Abstract
The integral V0 domain of the vacuolar (H+)-ATPases (V-ATPases) provides the pathway by which protons are transported across the membrane. Subunit a is a 100-kDa integral subunit of V0 that plays an essential role in proton translocation. To better define the membrane topology of subunit a, unique cysteine residues were introduced into a Cys-less form of the yeast subunit a (Vph1p) and the accessibility of these cysteine residues to modification by the membrane permeant reagent N-ethylmaleimide (NEM) and the membrane impermeant reagent polyethyleneglycol maleimide (PEG-mal) in the presence and absence of the protein denaturant SDS was assessed. Thirty Vph1p mutants containing unique cysteine residues were constructed and analyzed. Cysteines introduced between residues 670 and 710 and between 807 and 840 were modified by PEG-mal in the absence of SDS, indicating a cytoplasmic orientation. Cysteines introduced between residues 602 and 620 and between residues 744 and 761 were modified by NEM but not PEG-mal in the absence of SDS, suggesting a lumenal orientation. Finally, cysteines introduced at residues 638, 645, 648, 723, 726, 734, and at nine positions between residue 766 and 804 were modified by NEM and PEG-mal only in the presence of SDS, consistent with their presence within the membrane or at a protein-protein interface. The results support an eight transmembrane helix (TM) model of subunit a in which the C terminus is located on the cytoplasmic side of the membrane and provide information on the location of hydrophilic loops separating TM6, 7, and 8.
The vacuolar (H+)-ATPases (V-ATPases)2 are ATP-dependent proton pumps present in a variety of intracellular compartments, including endosomes, lysosomes, Golgi-derived vesicles, and secretory vesicles (1–3). Acidification of intracellular compartments is important for membrane traffic processes, protein degradation and processing, coupled transport of small molecules, and the entry of various pathogens, including envelope viruses like influenza virus and bacterial toxins like anthrax toxin (4). V-ATPases are also present at the plasma membrane of a variety of cell types, including renal-intercalated cells, osteoclasts, macrophages, and neutrophils, epididymal clear cells, insect goblet cells, and certain tumor cells (5–10). Plasma membrane V-ATPases play a critical role in processes such as urinary acidification, bone resorption, sperm maturation, pH homeostasis, coupled transport, and tumor metastasis.
The V-ATPases are multisubunit complexes containing two domains (1–3). The V1 domain is peripheral to the membrane, contains eight different polypeptides (subunits A–H) and carries out ATP hydrolysis. The V0 domain is membrane-integral, contains subunits a, c, c′, c″, d, and e (in yeast), and is responsible for proton transport. Both the proteolipid subunits (c, c′, and c″) and subunit a contain residues that are essential for proton translocation (11, 12). The proteolipid subunits form a ring containing buried glutamic acid residues (13, 14) that are thought to undergo reversible protonation during proton transport. Subunit a is a 100-kDa integral membrane protein that contains two domains. The N terminus is a 50-kDa, hydrophilic domain present on the cytoplasmic side of the membrane (15). It functions as part of the peripheral stalk (or stator) that connects the V1 and V0 domains (16, 17) and stabilizes the complex during rotary catalysis (see below). It also contains the information necessary for intracellular targeting of V-ATPases (18). The C terminus is a 50-kDa, hydrophobic domain and contains multiple transmembrane helices (15). Site-directed mutagenesis has shown that the C-terminal domain of subunit a contains several charged amino acid residues buried in the membrane that are important for proton transport, including Arg-735 in TM7 that is absolutely required (12, 19, 20).
The V-ATPases operate by a rotary mechanism in which ATP hydrolysis in the V1 domain causes the rotor domain (including the ring of proteolipid subunits) to rotate relative to subunit a (21, 22). Rotation of the proteolipid ring past subunit a in turn causes proton translocation, as first proposed for the related family of F1F0 ATP synthases (23). Subunit a is thought to serve two essential functions in this mechanism, as described in detail under “Discussion.” Briefly, these include conduction of protons to and from the protonatable carboxyl groups on the proteolipid ring and stabilization of these groups in their charged state (1). To understand how subunit a performs these functions, it is necessary to define the structure of this polytopic membrane protein. Previous data from our laboratory analyzing the membrane topology of subunit a led to a model of the protein in which the cytoplasmic N-terminal domain was followed by a C-terminal domain containing nine transmembrane helices, placing the C terminus on the lumenal side of the membrane (15). Other data, however, have suggested that the C terminus is located on the cytoplasmic rather than the lumenal side of the membrane (24, 25). To further address the membrane topology of subunit a, we have employed a combination of cysteine-scanning mutagenesis and chemical modification with membrane permeant and impermeant reagents. The resulting model contains eight transmembrane helices with both the N and C terminus located on the cytoplasmic side of the membrane.
