Abstract
Synaptic vesicle protein 2 (SV2) is required for normal calcium-regulated secretion of hormones and neurotransmitters. Neurons lacking the two most widely expressed isoforms, SV2A and SV2B, have a reduced readily releasable pool of synaptic vesicles, indicating that SV2 contributes to vesicle priming. The presence of putative ATP-binding sites in SV2 suggested that SV2 might be an ATP-binding protein. To explore this, we examined the binding of the photoaffinity reagent 8-azido-ATP[γ] biotin to purified, recombinant SV2 in the presence and absence of other nucleotides. Our results indicate that SV2A and SV2B bind nucleotides, with the highest affinity for adenine-containing nucleotides. SV2A contains two binding sites located in the cytoplasmic domains preceding the first and seventh transmembrane domains. These results suggest that SV2-mediated vesicle priming could be regulated by adenine nucleotides, which might provide a link between cellular energy levels and regulated secretion.
Calcium-regulated secretion is an adaptation of generic soluble NSF attachment protein receptor (SNARE)-mediated2 membrane trafficking. Its unique properties depend on specialized proteins that control the rate at which vesicles become competent for fusion. One of these proteins, SV2 (1), is specifically expressed in neurons and endocrine cells, where it is localized to vesicles that undergo regulated secretion. Mammalian genomes contain three isoforms of SV2 (2–5), termed SV2A, SV2B, and SV2C. Of these, SV2A is the most widely expressed (6) and is the isoform whose expression is required for survival (7, 8). SV2A is also the binding site of the anti-epileptic drug levetiracetam (9) and thus the only known synaptic vesicle target of a central nervous system-directed therapy.
When SV2 expression is knocked out or reduced in neurons, chromaffin cells, or pancreatic islet cells, regulated secretion is decreased because of a reduction in the number of vesicles primed for release (10–12). Synapses from SV2 knock-out mice have equal numbers of vesicles attached to the plasma membrane (10) and fewer SNARE complexes (12). This suggests that SV2 acts after vesicle docking and before SNARE complex assembly, which occurs just prior to fusion. How SV2 contributes to vesicle priming remains unknown. All SV2 proteins are predicted to have 12 transmembrane domains, and all contain the signature motifs of major facilitator transporters (13, 14). Loss of SV2 does not affect vesicular uptake of calcium (11)3 or neurotransmitters.4 It is therefore unclear whether SV2 acts as a transporter or, like adenylate cyclase, has a transporter-like structure but performs a nontransport function (15).
Several major facilitator transporters contain nucleotide-binding sites and in some cases ATP binding regulates transporter activity. For example, ATP inhibits glucose transport by the human erythrocyte glucose transporter (16–18). SV2A contains two weak Walker ATP-binding domains (19) located in the cytoplasmic N terminus (a.a. 129–143) and a region that spans the fourth transmembrane domain through the loop between transmembrane domains 4 and 5 (a.a. 266–288). To explore the possibility that SV2 action requires or is regulated by nucleotide binding, we examined the ability of the photoaffinity reagent 8-azido-ATP[γ] biotin to bind to recombinant SV2 and the ability of other nucleotides to compete for binding.
MATERIALS AND METHODS
Plasmids—cDNA encoding rat SV2A and SV2B with the FLAG epitope (DYKDDDK) fused to the C terminus was subcloned into the mammalian expression vector pIRES2-EGFP (Clontech). Truncated and mutated SV2 proteins were generated by PCR amplification of rat SV2A cDNA and subcloned into FLAG-pIRES2-EGFP. DNA sequencing was performed to verify that all of the sequences were correct.
Cell Culture and Transfection—HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml) at 37 °C in 5% CO2. Transfection of HEK293 cells was performed using Lipofectamine™ 2000 (Invitrogen) following the manufacturer's protocol. The cells were harvested 32–48 h after transfection.
Preparation of Recombinant FLAG Fusion Proteins—Transfected HEK293 cells were harvested and washed twice with ice-cold phosphate-buffered saline. The cells were resuspended in extraction buffer without detergent (150 mm potassium acetate, 10 mm HEPES-KOH (pH 7.4), 1× protease inhibitor mixture (Roche Applied Science)) and disrupted by sonication. Cell debris was removed by centrifugation at 800 × g for 10 min. The supernatant was collected and added to an equal volume of extraction buffer with 2% Triton X-100. The extraction mixture was incubated at 4 °C for 1 h and centrifuged at 19,000 × g for 20 min to remove insoluble material. The resulting extract was mixed with prewashed and equilibrated Anti-FLAG M2 affinity gel (Sigma) and incubated end-over-end at 4 °C for 3–4 h. The beads were washed four times with 20 volumes of extraction buffer containing 0.5% Triton X-100. The second of these washes contained 500 mm potassium acetate. FLAG fusion protein was eluted from the beads with 3× FLAG peptide (Sigma) in buffer containing 150 mm potassium acetate, 10 mm HEPES-KOH (pH 7.4), and 0.5% Triton X-100. Protein concentration was determined using the BCA assay (Pierce) using bovine serum albumin as a standard. The resulting preparation was assessed by SDS-PAGE followed by silver staining and also by immunoblot analysis using anti-SV2 and anti-FLAG antibodies.
