | F | E | T | S | A | A | K | L | K | R | K | Y | W | W | K | ||||||||||||||||||||||||||
| Mutations | A | S | A | S | A | A | S | A | S | A | A | A | A | A | A | S | S | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||
| LC/B Cleavage | 4 | 8 | 8 | 1 | 240 | 16 | 1 | 32 | 1 | 48 | 12 | 320 | 8 | 0.2 | 4 | 0.4 | 2 | 8 | 1 | 8 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||
| LC/TeNT Cleavage | 3 | 1 | 1 | 1 | 320 | 6 | 6 | 64 | 16 | 1 | 8 | >320 | 240 | 1 | 8 | 1 | 1 | 1 | 12 | 4 | 4 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||
N-terminal region (NT), cleavage region (CR), C-terminal region (CT), and SNARE motifs V1-(38-47) and V2-(62-71) are shown.
The ratio of LCs needed for 50% cleavage of VAMP-2-(1-97) containing the indicated point mutation and VAMP-2-(1-97)-WT. For example, 4 means that 4-fold more LC/B is required to cleave VAMP-2(D40A) than wild type VAMP-2; mutations that had a >4-fold increase in the amount of LC required for cleavage are shown in boldface type.
1 designates that the activity was within 2-fold of WT.
320-fold is the limit of resolution of the assay.
FIGURE 3.
Partial trypsin digestion of wild type VAMP-2 and derivatives. 120 μm GST-VAMP-2-(1-97)-WT and VAMP-2 derivatives were incubated with 20 nm trypsin for 20 min at 37 °C. Reactions were stopped with SDS-PAGE sample buffer, and proteins were resolved by 12% SDS-PAGE. A Coomassie-stained gel is shown. *, a fast moving band that is not present in the VAMP-2 proteins that are mutated within the SNARE motif V1.
In addition to the SNARE recognition region, other individual amino acid mutations within VAMP-2 were localized to three regions that affected the efficiency of LC/B cleavage: an N-terminal region (residues 63-68), the cleavage region (residues 70-80), and a C-terminal region (residues 83-85) (Table 1). In the N-terminal region (residues 63-68), individual mutations reduced the efficiency for LC/B cleavage of VAMP-2 4-32-fold. In the cleavage region (residues 70-80), individual mutations reduced the efficiency of LC/B substrate cleavage 8-320-fold, with the most inhibition upon mutation of the P7-Leu-70 and P1′-Phe-77 residues. In the C-terminal region, mutations K83A and K85A) reduced LC/TeNT cleavage of VAMP-2 8-fold (Table 1). Unexpectedly, some mutations within VAMP-2 enhanced LC/B cleavage such as T79A and A81S, suggesting that these residues might also contribute to substrate recognition (Table 1). This has implications for future engineering of “custom” BoNT therapeutic agents.
LC/TeNT Cleavage of VAMP-2—Mutations at residues 41-51 showed dramatic effects on LC/TeNT cleavage with ∼320-fold inhibition by the individual mutations, E41A, V42A, V43A, and R47A (Table 1). This inhibition was greater than anticipated from the deletion mapping experiments, since deletion of the N-terminal 50 residues of VAMP-2 caused only a 500-fold reduction in LC/TeNT efficiency. One of the VAMP V1 mutated derivatives, VAMP-2-D44A was subjected to a kinetic analysis using LC/TeNT and observed to have a 10-fold higher Km (46 μm) and 10-fold lower kcat (1.1 ± 0.5 min-1) than VAMP-WT cleaved by LC/TeNT, suggesting a pleomorphic change in protein structure. VAMP mutated within residues 40-52 also possessed a different trypsin digestion pattern than VAMP-WT, precluding their further analysis (Fig. 3).
In addition to the SNARE recognition region, other individual amino acid mutations of VAMP-2 were localized to two regions that affected the efficiency of LC/B cleavage; the cleavage region (residues 70-80) and a C-terminal region (residues 84-87) reduced LC/TeNT cleavage efficiency up to >320-fold (Table 1). Within the cleavage region, individual mutations reduced the efficiency of LC/TeNT ∼6- to >320-fold with mutations at the P7, P1′, and P2′ having the greatest inhibition of LC/TeNT activity. In the C-terminal region, mutations L84A, K85A, and R86A reduced LC/TeNT cleavage of VAMP-2 4-12-fold (Table 1).