EXPERIMENTAL PROCEDURES
Materials—Zymolyase 100T was obtained from Seikagaku America, Inc. Concanamycin A was purchased from Fluka. Protease inhibitors were from Roche Applied Science. The mouse monoclonal antibody 8B1-F3 against the yeast V-ATPase A subunit and the mouse monoclonal antibody 10D7 against the 100-kDa subunit a were from Invitrogen. Escherichia coli and yeast culture media were purchased from Difco. Restriction endonucleases, T4 DNA ligase, and other molecular biology reagents were from Invitrogen, Promega, and New England Biolabs. Polyethyleneglycol maleimide (PEG-Mal, 5 kDa) was from SunBio Inc. ATP, phenylmethylsulfonyl fluoride and most other chemicals were purchased from Sigma.
Strains and Culture Conditions—Yeast strain MM112 (MATa Δvph1::LEU2 Δstv1::LYS2 his3-Δ200 leu2 lys2 ura3-52) lacking the endogenous Vph1p and Stv1p a subunit isoforms was used to study all Vph1p mutants. All yeast strains were cultured in Ura-S.D. minimal media or YEPD medium buffered to pH 5.5 using 50 mm succinate/phosphate.
Transformation and Selection—Site-directed mutants of Vph1p were constructed using the Altered Sites II in vitro mutagenesis system (Promega), and the presence of the mutations was verified by sequencing the entire length of subcloned DNA. The mutant forms of subunit a in the pRS316 expression plasmid were transformed into yeast strain MM112 by the lithium acetate method (26). The transformants were selected on uracil minus plates and growth phenotypes of the mutants were assessed on YEPD plates buffered with 50 mm KH2PO4 or 50 mm succinic acid to either pH 7.5 or pH 5.5, respectively.
Protein Preparation, SDS-PAGE, and Immunoblot Analysis—Vacuolar membrane vesicles were isolated as described previously (27). Vacuolar membranes were separated by SDS-PAGE on 7.5% acrylamide gels (28). The presence of Vph1p (subunit a) or Vma1p (subunit A) were detected by Western blotting using the monoclonal antibodies 10D7 and 8B1-F3, respectively, followed by a horseradish peroxidase-conjugated secondary antibody, as described previously (29). Blots were developed using a chemiluminescent detection method obtained from Kirkegaard & Perry Laboratories.
Chemical Blocking and Detergent Solubilization—200 μg of vacuolar membrane vesicles were diluted into 1 ml of ice-cold phosphate-buffered saline (PBS) containing 137 mm NaCl, 1.2 mm KH2PO4, 15.3 mm Na2HPO4, 2.7 mm KCl, 2 mm EDTA, 1 mm dithiothreitol, pH 7.2 and then centrifuged at 16,000 × g for 5 min. Pellets were resuspended in 400 μl of ice-cold PBS containing protease inhibitors aprotinin (2 μg/ml), leupeptin (5 μg/ml), pepstatin (0.7 μg/ml), and phenylmethylsulfonyl fluoride (1 mm) and divided into two tubes. Samples were incubated either in the absence or presence of 5 mm N-ethylmaleimide (NEM) for 30 min at room temperature. Samples were then washed by sedimentation with 1 ml of ice-cold PBS 5 times, and pellets were resuspended in 100 μl of cold PBS with protease inhibitors and again divided into two tubes. Samples were incubated with or without 2% SDS for 20 min at room temperature.
Sulfhydryl Modification with PEG-maleimide—To determine the accessibility of the cysteine residues in Vph1p in the various mutants, the samples that had been treated with or without NEM and with or without SDS as described above were incubated with 1 mm polyethylene glycol maleimide (PEG-Mal) in PBS with protease inhibitors for 1 h on ice (for samples in the absence of SDS) or room temperature (for samples in the presence of SDS). Room temperature was employed for the samples reacted in the presence of SDS to avoid precipitation of detergent, but incubation in the absence of SDS was done on ice to minimize any permeation of PEG-Mal into the vesicle interior. Samples were quenched with 10 mm dithiothreitol for 10 min and then mixed with sample buffer and separated by SDS-PAGE on 7.5% acrylamide gels. Samples were transferred to nitrocellulose and blotted with the monoclonal antibody against subunit Vph1p (10D7). Reaction of an accessible single cysteine residue on Vph1p with PEG-maleimide shifts the mobility of the protein by ∼10–15 kDa (30).
ATPase and Proton Transport Activity—ATPase activity was measured using a coupled spectrophotometric assay as previously described (31). The reactions were carried out at 30 °C, and vacuolar membrane vesicles were incubated with DMSO or 1 μm concanamycin A (in DMSO) for 5 min prior to measurement of ATPase activity or proton transport activity. Proton transport was measured by the initial rate of ATP-dependent fluorescence quenching using the fluorescence dye ACMA, as previously described (31).
Other Procedures—Protein concentrations were determined by the method of Lowry et al. (32).