Preparation of Recombinant FLAG Fusion Peptides—Plasmids encoding the N terminus of SV2A (Met1–Arg163) fused to the FLAG epitope in the pCTC expression vector (Sigma) were grown in the protease-deficient BL21 strain of Escherichia coli. Protein expression was induced by the addition of 200 μm isopropyl β-d-thiogalactopyranoside for 3 h at 25°C. The bacteria were harvested; resuspended in 150 mm potassium acetate, 2 mm EDTA, 0.05% Triton X-100, 10 mm HEPES, pH 7.4, 0.1% 2-mercaptoethanol, and 200 μm phenylmethylsulfonyl fluoride; and lysed by sonication. After sonication Triton X-100 was added to a final concentration of 0.5%. The resulting extract was centrifuged at 10,000 × g for 10 min to remove insoluble material. Fusion peptides were isolated using anti-FLAG M2 affinity gel as described above except that the wash elution buffers had no detergent.
Photoaffinity Labeling—A methanolic solution of 8-azidoATP[γ] biotin (Affinity Labeling Technologies Inc., Lexington, KY) was dried down under a gentle stream of nitrogen or in a SpeedVac. The dried photoreactive ATP analogue was suspended in 150 mm potassium acetate, 10 mm HEPES-KOH (pH 7.4) and incubated with purified SV2 at the indicated concentration. The reactions were performed on ice in the dark for 2 min. Generally, ∼5 μg of SV2-FLAG protein was used in 50-μl reactions. After incubation, the samples were irradiated with a hand-held UV lamp at 254 nm (UVP, Inc., San Gabriel, CA) for 2 min. The UV-irradiated samples were immediately diluted with SDS-PAGE sample buffer containing β-mercaptoethanol. For substrate competition studies, the indicated nucleotides were added to labeling reactions.
SDS-PAGE and Western Analysis—SDS-PAGE was performed using either 4–15% gradient or 10% gels. The proteins were stained with SilverSNAP kit (Pierce) or transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA) for Western blot analysis. SV2 was detected using a monoclonal antibody that recognizes all SV2 isoforms (20). FLAG fusion proteins were detected with a monoclonal anti-FLAG M2 antibody (Sigma). Antibody binding was detected with ECL reagent (Pierce). 8-Azido-ATP[γ] biotin binding was detected with ExtrAvidin-HRP (Sigma) and visualized with ECL reagent. The net intensity of labeled bands was quantified using a Kodak Image Station 440, and only nonsaturated images were used for quantification analysis.
Proteolytic Digestion of Photolabeled Protein—Purified recombinant proteins were labeled with 8-azido-ATP[γ] biotin as described above except that the labeling reaction was stopped with 40 mm dithiothreitol. Labeled protein was then incubated at 37 °C in the presence of sequencing grade trypsin (Sigma), 15:1 protein:enzyme by weight. At the indicated times, an aliquot was withdrawn, and the digestion was terminated by adding 1 mm phenylmethylsulfonyl fluoride. The sample was subjected to SDS-PAGE and Western blot analysis as described above.
Microsome Preparation and ATP Transport Assay—HEK293 cells transfected with pIRES2-EGFP or SV2A-pIRES2-EGFP were harvested 24–48 h after transfection. The cells were washed with ice-cold phosphate-buffered saline, suspended in homogenization buffer (320 mm sucrose, 10 mm HEPES, pH 7.4, 1× protease inhibitor mixture), and disrupted with a Dounce homogenizer using a total of 20 stokes. Cell debris and heavy membranes were removed by centrifugation at 15,000 × g for 20 min. Supernatants were used in ATP transport assays.