Individual VAMP-2 Residues Influence the Kinetics of LC/B and LC/TeNT Catalysis—Kinetic constants were obtained for several point mutations within VAMP-2 that reduced the cleavage efficiency of LC/B or LB/TeNT.
Kinetic Constants for LC/B Cleavage of Mutated VAMP-2—The primary effects of the N-terminal region mutations (L63A, A67S, and D68A) were to increase the Km for LC/B cleavage of VAMP-2 with a lesser effect on the reduction of the kcat (Table 2). In the cleavage region, two types of mutations were observed: the P7 site (L70A) and P4 site (G73A) mutations, which increased the Km by 4- and 2-fold and caused a ∼60 and ∼14-fold slower kcat, respectively, for LC/B cleavage of VAMP-2. Other mutations within the cleavage region reduced only the kcat by 48-fold for P2-S75A, 8-fold for P1-Q76A, 320-fold for P1′-F77A, 8-fold for P2′-E78A, and 4-fold for P4′-S80A (Table 2). In the C-terminal region, the K83A and K85A mutations of VAMP-2 increased the Km, by ∼8-fold for LC/B cleavage (Table 2). Thus, three regions were detected within VAMP-2 that influenced LC/B activity: a cleavage region (residues 70-80), which contributed primarily to catalysis, and an N-terminal (residues 63-68) and C-terminal (residues 83-85) region that decreased the affinity of LC/B for VAMP-2 (Fig. 4A).
FIGURE 4.
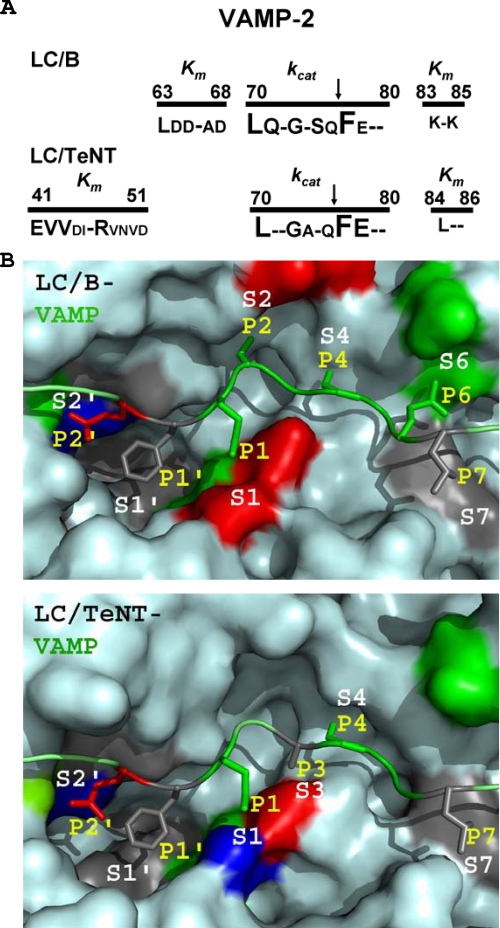
Alignment of VAMP-2 within the active sites of LC/B and TeNT/LC. A, domains and residues required for LC/B and LC/TeNT substrate binding and catalysis. The letter size corresponds to the contribution to catalysis, from 8-fold (the smallest) to ≥320-fold (the largest). A minus sign indicates <8-fold effect on catalysis. B, modeling the active site residues of LC/B and LC/TeNT. P site residues of VAMP were aligned to proposed S pockets in the active sites of LC/B and LC/TeNT. P site residues that when mutated yielded a ≥8-fold decrease in cleavage by the respective LC are shown. Each P site and S pocket is colored based on the side chain of the amino acid residues: acidic (red), basic (blue), polar neutral (green), or apolar (gray). Top, LC/B-VAMP-2 cleavage region model; bottom, LC/TeNT-VAMP-2 cleavage region model. Modeling predicts that the P1′ and P7 are recognized by hydrophobic pockets, S1′ and S7, and that the unique active site shapes of LC/B and LC/TeNT determine the different requirement of P site residues for substrate recognition.
