Abstract
The androgen receptor (AR) is required for prostate cancer development and contributes to tumor progression after remission in response to androgen deprivation therapy. Epidermal growth factor (EGF) increases AR transcriptional activity at low levels of androgen in the CWR-R1 prostate cancer cell line derived from the castration-recurrent CWR22 prostate cancer xenograft. Here we report that knockdown of AR decreases EGF stimulation of prostate cancer cell growth and demonstrate a mechanistic link between EGF and AR signaling. The EGF-induced increase in AR transcriptional activity is dependent on phosphorylation at mitogen-activated protein kinase consensus site Ser-515 in the AR NH2-terminal region and at protein kinase C consensus site Ser-578 in the AR DNA binding domain. Phosphorylation at these sites alters the nuclear-cytoplasmic shuttling of AR and AR interaction with the Ku-70/80 regulatory subunits of DNA-dependent protein kinase. Abolishing AR Ser-578 phosphorylation by introducing an S578A mutation eliminates the AR transcriptional response to EGF and increases both AR binding of Ku-70/80 and nuclear retention of AR in association with hyperphosphorylation of AR Ser-515. The results support a model in which AR transcriptional activity increases castration-recurrent prostate cancer cell growth in response to EGF by site-specific serine phosphorylation that regulates nuclear-cytoplasmic shuttling through interactions with the Ku-70/80 regulatory complex.
The androgen receptor (AR)3 is required for normal prostate development and the onset and progression of prostate cancer. AR has a modular structure characteristic of steroid hormone receptors, with an NH2-terminal transcriptional activation domain, DNA binding domain, hinge region, and carboxyl-terminal ligand binding domain (1, 2). AR mediates the biological effects of androgens by binding testosterone and dihydrotestosterone (DHT) with high affinity (3). Androgen binding in the ligand binding domain stabilizes AR through the NH2- and carboxyl-terminal N/C interaction that increases AR transcriptional activity (4). Androgen deprivation by surgical or chemical castration to treat advanced prostate cancer reduces AR transcriptional activity and promotes tumor regression.
Several mechanisms have been proposed to explain the emergence of castration-recurrent prostate cancer during androgen deprivation therapy (for review, see Ref. 5). AR transcriptional activity and CWR-R1 human prostate cancer cell proliferation are hypersensitive to DHT (6). AR localizes in the nuclei of prostate cancer cells despite low levels of circulating androgen and appears to mediate recurrent growth after androgen deprivation (7). This could be explained by the presence of sufficient testosterone or DHT to activate AR in the microenvironment of castration-recurrent prostate cancer tissue (8, 9). On the other hand, cell culture studies suggest that AR transcriptional activity involves growth factor signaling under conditions of androgen deprivation.
HER2/neu, keratinocyte growth factor, insulin-like growth factor-1, and interleukin-6 have been reported to activate AR in the absence of androgen (10–12). Epidermal growth factor (EGF)-dependent phosphorylation of transcriptional intermediary factor 2 and heregulin signaling through the HER2 and HER3 tyrosine kinases increase AR transactivation and alter the growth of CWR-R1 prostate cancer cells in response to low levels of androgen (13). HER2 and HER3 activation, possibly through autocrine signaling, contributes to cell proliferation during prostate cancer recurrence. Growth factor signaling contributes to the onset of castration-recurrent prostate cancer through cross-talk between AR and autocrine loops that drive prostate cancer growth in the androgen-deprived patient.
Despite evidence for AR transcriptional activity in castration-recurrent prostate cancer, there is little consensus regarding the mechanisms involved in growth factor-mediated AR phosphorylation (14). The present study determined the effects of EGF on AR phosphorylation, nuclear localization, gene transactivation, and prostate cancer cell proliferation. We demonstrate that AR is required for CWR-R1 prostate cancer cell growth in response to androgen or EGF and that DHT and EGF act synergistically to increase cell growth. EGF-dependent phosphorylation at MAP kinase consensus site Ser-515 in the AR NH2-terminal domain and protein kinase C consensus site Ser-578 in the AR DNA binding domain regulate AR nuclear-cytoplasmic shuttling through interactions with the Ku-70/80 regulatory subunits of DNA-dependent protein kinase (DNA-PK). The studies suggest that AR phosphorylation in the P-box of the DNA-binding-domain first zinc module regulates AR transactivation in response to EGF signaling.