RESULTS
Growth Phenotype of Yeast Strains Expressing Single Cysteine-containing Mutants of Subunit a—To further characterize the topology of the C-terminal domain of subunit a of the V-ATPase, a chemical modification approach was employed utilizing single cysteine-containing mutants of Vph1p, one of two isoforms of subunit a in yeast. We have previously shown that a Cys-less form of Vph1p in which the seven endogenous cysteine residues were replaced with serine was able to complement the vma– growth phenotype of a yeast strain (MM112) in which the genes encoding the two endogenous isoforms of subunit a in yeast (VPH1 and STV1) had been disrupted (15). That is, cells expressing the Cys-less form of Vph1p were able to grow at pH 7.5 as well as pH 5.5, whereas the parental MM112 strain grew only at pH 5.5. Starting with this Cys-less form of Vph1p, thirty single cysteine-containing mutants were constructed by site-directed mutagenesis. Because the topology of the C-terminal most portion of Vph1p was in dispute (15, 24, 25), and because all of the residues shown to be important for proton transport had been localized to this region (12, 19, 20), we focused our mutagenesis beginning at residue Gly-602 in the putative loop between TM5 and TM6 and continuing to the C-terminal residue Ser-840. Each of the single cysteine containing mutants of Vph1p was expressed in MM112 using the single copy plasmid pRS316 and the growth phenotype of the cells analyzed at pH 7.5. As shown in Table 1, each of the mutant strains showed growth at pH 7.5 that was at or near wild-type levels, suggesting that the mutant forms of Vph1p were able to complement the vma– phenotype.
TABLE 1.
Growth phenotype of yeast strains expressing wild type and mutant forms of Vph1p
Growth phenotype of yeast strains was assessed by colony size after overnight growth at 30 °C on plates buffered to pH 7.5.
| Strain | Growth (pH 7.5) | Strain | Growth (pH 7.5) |
|---|---|---|---|
| Wild type | +++a | T752C | +++ |
| Vector | -b | Q756C | +++ |
| Cys-less | +++ | F761C | +++ |
| G602C | +++ | G766C | +++ |
| G620C | +++ | T770C | +++ |
| L638C | ++c | A779C | +++ |
| I645C | +++ | T781C | +++ |
| L648C | +++ | A783C | +++ |
| S670C | +++ | L787C | +++ |
| S703C | +++ | M794C | ++ |
| D707C | +++ | R799C | ++ |
| D710C | +++ | E804C | +++ |
| C723 | +++ | S807C | +++ |
| C726 | +++ | G814C | +++ |
| L734C | +++ | S833C | +++ |
| A744C | ++ | S840C | +++ |
| S748C | +++ |
+++, corresponds to wild-type growth.
-, no growth.
++, corresponds to good growth.
Assembly of V1V0 Complexes Containing Single Cysteine-containing Mutants of Subunits a—To assess the effects of the mutations in subunit a on assembly of the V-ATPase complex, vacuolar membranes isolated from each strain were subjected to SDS-PAGE, and Western blot analysis was performed using antibodies against both Vph1p (as a measure of the presence of the V0 domain) and subunit A (as a measure of the presence of the V1 domain). It has previously been shown that mutations that disrupt assembly of the V-ATPase complex typically lead to the absence of subunit A in vacuolar membranes due to the inability of V1 to assemble with V0, whereas mutations that interfere with assembly of the V0 domain or normal targeting of the complex result in the absence of subunit a from vacuolar membranes (19). As can be seen in Fig. 1, vacuolar membranes isolated from each of the strains expressing the single cysteine-containing mutants of Vph1p showed near wild-type levels of both Vph1p and subunit A, suggesting normal assembly and targeting of the V-ATPase complex.
FIGURE 1.
Analysis of stability of V-ATPase complexes containing single cysteine mutants of Vph1p by Western blot of isolated vacuolar membranes using antibodies against subunit a (part of V0) and subunit A (part of V1). Vacuolar membranes were isolated from the yeast strain MM112 (disrupted in the endogenous VPH1 and STV1 genes) expressing either a wild-type form of Vph1p (WT), the pRS316 vector alone (Vector), the Cys-less form of Vph1p (cysless) or the indicated single cysteine-containing mutants of Vph1p. Samples of vacuolar membranes (2 μg of protein) were separated by SDS-PAGE followed by transfer to nitrocellulose and Western blot using mouse monoclonal antibodies against subunit a (10D7) or subunit A (8B1-F3) as described under “Experimental Procedures.”
ATPase and Proton Transport Activity—Because yeast mutants possessing as little as 20–25% of wild type levels of V-ATPase activity can still display a wild-type growth phenotype at pH 7.5 (33, 34), it is necessary to measure the effect of the introduced mutations on ATPase activity and proton transport by isolated vacuolar membranes. Concanamycin-sensitive ATP hydrolysis was measured by a continuous spectrophotometric assay, and concanamycin-sensitive proton transport was measured by the rate of ATP-dependent fluorescence change using the pH-sensitive dye ACMA, as described under “Experimental Procedures.” As can be seen from the results in Fig. 2, vacuolar membranes isolated from each of the mutant strains displayed at least 20% of wild-type levels of both ATPase activity and proton transport, consistent with the observed growth phenotype. It should also be noted that there is no correlation between lower activity and the slight differences observed in growth for some mutants. Activity data had previously been reported for four of the mutants analyzed in this report, including G602C, S703C, F761C, and S840C (15). Bafilomycin-sensitive, ATP-dependent proton transport activities for these mutants were all at least 70% of wild type (15).
FIGURE 2.