Kinetic Constants for LC/TeNT Cleavage of Mutated VAMP-2—For LC/TeNT cleavage of VAMP-2, in the cleavage region, the P7 mutation (L70A) decreased the kcat by 80-fold and increased the Km by 4-fold, whereas the P4 mutation (G73A) increased the Km by 2-fold and decreased the kcat by 30-fold (Table 2). Other mutations within the cleavage region only reduced the kcat: 16-fold for P3-A74S, 8-fold for P1-Q76A, >320-fold for P1′-F77A, 240-fold for P2′-E78A, and 8-fold for P4′-S80A (Table 2). In the downstream region, L84A, K85A, and R86A mutations in VAMP-2 increased the Km, by 13-, 4-, and 4-fold for LC/TeNT, respectively (Table 2). Thus, for LC/TeNT cleavage of VAMP-2, residues within the cleavage region of VAMP-2 (residues 70-80) contribute to substrate catalysis, whereas residues within the C-terminal region (residues 84-86) contribute primarily to substrate binding (Fig. 4A).
DISCUSSION
The prolific utilization of BoNTs in human therapy makes understanding the molecular basis for substrate recognition and specificity fundamental to developing novel BoNT-derived pharmaceuticals (4). The current study utilized biochemical approaches to investigate VAMP-2 recognition and specificity by BoNT/B and TeNT. These toxins were chosen because BoNT/B and TeNT share sequence and structural homology and cleave the same substrate, VAMP-2, at the same scissile bond, Gln-76-Phe-77. Three regions of VAMP-2 were observed to be recognized by LC/B and LC/TeNT for optimal VMAP-2 cleavage: a cleavage region (residues 70-80) that surrounds the site of scissile bond cleavage and contributes to substrate catalysis and two regions that contribute to high affinity binding to VAMP-2, an N-terminal region (determined directly for LC/B) and a C-terminal region.
Residues within the Cleavage Region of VAMP-2 That Are Recognized by LC/B and LC/TeNT—Previous studies implicated several residues in VAMP-2 that were important for cleavage by LC/B (16). An earlier study by Binz and co-workers (16) used substitution mutagenesis of nonconserved residues between TI-VAMP, an LC/B-insensitive VAMP isoform, and human VAMP-2 to show that Asp-64 to Ser-67, Gln-71 to Gln-76, Glu-78, Ser-80, and Lys-85 of human VAMP-2 contributed to LC/B cleavage. These studies explained why TeNT-insensitive VAMP was insensitive to LC/B. The current study corroborated the effect of these mutations of LC/B cleavage of VAMP-2 and extended these findings to provide a complete analysis of the recognition of VAMP-2 by LC/B. LC/B recognized several residues within the cleavage region of VAMP-2 (residues 70-80), including P2-S75A, P1-Q76A, P1′-F77A, P2′-E78A, and P4′-S80A, which lowered the kcat for LC/B cleavage of VAMP-2, with Leu70 and Phe77 contributing a major role for LC/B catalysis. LC/TeNT also recognized several residues within the cleavage region in VAMP-2 for efficient substrate catalysis, including P7-L70A, P4-A73S, P3-A74S, P1-Q76A, P1′-F77A, P2′-E78A, and P4′-S80A, with Leu-70, Phe-77, and Glu-78 contributing a major role in LC/TeNT catalysis.
Structural modeling of LC/B-VAMP-2 and LC/TeNT-VAMP-2 (Fig. 4B) based on the kinetic properties of mutated VAMP-2 proteins indicates that both LCs recognize a similar pair of P site residues. The P7-Leu-70 and P1′-Phe-77 residues can be aligned to hydrophobic S7 and S1′ pockets at the periphery of the active sites of both LC/B and LC/TeNT (Fig. 4B). Residue compositions of the S1′ and S7 pockets of LC/B and TeNT are similar and compatible with the hydrophobic nature of both P sites. The aromatic residue in the P1′-position appears to be required for LC/B S1′ pocket recognition, which is supported by the observation that replacement of P1′ Phe-77 with Tyr-77 had little affect on substrate cleavage, whereas replacement with smaller hydrophobic residues diminished the hydrolytic activity of LC/B (24). Recognition of the P7 and P1′ residues may facilitate or be facilitated by alignment of other P-site residues into their respective S pockets of LC/B and LC/TeNT through hydrophobic or polar interactions (Fig. 4B). There are subtle differences of the active site cavity of LC/B and LC/TeNT, which may account for the different P2 and P3 requirements for optimal substrate recognition and cleavage (Table 1). The putative S2′ pockets of LC/B and LC/TeNT showed similar composition, an R residue in a hydrophobic environment (Fig. 4B), and thus did not explain why LC/TeNT had a greater requirement for Glu-78 relative to LC/B. Although P2′ recognition may represent the outermost residue of the cleavage region of VAMP-2 for LC/TeNT, the limited effect of a P2′ mutation on LC/B catalysis may reflect the observation that P3′ and P5′ mutations had an unexpected elevation on LC/B cleavage for VAMP-2 and contribute to catalysis.