30 μg of microsomes were preincubated in 320 mm sucrose, 4 mm KCl, 4 mm MgSO4, 10 mm HEPES (pH 7.4) at 30 °C for 1 min. ATP transport was initiated by the addition of 50 μm [3H]ATP (0.1 Ci/mmol). The reactions were allowed to proceed at 30 °C for 5 min and then terminated by adding 3 ml of cold wash buffer (150 mm KCl, 10 mm HEPES, pH 7.4), after which they were quickly filtered through a prewetted HAWP filter (Millipore, Billerica, MA) followed by 3× 3 ml of ice-cold wash buffer. The filters were suspended in Ecoscint A scintillation mixture (National Diagnostics, Atlanta, GA), and the [3H]ATP bound was measured by scintillation counting. Background values were determined with 0-min reactions and subtracted from experimental values. The experiments were performed in duplicate and repeated four times. To distinguish transport from binding, a series of reactions was washed with wash buffer containing 2% Triton X-100 to disrupt membranes.
RESULTS
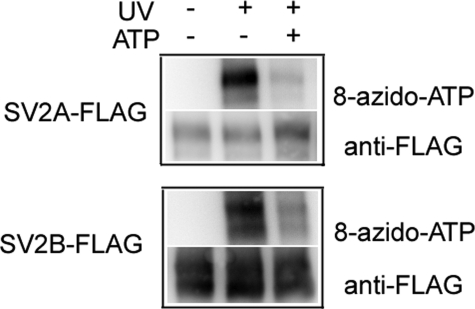
SV2 Binds ATP with Submillimolar Affinity—To determine whether SV2 binds ATP, we performed photoaffinity labeling with 8-azido-ATP[γ] biotin. Affinity-purified, recombinant SV2A-FLAG and SV2B-FLAG fusion proteins were incubated with the photoaffinity reagent in the presence or absence of UV irradiation. As shown in Fig. 1, both SV2 isoforms demonstrated UV-dependent incorporation of 8-azido-ATP. Bands labeled with either anti-FLAG (Fig. 1) or anti-SV2 (not shown) co-migrated with bands incorporating the photoaffinity label. The apparent doublet is likely due to differences in SV2 glycosylation because treatment with the glycosidase peptide N-glycosidase F resulted in a single band in both blots (not shown). Inclusion of excess ATP in the reaction decreased the binding of 8-azido-ATP, indicating that labeling was not due to nonspecific adherence of the photoaffinity reagent to SV2 protein. Thus SV2 is an ATP-binding protein.
FIGURE 1.
Purified SV2A-FLAG and SV2B-FLAG bind 8-azido-ATP-biotin. Recombinant SV2A-FLAG and SV2B-FLAG fusion proteins were purified from transfected HEK293 cells as described under “Materials and Methods.” Labeling reactions contained ∼5 μg of protein and 100 μm 8-azido-ATP in the presence or absence of 1 mm nonphotoreactive ATP. Control reactions that did not receive UV irradiation were run in parallel. The samples were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Bound 8-azido-ATP was visualized by ExtrAvidin-HRP and SV2-FLAG fusion proteins were detected with anti-FLAG antibody.
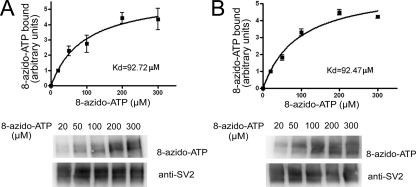
To determine the affinity of SV2 for ATP, we measured binding in the presence of 20–300 μm 8-azido-ATP. Binding saturated and displayed apparent Kd values of 93 and 92 μm for SV2A and SV2B, respectively (Fig. 2). These concentrations are within the physiological range of ATP in cells.
FIGURE 2.
SV2A and SV2B demonstrate sub-millimolar affinity for ATP. Purified SV2A-FLAG (A) and SV2B-FLAG (B) were labeled with the indicated concentrations of 8-azido-ATP-biotin and subjected to SDS-PAGE and Western analysis. The net intensity of labeled bands was quantified as described under “Materials and Methods.” The data are expressed as the intensity of 8-azido-ATP labeling normalized to SV2 protein detected by Western analysis. Representative Western blots are shown for each series. The error bars represent the S.E. (n = 3).