Effect of mutations in Vph1p on concanamycin-sensitive ATPase activity and concanamycin-sensitive, ATP-dependent proton transport in isolated vacuolar membranes. Vacuolar membranes were isolated from the yeast strain MM112 (disrupted in the endogenous VPH1 and STV1 genes) expressing either a wild-type form of Vph1p (WT), the pRS316 vector alone (Vector), the Cys-less form of Vph1p (Cysless) or the indicated single cysteine-containing mutants of Vph1p. ATPase activity (solid bars) was measured using a continuous spectrophotometric assay, and ATP-dependent proton transport (open bars) was measured as the rate of change of fluorescence using the pH-sensitive dye ACMA in the presence or absence of 1μm concanamycin as described under “Experimental Procedures.” Values are expressed relative to vacuolar membranes expressing wild-type Vph1p. Concanamycin-sensitive ATPase activity for wild-type vacuolar membranes was 1.12 μg/min/mg protein. Values represent the average of at least two measurements on two independent vacuolar membrane preparations, with the error bars corresponding to the S.E.
Reactivity of Single Cysteine-containing Mutants of Subunit a to NEM and PEG-Mal in the Presence and Absence of SDS—To analyze the topology of subunit a, the accessibility of the introduced cysteine residues to NEM and PEG-Mal in the presence and absence of SDS was assessed. As previously shown, NEM can readily cross membranes and modify cysteine residues that are exposed on either side of the membrane (30). Because reaction with NEM and other maleimides requires the thiolate form of cysteine, NEM does not react with cysteine residues within the hydrophobic phase of the bilayer (35). PEG-Mal, by virtue of the large carbohydrate moiety, is membrane impermeant and can only modify cysteine residues exposed on the same side of the membrane as that to which it is added (30). Because the vacuolar membrane preparation has previously been shown to have an almost exclusively cytoplasmic-outward orientation (15, 36), PEG-Mal modification in the absence of SDS would indicate a cytoplasmic orientation of the modified cysteine residue. If a cysteine residue cannot be modified by PEG-Mal in the absence of SDS but can be modified by NEM in the absence of SDS, this suggests the cysteine is exposed on the lumenal side of the membrane (30). If a cysteine residue cannot be modified by either PEG-Mal or NEM in the absence of SDS, this suggests that the cysteine residue is present within a transmembrane segment, at a protein-protein interface or buried within the tertiary structure of the protein. Modification by PEG-Mal is detected by a shift in mobility of the Vph1p band due to the attachment of the PEG moiety (30), which in turn is detected by Western blot analysis using the monoclonal antibody specific for Vph1p. The predicted labeling pattern that should be obtained by first treating vacuolar membranes in the absence or presence of NEM, followed by incubation with or without SDS and concluding with treatment by PEG-Mal is shown in Fig. 3.
FIGURE 3.
Theoretical Western blot pattern predicted for the cysteine accessibility assay. Vph1p mutants containing single cysteine residues either exposed to the cytoplasm (left panel), sequestered within the membrane (center panel), or exposed to the lumen (right panel) of sealed vacuolar membranes are predicted to show the indicated labeling patterns following sequential treatment in the presence or absence of NEM, the presence or absence of SDS and the presence of PEG-mal. Cysteines exposed to the cytoplasmic surface are modified by PEG-mal (and hence show a shift to lower mobility) in the absence of SDS and NEM but do not show this shift on pretreatment with NEM. Cysteines buried within the membrane (or otherwise shielded from modification) cannot be modified by either PEG-mal or NEM in the absence of SDS. Cysteines exposed on the lumenal surface of the membrane cannot be modified by PEG-mal in the absence of SDS but can be modified by NEM in the absence of SDS, resulting in pretreatment with NEM preventing the shift in mobility observed with PEG-mal in the presence of SDS.
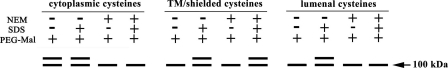
As can be seen from the results presented in Fig. 4, each of the single cysteine-containing mutants of Vph1p show one of the three predicted labeling patterns shown in Fig. 3. For the mutants S670C, S703C, D707C, and D710C, a cytoplasmic-labeling pattern is observed (Fig. 4a), consistent with the loop between TM6 and TM7 having a cytoplasmic orientation (Fig. 5). Similarly, for mutants S807C, G814C, S833C, and S840C, a cytoplasmic labeling pattern is also observed (Fig. 4a), consistent with the C-terminal tail of Vph1p having a cytoplasmic orientation. By contrast, for mutants G602C, G620C, A744C, S748C, T752C, Q756C, and F761C, a lumenal pattern of labeling is observed (Fig. 4c), suggesting the presence within the vacuole of both the loop between TM5 and TM6 and the loop between TM7 and TM8. Finally, for mutants L638C, I645C, L648C, C723, C726, L734C, G766C, T770C, A779C, T781C, A783C, L787C, M794C, R799C, and E804C, a labeling pattern consistent with their presence within the membrane or in an otherwise sequestered location is observed (Fig. 4b). These results are summarized in the topological model of subunit a shown in Fig. 5.
FIGURE 4.