Although the overall organization of the residues in the cleavage regions of SNAP-25 that are recognized by LC/A and LC/E (6) is less complex than the residues in the cleavage region of VAMP-2 recognized by LC/B and LC/TeNT (where LC/A and LC/E required P4′,P1′, P3, and P5 and P1′, P2, and P3 for substrate recognition, respectively, whereas LC/B and LC/TeNT required 7 or 8 residues between P7 and P2′ for substrate recognition), there is a common feature at the N terminus of the cleavage regions recognized by these LCs. The outermost residue of cleavage regions recognized by these LCs is either leucine or iosleucine, where mutation to an alanine reduced both the Km and kcat for substrate cleavage. Thus, the outermost residue of the cleavage regions contributes to substrate affinity as well as orienting the scissile for cleavage. This suggests that these LCs utilize a common mechanism for substrate recruitment and optimal alignment of the scissile bond for cleavage.
Residues within the cleavage region of VAMP-2 recognized by LC/B that were identified in the current study differ from those previously predicted in a LC/B-VAMP-2 complex structure (30). The validity of this complex structure has been questioned (31), primarily due to the absence of electron density for the substrate and the orientation of the VAMP-2 within LC/B. In addition, the kinetic constants for wild type VAMP derived in the current study differ from those reported previously (18), where in the current study the Km values are lower, and the kcat values are faster for LC/B and LC/TeNT cleavage of VAMP-2. The reason for these differences may be the use of recombinant LCs in the current study rather than full-length holotoxins, which need to be activated.
N-terminal and C-terminal Regions Contribute to High Affinity Binding of VAMP-2 by LC/B and LC/TeNT—Earlier studies identified regions N-terminal of the cleavage region that contributed to efficient substrate cleavage by clostridial neurotoxins, termed the SNARE motifs. In cultured cells, SNARE-specific peptides and antibodies against the VAMP-2 V2 motif (32) blocked BoNT/A, B, and C cleavage of substrates, and mutations of charged residues within the SNARE motifs in VAMP-2 and SNAP-25 also inhibited substrate hydrolysis (33, 34). The current mutational analysis observed that mutations of individual amino acids in the V1 SNARE motif of VAMP-2 inhibited LC/TeNT cleavage, whereas mutations of individual amino acids in the V2 SNARE motifs inhibited LC/B cleavage. In a biological setting, the V1 region appears to contribute to SNARE complex formation (35). In addition, a peptide containing the V1 motif decreased the Km of LC/TeNT to cleave VAMP, suggesting that the V1 motif is contributing to LC/TeNT substrate binding (8).
An earlier study (6) for LC/A and LC/E and the current study for LC/B and LC/TeNT show that LCs recognize an N-terminal region of VAMP-2 or SNAP-25 for high affinity binding. The physical locations of the N-terminal regions recognized by LC/E and LC/B are similar, being located nearly contiguous with the cleavage region, and include a similar recognition profile, where the N-terminal region is dominant for recognition, and the region includes charged and hydrophobic amino acids. In the six-residue N-terminal region of VAMP-2 recognized by LC/B, three are charged residues, whereas of the seven-residue N-terminal region of SNAP-25 recognized by LC/E, two are charged residues. The N-terminal region of SNAP-25 recognized by LC/A is unique, being extended and including numerous charged and hydrophobic residues. The N-terminal V1 SNARE motif of VAMP-2 recognized by LC/TeNT is also unique, where mutations appear to have a physical effect on substrate structure.