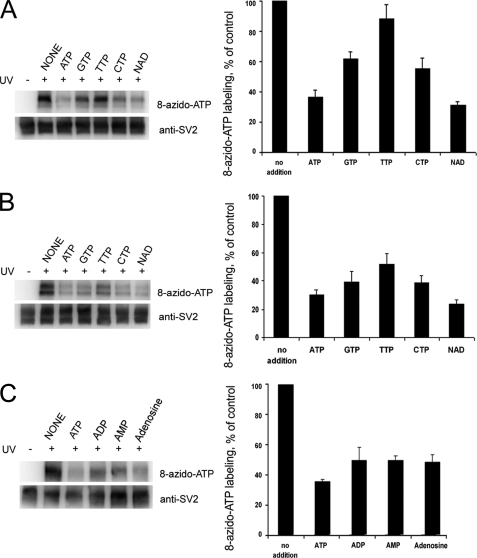
SV2 Preferentially Binds ATP and NAD—To determine the specificity of nucleotide binding by SV2, we tested the ability of a 10-fold excess of various nucleotides to compete with 8-azido-ATP binding to SV2. All of the nucleotides tested (ATP, GTP, TTP, CTP, and NAD) decreased labeling of both SV2A and SV2B. Although the pattern of inhibition was similar for both isoforms, SV2B was inhibited to a greater degree by the presence of other nucleotides. For both isoforms, ATP and NAD produced the greatest inhibition (Fig. 3, A and B), suggesting that SV2 preferentially binds adenine nucleotides.
FIGURE 3.
ATP and NAD are the most effective at inhibiting SV2 binding to 8-azido-ATP. The ability of different nucleotides to compete with 8-azido-ATP binding was assessed. SV2A-FLAG (A) and SV2B-FLAG (B) were labeled with 100μm 8-azido-ATP in the absence or presence of 1 mm of the indicated nucleotide. The samples were subjected to SDS-PAGE and Western analysis and quantified as described under “Materials and Methods.” The results represent the means ± S.E. of six experiments for SV2A and five experiments for SV2B. C shows the specificity of different adenine nucleotides binding to SV2A-FLAG. The error bars represent S.E. (n = 4).
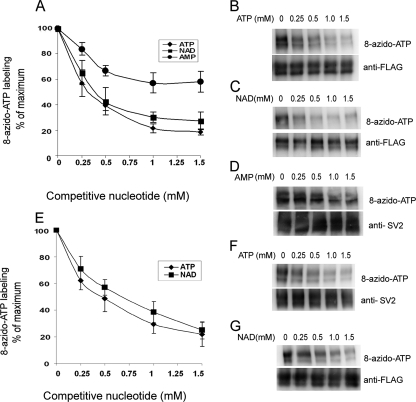
ATP, ADP, AMP, and adenosine all inhibited 8-azido-ATP binding to SV2A when present in 10-fold excess (Fig. 3C), although inhibition by ATP was greater (65% inhibition by ATP versus 50% inhibition by ADP, AMP, and adenosine). To assess differences in affinity, we compared dose-dependent inhibition by ATP and NAD. As shown in Fig. 4, ATP and NAD produced similar inhibition curves. For SV2A, the IC50 was 0.3–0.4 mm for ATP and ∼0.4 mm for NAD. For SV2B, ATP inhibited 8-azido-ATP binding with an IC50 of 0.4–0.5 mm, whereas NAD showed half-maximum inhibition at 0.6–0.7 mm. AMP inhibited 8-azido-ATP binding to SV2A in a dose-dependent manner; however, at the highest concentration tested (1.5 mm), inhibition was only half that of ATP and NAD.
FIGURE 4.
NAD and ATP demonstrate similar affinities for SV2. A–D, SV2A-FLAG was labeled with 100 μm 8-azido-ATP in the absence or presence of ATP, NAD or AMP (0.25–1.5 mm). The data were expressed as the percentage of 8-azido-ATP labeling in control reactions. Representative blots of SV2A labeling in the presence of different concentrations of ATP (B), NAD (C), or AMP (D) are shown. E–G, similar studies were done with SV2B-FLAG. The error bars represent the S.E. (n = 3).
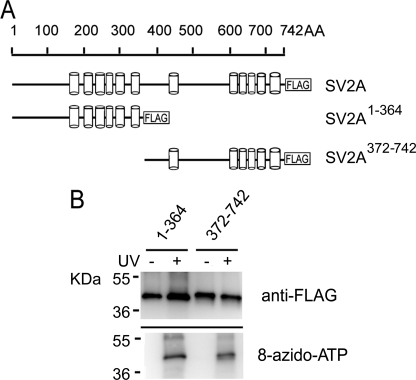
SV2A Contains Two Nucleotide-binding Domains—To identify the location of the nucleotide-binding site, we first assessed binding by the N- and C-terminal halves of SV2A. Recombinant FLAG fusion proteins containing either the N-(SV2A1–364) or C-terminal (SV2A372–742) half of SV2A were assessed for binding to 8-azido-ATP. Both of these proteins bound ATP (Fig. 5), suggesting that SV2A contains at least two nucleotide-binding sites.