Modification of Vph1p mutants containing single cysteine residues by PEG-mal following sequential treatment in the presence or absence of NEM and the presence or absence of SDS. Vacuolar membranes were isolated from the yeast strain MM112 (disrupted in the endogenous VPH1 and STV1 genes) expressing either the Cys-less form of Vph1p (Cysless) or the indicated single cysteine-containing mutants of Vph1p. 200 μg of vacuolar membrane protein were split into two samples and treated sequentially in the presence or absence of NEM, the NEM removed, and the samples split again and treated in the presence or absence of SDS followed finally by treatment with PEG-mal, separation of samples on SDS-PAGE and Western blotting using the monoclonal antibody 10D7 against Vph1p as described under “Experimental Procedures.” Modification of Vph1p by PEG-mal results in a shift in a portion of the Vph1p band by 5–10 kDa. Panel a, the Cys-less Vph1p and cysteine mutants of Vph1p showing a cytoplasmic labeling pattern (see Fig. 3). Panel b, cysteine mutants of Vph1p showing a labeling pattern consistent with localization in a transmembrane segment or in some other location inaccessible to modification by either NEM or PEG-mal in the absence of SDS. Panel c, cysteine mutants of Vph1p showing a lumenal labeling pattern.
FIGURE 5.
Topological model of the yeast subunit a (Vph1p). Folding diagram of the yeast V-ATPase subunit a (Vph1p) based upon previous data (15), hydropathy analysis, and the results presented in the current work. Residues shown in open squares correspond to those cysteines showing a cytoplasmic labeling pattern (Fig. 4a). Residues shown in open circles correspond to cysteines whose labeling pattern is consistent with their presence in a transmembrane segment or in a location which prevents their reaction with NEM or PEG-mal in the absence of SDS (Fig. 4b). Residues shown in black boxes correspond to cysteines showing a lumenal labeling pattern (Fig. 4c). Residues shown in shaded boxes correspond to residues previously shown to have a cytoplasmic orientation (15). Residues shown in shaded circles correspond to buried charged residues whose mutation reduces (but does not eliminate) ATP-dependent proton transport by the V-ATPase. Arg-735 (shown as a black circle) corresponds to the buried arginine residue that is essential for ATP-dependent proton transport. Also shown is the site at position 560 where introduction of a factor Xa cleavage site resulted in sensitivity of the mutant Vph1p to cleavage by factor Xa protease from the cytoplasmic side of the membrane (15). The resulting model shows eight transmembrane segments with both the N and C terminus of the protein exposed to the cytoplasmic side of the membrane.
DISCUSSION
Subunit a is thought to play two crucial roles in the mechanism by which V-ATPases transport protons (1). First, it allows protons to gain access to buried glutamic acid residues on the ring of proteolipid subunits from the cytoplasmic side of the membrane and to exit from these sites to the lumenal (or extracellular) side of the membrane via “hemi-channels.” Originally proposed for the a subunit of the F-ATPase (23, 37–39, 47), each hemi-channel extends only partway across the membrane, necessitating rotation of the proteolipid ring relative to subunit a to complete the proton conductance pathway. Second, subunit a contains a key buried arginine residue (Arg-735 in Vph1p) that is proposed to interact with the buried glutamic acid residues in the proteolipid ring, stabilizing them in the negatively charged state, and thus displacing the previously bound protons into the lumenal hemi-channel (1, 12). Mutation of Arg-735 to any residue, including lysine, results in complete loss of proton transport (12), suggesting that it plays a role similar to that of Arg-210 in the E. coli F-ATPase a subunit (37–39).
Mutagenesis studies of Vph1p also identified a number of charged residues in the C-terminal domain of subunit a whose mutation, while not completely inhibiting proton transport, nevertheless significantly reduced it (12, 19, 20). These residues included His-729, His-743, Glu-789, and Arg-799. Interestingly, the R799C mutation constructed in the present study gives a complex with wild-type levels of proton transport (Fig. 2), although mutation to lysine gave a complex possessing only 5% of wild-type proton transport activity (12). Mutation to other residues (Ala and Leu) disrupted assembly of the complex (12). These results suggest that a charged residue at this position is not essential and can even be detrimental to proton transport. To function in the proposed hemi-channels, these charged residues would need to be located within membrane spanning segments, but evidence for their presence within the membrane was largely lacking.
Our current model for the topology of subunit a, shown in Fig. 5, places His-729 in TM7 and Glu-789 and Arg-799 in TM8 within the membrane and on the cytoplasmic side of the critical Arg-735. These residues may therefore contribute to a cytoplasmic hemi-channel. Other charged residues that might contribute to such a cytoplasmic hemi-channel or be located near its opening would be His-718, Glu-721, and His-796. By contrast, His-743 is predicted to be near the lumenal border of TM7 and may therefore contribute to a lumenal hemi-channel. The location of Arg-735 within the membrane is supported by the ability to form zero-length cross-links between cysteine residues introduced near Arg-735 in subunit a and cysteine residues near the critical buried glutamic acid residues in TM4 of subunit c′ or TM3 of subunit c″ (40, 41).