Neither LC/A nor LC/E recognized a region C-terminal of the cleavage region for SNAP-25 that enhanced the affinity for substrate (6). Thus, the C-terminal regions of VAMP-2 recognized by LC/B and LC/TeNT are unique. The C-terminal regions of VAMP-2 recognized by LC/B and LC/TeNT overlap and constitute a basic series of amino acids. Within this C-terminal region, the residues recognized by LC/B and LC/TeNT are both common and unique, which may reflect the specific orientations of the LCs with VAMP-2. The need for additional contacts C-terminal of the cleavage site in VAMP-2 cleaving LC/B and LC/TeNT may reflect that the cleavage site is adjacent to the vesicle membrane binding region within VAMP-2 (Fig. 1). In contrast, LC/A and LC/E cleave near the C terminus of SNAP-25 at sites that are distanced from the membrane-spanning domain (6).
These studies provide new information on how the clostridial neurotoxins recognize their substrates and provide a basis for the development of reagents to optimize therapeutic utilization of the toxins and to impede malicious toxin applications.
Acknowledgments
We acknowledge members of the Barbieri laboratory for helpful comments; Na Li for the construction of the VAMP-2 mutations; and John Savaryn, Abby Kroken, and Josha Ziarek, who conducted pilot studies on the cleavage region mutations of VAMP-2.
This work was supported, in whole or in part, by National Institutes of Health, NIAID, Grant U54 AI057153 (Great Lakes Regional Center of Excellence). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: BoNT, botulinum neurotoxin; LC, light chain; TeNT, tetanus neurotoxin; VAMP, vesicle-associated membrane protein; WT, wild type; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor.
References
- 1.Schiavo, G., Rossetto, O., Tonello, F., and Montecucco, C. (1995) Curr. Top. Microbiol. Immunol. 195257 -274 [DOI] [PubMed] [Google Scholar]
- 2.Arnon, S. S., Schechter, R., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Hauer, J., Layton, M., Lillibridge, S., Osterholm, M. T., O'Toole, T., Parker, G., Perl, T. M., Russell, P. K., Swerdlow, D. L., and Tonat, K. (2001) J. Am. Med. Assoc. 2851059 -1070 [DOI] [PubMed] [Google Scholar]
- 3.Humeau, Y., Dou ssau, F., Grant, N. J., and Poulain, B. (2000) Biochimie 82 427-446 [DOI] [PubMed] [Google Scholar]
- 4.Klein, A. W. (2004) Dermatol. Clin. 22197 -205 [DOI] [PubMed] [Google Scholar]
- 5.Zuber, M., Sebald, M., Bathien, N., de Recondo, J., and Rondot, P. (1993) Neurology 431715 -1718 [DOI] [PubMed] [Google Scholar]
- 6.Montecucco, C., and Schiavo, G. (1994) Mol. Microbiol. 131 -8 [DOI] [PubMed] [Google Scholar]
- 7.Brandt, F. S., and Bellman, B. (1998) Dermatol. Surg. 241232 -1234 [DOI] [PubMed] [Google Scholar]
- 8.Purcell, A. L., and Hoard-Fruchey, H. M. (2007) Anal. Biochem. 366207 -217 [DOI] [PubMed] [Google Scholar]
- 9.Foran, P., Lawrence, G. W., Shone, C. C., Foster, K. A., and Dolly, J. O. (1996) Biochemistry 352630 -2636 [DOI] [PubMed] [Google Scholar]
- 10.Tonello, F., Morante, S., Rossetto, O., Schiavo, G., and Montecucco, C. (1996) Adv. Exp. Med. Biol. 389251 -260 [PubMed] [Google Scholar]
- 11.Schiavo, G., Malizio, C., Trimble, W. S., Polverino de Laureto, P., Milan, G., Sugiyama, H., Johnson, E. A., and Montecucco, C. (1994) J. Biol. Chem. 26920213 -20216 [PubMed] [Google Scholar]