FIGURE 5.
SV2A contains nucleotide binding sites on both N- and C-terminal halves. A, schematic structures of SV2A and its mutants used for mapping studies. Cylinders represent transmembrane domains. A FLAG epitope was attached to the C termini of the proteins. B, both the N-terminal and C-terminal halves of SV2A contain ATP-binding sites. N- and C-terminal halves of recombinant SV2A-FLAG were subjected to photoaffinity labeling as described under “Materials and Methods.”
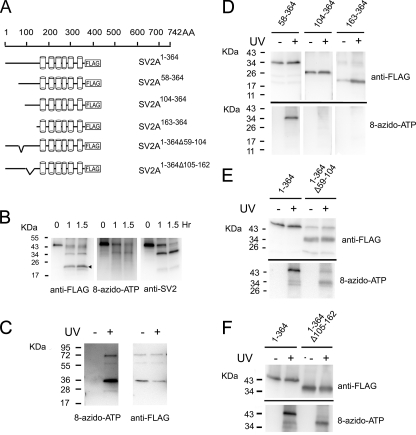
To further define the nucleotide-binding site in the first half of SV2A, we utilized trypsin digestion of photoaffinity-labeled SV2A1–364 (similar studies with SV2A372–742 did not yield interpretable results). These studies took advantage of the fact that SV2A1–364 can be labeled on its C terminus with anti-FLAG and on its N terminus with anti-SV2. As shown in Fig. 6B, full-length SV2A1–364 (0 h trypsin treatment) produced a single band that labeled with 8-azido-ATP, anti-FLAG, and anti-SV2. Trypsin digestion produced a 21-kDa fragment that was detected only with anti-FLAG and was not labeled with 8-azido-ATP. This suggests that the nucleotide-binding site is in the N terminus of SV2A, roughly between a.a. 1–170. This region corresponds to a cytoplasmic domain prior to the first transmembrane domain. To test this further, we generated a bacterially expressed recombinant FLAG fusion peptide containing SV2A a.a. 1–163 and performed photoaffinity labeling on the purified fusion peptides. As predicted by the trypsin digestion studies, this peptide bound 8-azido-ATP (Fig. 6C). We next generated a series of SV2A1–364 proteins in which the N terminus was truncated in steps. We found that proteins containing a.a. 58–364 retained nucleotide binding, whereas peptides containing a.a. 104–364 or 163–364 did not (Fig. 6D). These results indicate that a.a. 58–104 are required for nucleotide binding.
FIGURE 6.
Mapping the nucleotide binding site in the N terminus of SV2A. A, schematic of SV2A1–364 FLAG fusion proteins and truncation and deletion mutants. The cylinders represent transmembrane domains. Discontinuous lines represent deleted residues. B, an ATP-binding site located in the first 170 amino acids of SV2A. Shown are the results of trypsin digestion of 8-azido-ATP labeled SV2A1–364. Labeled protein was digested at 37 °C in the presence of trypsin. At the time periods indicated, an aliquot was withdrawn and subjected to analysis as described under “Materials and Methods.” The arrowhead indicates a 21-kDa proteolytic fragment, which is recognized by anti-FLAG but was not labeled by 8-azido-ATP. C, a FLAG fusion peptide containing SV2A a.a. 1–163 binds ATP. Photoaffinity labeling reactions were performed with 1 μg of purified peptide and analyzed as described under “Materials and Methods.” The predominant band at ∼35 kDa is larger than the predicted molecular mass of the peptide (19 kDa) and most likely represents a dimer. D, amino acids 58–104 are required for nucleotide binding. Serial truncations of SV2A1–364 were subjected to photoaffinity labeling as described under “Materials and Methods.” Only SV2A1–364 and SV2A58–364 bound 8-azido-ATP[γ] biotin indicating that a.a. 58–104 are critical for nucleotide binding. We note that the apparent molecular masses of SV2A104–364 and SV2A163–364 are about 25.5 and 19.5 kDa, respectively, which is smaller than the calculated molecular mass based on the amino acids sequences (29.6 and 23.4 kDa, respectively). We interpret this difference to be due to the highly hydrophobic nature of these proteins, which is likely to influence their migration in gel electrophoresis. E, amino acids 58–104 contribute to nucleotide binding. SV2A1–364 lacking residues 59–104 was subjected to photoaffinity labeling. 8-azido-ATP labeling was decreased by an average of 36% (n = 3). A representative Western blot is shown. F, amino acids 105–162 are required for nucleotide binding. SV2A1–364 lacking residues 105–162 was subjected to photoaffinity labeling. Deletion of these residues reduced binding by an average of 65%. Shown is a representative example of three independent experiments.