It is of interest to compare the location of putative hemi-channel residues in the V- and F-ATPase subunit a. For the F-ATPase subunit a, the critical arginine residue (Arg-210) is closer to the cytoplasmic than the lumenal end of the penultimate transmembrane helix (TM4) (42, 43) A negatively charged residue in the same helix (Glu-219 in TM4) and a positively charged residue (His-245) in TM5, as well as a number of polar residues (for example Ser-144 in TM3) appear to contribute to the lumenal hemi-channel (42, 43). The cytoplasmic hemi-channel has been proposed to be formed largely by residues at the external surface of TM4, including Arg-210, Ser-206, and Lys-203 (43). By contrast, Arg-735 of Vph1p is slightly closer to the lumenal than the cytoplasmic end of TM7 (Fig. 5). In addition, both His-729 in TM7 as well as Glu-789 and Arg-799 in TM8 are located on the cytoplasmic side of Arg-735. Of the important buried charged residues, only His-743 in TM7 may reside between Arg-735 and the lumenal surface. This suggests that the longer and more diversely charged hemi-channel in the V-ATPase subunit a may be the cytoplasmic hemi-channel, in contrast to the F-ATPase subunit a.
In addition to the above differences, TM8 of the V-ATPase subunit a, which is nearly forty residues in length, is much longer than the longest of the F-ATPase subunit a transmembrane helices, which is ∼25 residues in length (42). This suggests that either TM8 has a substantial tilt relative to perpendicular to the membrane or that the cytoplasmic or lumenal ends of TM8 are sequestered from contact with the aqueous phase. If TM8 extends into the cytoplasmic space, it may interact with the V1 domain. One precedent for an ATP-driven ion pump that contains a transmembrane helix possessing a long cytoplasmic extension is the (Ca2+)-ATPase. In that case, one of the critical transmembrane segments that contains residues essential for Ca2+ binding (TM5) extends ∼60 Å (40 residues) from the lumenal side of the membrane to well into the cytoplasmic space (44). TM5 of the (Ca2+)-ATPase functions to convey conformational information between the Ca2+ binding sites located within the membrane, and the ATP binding sites located in the cytoplasmic domains. It is possible that TM8 of subunit a of the V-ATPase may similarly convey information between the cytoplasmic V1 domain (or regulators that bind to it), and the proton translocation pathway that is in part formed by residues on TM8. The absence of such an extended helix in the F-ATPase subunit a is consistent with the more complex regulatory behavior observed for the V-ATPases (1).
Our previous model for the folding of subunit a was based upon hydropathy analysis and differential labeling of introduced cysteine residues to the membrane permeant reagent biotin-maleimide and the membrane impermeant reagent AMS (15). The model proposed nine transmembrane helices, with the large N-terminal domain located in the cytoplasm and the C terminus located in the lumen, differing from the current model in proposing the existence of a ninth transmembrane segment and in the location of the C terminus and the borders of TM7 and TM8. Accessibility of cysteines to AMS in intact vacuoles indicated a cytoplasmic orientation for the large N-terminal domain, the loop between TM2 and TM3 that included residues Asn-447 and Lys-450 and the large loop preceding TM7 that included Ser-703 (see Fig. 5). Consistent with the latter assignment, trypsin was shown to cleave the bovine-coated vesicle V-ATPase a subunit in the same hydrophilic loop preceding TM7.3 In addition, the accessibility of tandem factor Xa protease sites introduced after residue 560 to factor Xa cleavage in intact vacuolar membranes indicated the loop between TM4 and TM5 is also cytoplasmic (15). Two cysteine residues showed a labeling pattern consistent with a lumenal orientation. Both G602C and S840C reacted poorly with AMS unless the membrane had first been permeabilized with a low concentration of Zwittergent. In the presence of Zwittergent, both residues readily reacted with AMS (15). While the results of the current study are in agreement on the lumenal location of Gly-602, they differ in the location of the C terminus of subunit a. Four cysteine residues in this region, including S807C, G814C, S833C, and S840C show clear modification by PEG-Mal in intact vacuolar membranes (Fig. 4a), indicating a cytoplasmic orientation. A cytoplasmic orientation of the C-terminal tail is also in agreement with previous studies showing that the C terminus of subunit a is able to bind the glycolytic enzyme phosphofructokinase, as demonstrated by co-immunoprecipitation (24), and that a GFP tag attached at the C terminus of subunit a from Dictyostelium can be proteolytically removed in intact endosomes (25). The former result does not rule out an extracellular orientation for the C terminus of subunit a as a number of cytoplasmic proteins (such as HSP90) have been shown to be released into the extracellular medium (45). In addition, attachment of epitope tags has led to conflicting results on the topology of membrane proteins (for a discussion of the divergent results obtained from epitope-tagging studies of the F-ATPase subunit a, see reference 46). Nevertheless, the current results support a model in which the C terminus of subunit a is cytoplasmic. Such an orientation also more readily explains the observation that mutations at a variety of residues in the C-terminal tail, including Leu-800, Trp-802, Val-803, Phe-809, and Gly-814 appear to disrupt assembly of V1 and V0 (19, 20), suggesting that the C-terminal tail may function in the interaction between these two domains, as suggested above. Although the reason for the failure of AMS to react with the S840C mutant in intact vacuolar membranes is not clear, it is possible that because this is the C-terminal most residue in the protein, and therefore bears an additional negative charge, this may retard its reactivity toward the negatively charged reagent AMS. Nevertheless, the current data support a model of subunit a containing eight transmembrane helices.