- 12.Schiavo, G., Rossetto, O., Benfenati, F., Poulain, B., and Montecucco, C. (1994) Ann. N. Y. Acad. Sci. 71065 -75 [DOI] [PubMed] [Google Scholar]
- 13.Bohnert, S., and Schiavo, G. (2005) J. Biol. Chem. 28042336 -42344 [DOI] [PubMed] [Google Scholar]
- 14.Deinhardt, K., Berninghausen, O., Willison, H. J., Hopkins, C. R., and Schiavo, G. (2006) J. Cell Biol. 174459 -471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deinhardt, K., and Schiavo, G. (2005) Biochem. Soc. Symp. 139 -150 [DOI] [PubMed]
- 16.Sikorra, S., Henke, T., Swaminathan, S., Galli, T., and Binz, T. (2006) J. Mol. Biol. 357574 -582 [DOI] [PubMed] [Google Scholar]
- 17.Vaidyanathan, V. V., Yoshino, K., Jahnz, M., Dorries, C., Bade, S., Nauenburg, S., Niemann, H., and Binz, T. (1999) J. Neurochem. 72327 -337 [DOI] [PubMed] [Google Scholar]
- 18.Foran, P., Shone, C. C., and Dolly, J. O. (1994) Biochemistry 3315365 -15374 [DOI] [PubMed] [Google Scholar]
- 19.Breidenbach, M. A., and Brunger, A. T. (2004) Nature 432925 -929 [DOI] [PubMed] [Google Scholar]
- 20.Chen, S., and Barbieri, J. T. (2006) J. Biol. Chem. 28110906 -10911 [DOI] [PubMed] [Google Scholar]
- 21.Chen, S., Kim, J. J., and Barbieri, J. T. (2007) J. Biol. Chem. 2829621 -9627 [DOI] [PubMed] [Google Scholar]
- 22.Chen, S., and Barbieri, J. T. (2007) J. Biol. Chem. 28225540 -25547 [DOI] [PubMed] [Google Scholar]
- 23.Cornille, F., Martin, L., Lenoir, C., Cussac, D., Roques, B. P., and Fournie-Zaluski, M. C. (1997) J. Biol. Chem. 2723459 -3464 [DOI] [PubMed] [Google Scholar]
- 24.Shone, C. C., and Roberts, A. K. (1994) Eur. J. Biochem. 225263 -270 [DOI] [PubMed] [Google Scholar]
- 25.Wictome, M., Rossetto, O., Montecucco, C., and Shone, C. C. (1996) FEBS Lett. 386133 -136 [DOI] [PubMed] [Google Scholar]
- 26.Yamasaki, S., Baumeister, A., Binz, T., Blasi, J., Link, E., Cornille, F., Roques, B., Fykse, E. M., Sudhof, T. C., and Jahn, R. (1994) J. Biol. Chem. 26912764 -12772 [PubMed] [Google Scholar]
- 27.Dasgupta, B. R., and Sugiyama, H. (1972) Biochim. Biophys. Acta. 268719 -729 [DOI] [PubMed] [Google Scholar]
- 28.Siegel, L. S., and Metzger, J. F. (1979) Appl. Environ. Microbiol. 38 606-611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perpetuo, E. A., Juliano, L., Prado, S. M., Fratelli, F., Fernandes, I., and Lebrun, I. (2002) Biotechnol. Appl. Biochem. 36155 -161 [DOI] [PubMed] [Google Scholar]
- 30.Hanson, M. A., and Stevens, R. C. (2000) Nat. Struct. Biol. 7687 -692 [DOI] [PubMed] [Google Scholar]
- 31.Rupp, B., and Segelke, B. (2001) Nat. Struct. Biol. 8663 -664 [DOI] [PubMed] [Google Scholar]
- 32.Rossetto, O., Schiavo, G., Montecucco, C., Poulain, B., Deloye, F., Lozzi, L., and Shone, C. C. (1994) Nature 372415 -416 [DOI] [PubMed] [Google Scholar]
- 33.Washbourne, P., Pellizzari, R., Baldini, G., Wilson, M. C., and Montecucco, C. (1997) FEBS Lett. 418 1-5 [DOI] [PubMed] [Google Scholar]
- 34.Pellizzari, R., Rossetto, O., Lozzi, L., Giovedi, S., Johnson, E., Shone, C. C., and Montecucco, C. (1996) J. Biol. Chem. 27120353 -20358 [DOI] [PubMed] [Google Scholar]
- 35.Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998) Nature 395347 -353 [DOI] [PubMed] [Google Scholar]