To further define the binding site, we sought to generate an SV2A-FLAG protein with the minimal deletion required to abolish nucleotide binding. Mutants lacking a.a. 59–83 or 84–104 retained nucleotide binding (data not shown). Removal of a.a. 59–104, however, reduced nucleotide binding by an average of 36% (n = 3) compared with SV2A1–364 (Fig. 6E). This confirms the importance of this region but also suggests that other residues are involved. Indeed, deletion of a.a. 105–162 decreased 8-azido-ATP labeling by an average of 65% (n = 3) (Fig. 6F). Together these findings indicate that the nucleotide-binding domain in the N terminus of SV2A spans residues 59–162.
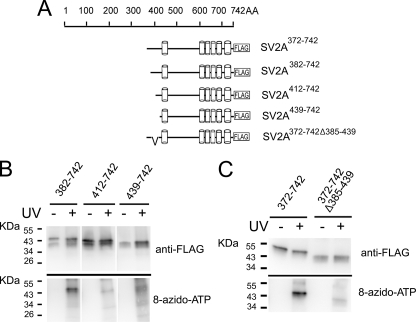
Major facilitator transporters are proposed to share a similar structure (21) composed of two symmetric six-helix lobes (22). Based on this predicted repeated structure, we hypothesized that the second nucleotide-binding site in SV2A would be located in the cytoplasmic loop between transmembrane domains 6 and 7. To test this, we generated a series of FLAG fusion proteins, illustrated in Fig. 7A, in which we progressively removed residues corresponding to the loop between transmembrane domains six and seven. As shown in Fig. 7B, SV2A382–742 demonstrated 8-azido-ATP binding, SV2A412–742 bound weakly, and SV2A439–742 did not demonstrate detectable binding. This suggests that the critical region for nucleotide binding in C-terminal half of SV2A is located between a.a. 382–439, a region analogous to the binding site in the N-terminal half of SV2A. Binding of 8-azido-ATP to a deletion mutant lacking a.a. 385–439 was reduced by an average of 64% (n = 3) compared with SV2A372–742 (Fig. 7C), supporting the conclusion that this region contains the binding site.
FIGURE 7.
Mapping the nucleotide binding site in the C-terminal half of SV2A. A, schematic structure of SV2A372–742 FLAG fusion proteins depicting truncation and deletion mutants. The cylinders represent transmembrane domains. The discontinuous lines in the N terminus of SV2A372–742 represent the deleted amino acids residues. B, the nucleotide-binding site in the C-terminal half of SV2A resides between a.a. 382–439. Serial truncations of the C-terminal half of SV2A were subjected to photoaffinity labeling as described under “Materials and Methods.” SV2A382–742 bound 8-azido-ATP[γ] biotin, whereas SV2A412–742 demonstrated reduced binding, and SV2A439–742 showed almost no binding indicating a binding site between a.a. 382–439. C, deletion of a.a. 385–439 decreased nucleotide binding by an average of 64%. Shown is a representative example of three independent experiments.
SV2A Does Not Affect ATP Transport—Although SV2 is structurally similar to MF transporter proteins, it has not been shown to have transporter activity. Given its ability to bind adenine nucleotides, we asked whether SV2 affects ATP transport. To test this, we expressed SV2A and a control vector in HEK293 cells and measured ATP uptake into microsomes. Expression of SV2A did not increase the ATP transport activity (Fig. 8). Although we did not consistently observe ATP transport into mouse brain synaptic vesicles, when we did there was no difference between vesicles from wild type and SV2A/B double knock-out mice (data not shown). Based on these findings, it is unlikely that SV2 is an ATP transporter, a conclusion consistent with the recent description of the SLC17A9 gene product as the vesicular nucleotide transporter (29).
FIGURE 8.
Expression of SV2A does not increase ATP transport into HEK293 cell microsomes. HEK293 cells were transfected with either pIRES2-EGFP (control) or SV2A-pIRES2-EGFP constructs. The cells were harvested 24–48 h after transfection, and transport of 3H-ATP into microsomal membranes was measured using a filter binding assay as described under “Materials and Methods.” A, results of four independent experiments done in duplicate. The error bars represent the S.E. B, transport was distinguished from binding by measuring [3H]ATP remaining after washing filters with buffer containing 2% Triton X-100, which disrupts membranes and displaces transported substrate. The data are from a single representative experiment. The error bars represent the standard deviation of duplicate reactions.