Acknowledgments
We thank Drs. Daniel Cipriano and Ayana Hinton, as well as Sarah Bond, Kevin Jeffries, and Jie Qi for many helpful discussions. We also thank Xing-Hong Leng and Shoko Kawasaki-Nishi for the construction of a number of single cysteine mutants of Vph1p. Finally, we thank Dr. Carol Deutsch (University of Pennsylvania), whose seminar suggested the use of PEG-mal.
This work was supported, in whole or in part, by National Institutes of Health Grants GM34478 (to M. F.) and DK34928 (for E. coli strains). This work was also supported by a Postdoctoral Fellowship from the NE Affiliate of the American Heart Association (to Y. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: V-ATPase, vacuolar proton translocating adenosine triphosphatase; F-ATPase, F1F0 ATP synthase; PEG-mal, polyethyleneglycol maleimide; NEM, N-ethylmaleimide; AMS, 4-acetamido-4′-male-imidylstilbene-2.2′-disulfonic acid; TM, transmembrane segment; ACMA, 9-amino-6-chloro-2-methoxyacridine; YEPD, yeast extract peptone dextrose; DMSO, dimethylsulfoxide; WT, wild type; PBS, phosphate-buffered saline.
I. Adachi and M. Forgac, unpublished observations.
References
- 1.Forgac, M. (2007) Nat. Rev. Mol. Cell. Biol. 8 917–929 [DOI] [PubMed] [Google Scholar]
- 2.Kane, P. M. (2006) Microbiol. Mol. Biol. Rev. 70 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson, N. (2003) J. Bioenerg. Biomembr. 35 281–289 [DOI] [PubMed] [Google Scholar]
- 4.Gruenberg, J., and van der Goot, F. G. (2006) Nat. Rev. Mol. Cell. Biol. 7 495–504 [DOI] [PubMed] [Google Scholar]
- 5.Wagner, C. A., Finberg, K. E., Breton, S., Marshansky, V., Brown, D., and Geibel, J. P. (2004) Physiol. Rev. 84 1263–1314 [DOI] [PubMed] [Google Scholar]
- 6.Toyomura, T., Murata, Y., Yamamoto, A., Oka, T., Sun-Wada, G. H., Wada, Y., and Futai, M. (2003) J. Biol. Chem. 278 22023–22030 [DOI] [PubMed] [Google Scholar]
- 7.Nanda, A., Brumell, J. H., Nordstrom, T., Kjeldsen, L., Sengelov, H., Borregaard, N., Rotstein, O. D., and Grinstein, S. (1996) J. Biol. Chem. 271 15963–15970 [DOI] [PubMed] [Google Scholar]
- 8.Pietrement, C., Sun-Wada, G. H., Silva, N. D., McKee, M., Marshansky, V., Brown, D., Futai, M., and Breton, S. (2006) Biol. Reprod. 74 185–194 [DOI] [PubMed] [Google Scholar]
- 9.Beyenbach, K. W., and Wieczorek, H. (2006) J. Exp. Biol. 209 577–589 [DOI] [PubMed] [Google Scholar]
- 10.Sennoune, S. R., Bakunts, K., Martinez, G. M., Chua-Tuan, J. L., Kebir, Y., Attaya, M. N., and Martinez-Zaguilan, R. (2004) Am. J. Physiol. Cell. Physiol. 286 C1443–C1452 [DOI] [PubMed] [Google Scholar]
- 11.Hirata, R., Graham, L. A., Takatsuki, A., Stevens, T. H., and Anraku, Y. (1997) J. Biol. Chem. 272 4795–4803 [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki-Nishi, S., Nishi, T., and Forgac, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12397–12402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkens, S., and Forgac, M. (2001) J. Biol. Chem. 276 44064–44068 [DOI] [PubMed] [Google Scholar]
- 14.Murata, T., Yamamoto, I., Kakinuma, Y., Leslie, A. G., and Walker, J. E. (2005) Science 308 654–659 [DOI] [PubMed] [Google Scholar]
- 15.Leng, X. H., Nishi, T., and Forgac, M. (1999) J. Biol. Chem. 274 14655–14661 [DOI] [PubMed] [Google Scholar]
- 16.Inoue, T., and Forgac, M. (2005) J. Biol. Chem. 280 27896–27903 [DOI] [PubMed] [Google Scholar]
- 17.Landolt-Marticorena, C., Williams, K. M., Correa, J., Chen, W., and Manolson, M. F. (2000) J. Biol. Chem. 275 15449–15457 [DOI] [PubMed] [Google Scholar]
- 18.Kawasaki-Nishi, S., Bowers, K., Nishi, T., Forgac, M., and Stevens, T. H. (2001) J. Biol. Chem. 276 47411–47420 [DOI] [PubMed] [Google Scholar]
- 19.Leng, X. H., Manolson, M., Liu, Q., and Forgac, M. (1996) J. Biol. Chem. 271 22487–22493 [DOI] [PubMed] [Google Scholar]
- 20.Leng, X. H., Manolson, M., and Forgac, M. (1998) J. Biol. Chem. 273 6717–6723 [DOI] [PubMed] [Google Scholar]
- 21.Imamura, H., Nakano, M., Noji, H., Muneyuki, E., Ohkuma, S., Yoshida, M., and Yokoyama, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2312–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata, T., Iwamoto-Kihara, A., Sun-Wada, G. H., Okajima, T., Wada, Y., and Futai, M. (2003) J. Biol. Chem. 278 23714–23719 [DOI] [PubMed] [Google Scholar]
- 23.Vik, S. B., and Antonio, B. J. (1994) J. Biol. Chem. 269 30364–30369 [PubMed] [Google Scholar]
- 24.Su, Y., Zhou, A., Al-Lamki, R. S., and Karet, F. E. (2003) J. Biol. Chem. 278 20013–20018 [DOI] [PubMed] [Google Scholar]
- 25.Clarke, M., Kohler, J., Arana, Q., Liu, T., Heuser, J., and Gerisch, G. (2002) J. Cell Sci. 115 2893–2905 [DOI] [PubMed] [Google Scholar]
- 26.Gietz, D., St Jean, A., Woods, R. A., and Schiestl, R. H. (1992) Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchida, E., Ohsumi, Y., and Anraku, Y. (1985) J. Biol. Chem. 260 1090–1095 [PubMed] [Google Scholar]
- 28.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 29.Arata, Y., Baleja, J. D., and Forgac, M. (2002) Biochemistry 41 11301–11307 [DOI] [PubMed] [Google Scholar]
- 30.Lu, J., and Deutsch, C. (2001) Biochemistry 40 13288–13301 [DOI] [PubMed] [Google Scholar]
- 31.Vasilyeva, E., Liu, Q., MacLeod, K. J., Baleja, J. D., and Forgac, M. (2000) J. Biol. Chem. 275 255–260 [DOI] [PubMed] [Google Scholar]
- 32.Lowry, O., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265–275 [PubMed] [Google Scholar]
- 33.Liu, J., and Kane, P. M. (1996) Biochemistry 35 10938–10948 [DOI] [PubMed] [Google Scholar]
- 34.MacLeod, K. J., Vasilyeva, E., Baleja, J. D., and Forgac, M. (1998) J. Biol. Chem. 273 150–156 [DOI] [PubMed] [Google Scholar]
- 35.Tamura, N., Konishi, S., Iwaki, S., Kimura-Someya, T., Nada, S., and Yamaguchi, A. (2001) J. Biol. Chem. 276 20330–20339 [DOI] [PubMed] [Google Scholar]
- 36.Kakinuma, Y., Ohsumi, Y., and Anraku, Y. (1981) J. Biol. Chem. 256 10859–10863 [PubMed] [Google Scholar]
- 37.Cain, B. D. (2000) J. Bioenerg. Biomemb. 32 365–371 [DOI] [PubMed] [Google Scholar]
- 38.Vik, S. B., Long, J. C., Wada, T., and Zhang, D. (2000) Biochim. Biophys. Acta 1458 457–466 [DOI] [PubMed] [Google Scholar]
- 39.Fillingame, R. H., Angevine, C. M., and Dmitriev, O. Y. (2002) Biochim. Biophys. Acta 1555 29–36 [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki-Nishi, S., Nishi, T., and Forgac, M. (2003) J. Biol. Chem. 278 41908–41913 [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., Inoue, T., and Forgac, M. (2004) J. Biol. Chem. 279 44628–44638 [DOI] [PubMed] [Google Scholar]
- 42.Vik, S. B., and Ishmukhametov, R. R. (2005) J. Bioenerg. Biomembr. 37 445–449 [DOI] [PubMed] [Google Scholar]
- 43.Angevine, C. M., Herold, K. A., Vincent, O. D., and Fillingame, R. H. (2007) J. Biol. Chem. 282 9001–9007 [DOI] [PubMed] [Google Scholar]
- 44.Toyoshima, C., and Nomura, H. (2002) Nature 418 605–611 [DOI] [PubMed] [Google Scholar]
- 45.Eustace, B. K., Sakurai, T., Stewart, J. K., Yimlamai, D., Unger, C., Zehetmeier, C., Lain, B., Torella, C., Henning, S. W., Beste, G., Scroggins, B. T., Neckers, L., Ilag, L. L., and Jay, D. G. (2004) Nat. Cell Biol. 6 507–514 [DOI] [PubMed] [Google Scholar]
- 46.Long, J. C., Wang, S., and Vik, S. B. (1998) J. Biol. Chem. 273 16235–16240 [DOI] [PubMed] [Google Scholar]
- 47.Junge, W., Sabbert, D., and Engelbrecht, S. (1996) Ber. Bunsenges. Phys. Chem. 100 2014–2019 [Google Scholar]