DISCUSSION
SV2 is a synaptic vesicle protein that contributes to vesicle priming. The studies reported here show that SV2 binds adenine nucleotides, suggesting that its priming activity could be modulated by synaptic energy levels. Purified SV2A and SV2B reacted with the photoaffinity reagent 8-azido-ATP-biotin. Binding was saturable and most efficiently inhibited by ATP and NAD, indicating that SV2 has highest affinity for these two adenine nucleotides. Although these studies were done with recombinant, purified SV2 protein in detergent micelles, we were also able to label SV2 in synaptic vesicles (data not shown). Labeling was not always detected, however, because of the much lower levels of SV2 in vesicles compared with recombinant protein preparations. Although we did not test ATP binding to recombinant SV2C, SV2C immunoisolated from synaptic vesicle extracts prepared from SV2A/B knock-out mice was labeled by 8-azido-ATP-biotin (data not shown). This suggests that SV2C also binds ATP and therefore that all SV2 proteins bind ATP.
Recent proteomic studies of synaptic vesicles have identified a number of glycolytic enzymes associated with vesicles (23, 24). These findings suggest cross-talk between energy production and the synaptic vesicle cycle. Synaptic secretion is an energy-intensive process. ATP is needed to maintain plasma membrane ion gradients to establish a proton gradient across the synaptic vesicles and to prime vesicles for exocytosis. Modulation of the activity of proteins involved in the synaptic cycle would ensure that the level of exocytosis does not surpass the energy available to support it.
Mapping studies revealed that SV2 contains two cytoplasmic nucleotide-binding sites: one that precedes the first transmembrane domain and one that precedes the seventh transmembrane domain. We note that both of these regions are predicted to form α helices (mGenTHREADER). The repetition of nucleotide-binding sites at the beginning of each six-transmembrane portion of SV2 is consistent with the hypothesis that it, like other MF transporters, is composed of two six-transmembrane domain repeats (25).
Our observation that SV2 binds adenine nucleotides raised the obvious question of whether it is an ATP transporter or contributes to ATP transport. Expression of SV2 in HEK293 fibroblasts did not increase ATP uptake into mitochondria-free membrane preparations, indicating that SV2 neither is an ATP transporter nor affects ATP transport. Our results can be contrasted to the effect of exogenous expression of the vesicular monoamine transporter (26, 27) or of the vesicular acetylcholine transporter (28), both of which increase uptake of their substrates into microsomes. We also examined ATP uptake into synaptic vesicle preparations. Vesicles from wild type and SV2A/B double knock-out mice showed equal levels of ATP uptake, although we note that transport levels were low and not consistently observed. We did observe increased NAD transport when SV2 was expressed in a single subclone of HEK293 cells, but this was not true when SV2 was expressed in other HEK cell clones or in PC12, COS, or Chinese hamster ovary cells.5 Therefore, the interpretation most consistent with the data is that nucleotide binding to SV2 modulates its function rather than that SV2 binds nucleotides as the first step in transporting them. This conclusion is consistent with a recent report demonstrating that the protein encoded by the SLC17A9 gene is the vesicular nucleotide transporter located on the synaptic vesicles or secretary granules (29).
Because the molecular action of SV2 remains unknown, it is difficult to know how ATP affects its functioning in the synapse. Although it remains possible that ATP regulates SV2 transport activity, it is worth noting that the ATP-binding sites in SV2 are not in the same regions as those identified in the glucose transporter. Therefore it remains just as likely that nucleotide binding regulates an action of SV2 other than transport.
Acknowledgments
We thank Dr. Ken Custer for making the SV2-FLAG pIRES2 construct, Dr. Neil Nathanson for comments on the manuscript, and Lisa Baldwin for editorial review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health, National Institute of Mental Health Grant R01MH059842 (to S. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SNARE, soluble NSF attachment protein receptors; SV2, synaptic vesicle protein 2; a.a., amino acid(s).
R. Bartlett and S. Bajjalieh, unpublished observations.
J. Yao, R. Bartlett, S. Bajjalieh, unpublished observations.
J. Yao and S. Bajjalieh, unpublished observations.
References
- 1.Buckley, K. M., Floor, E., and Kelly, R. B. (1987) J. Cell Biol. 105 2447–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajjalieh, S. M., Peterson, K., Linial, M., and Scheller, R. H. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 2150–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajjalieh, S. M., Peterson, K., Shinghal, R., and Scheller, R. H. (1992) Science 257 1271–1273 [DOI] [PubMed] [Google Scholar]
- 4.Feany, M. B., Lee, S., Edwards, R. H., and Buckley, K. M. (1992) Cell 70 861–867 [DOI] [PubMed] [Google Scholar]
- 5.Janz, R., Hofmann, K., and Sudhof, T. C. (1998) J. Neurosci. 15 9269–9281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajjalieh, S. M., Franz, G., Weimann, J. M., McConnell, S. K., and Scheller, R. H. (1994) J. Neurosci. 14 5223–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowder, K. M., Gunther, J. M., Jones, T. A., Hale, B. D., Zhang, H.-Z., Peterson, M. R., Scheller, R. H., Chavkin, C., and Bajjalieh, S. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 115268–115273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janz, R., Goda, Y., Geppert, M., Missler, M., and Sudhof, T. C. (1999) Neuron 24 1003–1016 [DOI] [PubMed] [Google Scholar]
- 9.Lynch, B. A., Lambeng, N., Nocka, K., Kensel-Hammes, P., Bajjalieh, S. M., Matagne, A., and Fuks, B. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9861–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Custer, K. L., Austin, N. S., Sullivan, J. M., and Bajjalieh, S. M. (2006) J. Neurosci. 26 1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iezzi, M., Theander, S., Janz, R., Loze, C., and Wollheim, C. B. (2005) J. Cell Sci. 118 5647–5660 [DOI] [PubMed] [Google Scholar]
- 12.Xu, T., and Bajjalieh, S. M. (2001) Nat. Cell Biol. 3 691–698 [DOI] [PubMed] [Google Scholar]
- 13.Henderson, P. J. (1993) Curr. Opin. Cell Biol. 5 708–721 [DOI] [PubMed] [Google Scholar]
- 14.Marger, M. D., and Saier, J. M. H. (1993) Trends Biochem. 18 13–20 [DOI] [PubMed] [Google Scholar]
- 15.Krupinski, J., Coussen, F., Bakalyar, H. A., Tang, W. J., Feinstein, P. G., Orth, K., Slaughter, C., Reed, R. R., and Gilman, A. G. (1989) Science 244 1558–1564 [DOI] [PubMed] [Google Scholar]
- 16.Carruthers, A., and Helgerson, A. L. (1989) Biochemistry 28 8337–8346 [DOI] [PubMed] [Google Scholar]
- 17.Levine, K. B., Cloherty, E. K., Fidyk, N. J., and Carruthers, A. (1998) Biochemistry 37 12221–12232 [DOI] [PubMed] [Google Scholar]
- 18.Levine, K. B., Cloherty, E. K., Hamill, S., and Carruthers, A. (2002) Biochemistry 41 12629–12638 [DOI] [PubMed] [Google Scholar]
- 19.Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J. (1982) EMBO J. 1 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckley, K., and Kelly, R. B. (1985) J. Cell Biol. 100 1284–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vardy, E., Arkin, I. T., Gottschalk, K. E., Kaback, H. R., and Schuldiner, S. (2004) Protein Sci. 13 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirai, T., Heymann, J. A., Maloney, P. C., and Subramaniam, S. (2003) J. Bacteriol. 185 1712–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burre, J., Beckhaus, T., Schagger, H., Corvey, C., Hofmann, S., Karas, M., Zimmermann, H., and Volknandt, W. (2006) Proteomics 6 6250–6262 [DOI] [PubMed] [Google Scholar]
- 24.Morciano, M., Burre, J., Corvey, C., Karas, M., Zimmermann, H., and Volknandt, W. (2005) J. Neurochem. 95 1732–1745 [DOI] [PubMed] [Google Scholar]
- 25.Pao, S. S., Paulsen, I. T., and Saier, M. H., Jr. (1998) Microbiol. Mol. Biol. Rev. 62 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merickel, A., Rosandich, P., Peter, D., and Edwards, R. H. (1995) J. Biol. Chem. 270 25798–25804 [DOI] [PubMed] [Google Scholar]
- 27.Peter, D., Jimenez, J., Liu, Y., Kim, J., and Edwards, R. H. (1994) J. Biol. Chem. 269 7231–7237 [PubMed] [Google Scholar]
- 28.Kim, M. H., Lu, M., Kelly, M., and Hersh, L. B. (2000) J. Biol. Chem. 275 6175–6180 [DOI] [PubMed] [Google Scholar]
- 29.Sawada, K., Echigo, N., Juge, N., Miyaji, T., Otsuka, M., Omote, H., Yamamoto, A., and Moriyama, Y. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5683–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]