EXPERIMENTAL PROCEDURES
RNA Interference and Transcription Assays—Duplex AR-siRNA-3 was ucaaggaacucgaucguauuu (sense) and auacgaucgaguuccuugauu (antisense) sequences, and AR-siRNA-4 was gaaaugauugcacuauugauu (sense) and ucaauagugcaaucauuucuu (antisense) sequences (NCBI M20132) (SMART selection designed siRNAs, Dharmacon Inc., Lafayette, CO). The control was siCONTROL nontargeting siRNA pool (Dharmacon Inc.). CWR-R1 prostate cancer cells derived from the CWR22 castration-recurrent prostate cancer xenograft (6, 15) were transfected using Effectene (Qiagen, Valencia, CA) with 10 nm AR siRNA or control siRNA duplex and 0.1 μg of mouse mammary tumor virus luciferase reporter (MMTV-Luc). CWR-R1 cells were plated (1.6 × 105 cells/well) in 12-well plates using Richter's improved minimal essential prostate growth medium (Irvine Scientific, Santa Ana, CA) or Dulbecco's modified Eagle's medium (DMEM, Invitrogen), each supplemented with 10 nm nicotinamide, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml selenium, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2% fetal bovine serum (FBS). DMEM growth medium was further supplemented with 15 mm HEPES, pH 7.2. DNA and siRNA (220 μl) containing per well 45 μl of EC buffer (Qiagen), 1 μl of enhancer, 1 μl of Effectene reagent (Qiagen), and 200 μl of 2% serum-containing medium was added to cell cultures containing 0.8 ml of fresh medium. The next day the medium was replaced with phenol red-free, serum-free Improved Minimal Essential Zinc Option medium (Invitrogen) with and without DHT, and EGF and incubations were continued for 24 h. For protein kinase C (PKC) inhibition studies cells were treated with and without calphostin (Calbiochem), a PKC inhibitor, 1 h before transfection. Cells were treated again the next day in serum-free medium with and without calphostin in the absence and presence of 10 ng/ml EGF. Human endometrial Ishikawa cells were transfected using FuGENE-6 (Roche Applied Science) as previously described (16) with 0.025 μg of pCMV-AR and 0.1 μl of PSA-Enh-Luc. Cells were harvested in 0.25 ml of lysis buffer containing 1% Triton X-100, 2 mm EDTA, and 25 mm Tris phosphate, pH 7.8 (17). Luciferase activity was measured using an automated LumiStar Galaxy multiwell plate reader luminometer (BMG Labtechnologies, Durham, NC). Transfection data are representative of at least three independent experiments.
Construction of Self-complementary Adeno-associated Virus (scAAV)-AR-siRNA and scAAV-control (CTR)-siRNA— pSilencer 1.0-U6 siRNA containing an RNA polymerase III promoter (18, 19) (Ambion Inc., Austin, TX) and the Insert Design Tool for the pSilencer Vectors (Ambion) were used to generate hairpin siRNA encoding DNA oligonucleotide sequences based on the siRNA-3 sequence. A central loop sequence ttcaagaga and single overhang strand were added for cloning. Oligonucleotides that target AR were aaggaactcgatcgtatcattcaagagatgatacgatcgagttccttgatttttt (sense, 55 nucleotides) and aattaaaaaatcaaggaactcgatcgtatcatctcttgaatgatacgatcgagttccttggcc (antisense, 63 nucleotides) with 5′-EcoRI and 3′-ApaI terminal restriction sites. Control scAAV duplex oligonucleotides were ttctccgaacgtgtcacgtttcaagagaacgtgacacgttcggagaatttttt (sense, 53 nucleotides) and aattaaaaaattctccgaacgtgtcacgttctcttgaaacgtgacacgttcgagaaggcc (antisense, 61 nucleotides). Oligonucleotides were annealed by incubating at 90 °C for 3 min in the presence of 46 μl of annealing buffer containing 0.1 m potassium acetate, 2 mm sodium acetate, and 0.03 m HEPES-KOH, pH 7.4, followed by 1 h of incubation at 37 °C. pSilencer 1.0-U6 was linearized with ApaI and EcoRI and ligated overnight at 25 °C to the siRNA insert. Type II scAAV expression vectors ptrs-U1a-RFP-U6 and ptrs-U1a-green fluorescent protein and helper plasmids pXX6 (adenoviral helper genes) and pXX2 (AAV helper genes) were generously provided by Douglas M. McCarthy and Jude R. Samulski (Gene Therapy Center, University of North Carolina at Chapel Hill). p-trs-U1a-RFP-U6 contains a small nuclear RNA U1a promoter to drive expression of red fluorescent protein derived from pDSRED2-C1 (Clontech, Palo Alto, CA). The U6 promoter and duplex sequences were excised from pSilencer 1.0-U6 and cloned into ptrs-U1a-RFP-U6 linearized with NotI and KpnI. In-frame ligation was confirmed by sequencing. scAAV vectors produced in human embryonic kidney 293 cells using three-plasmid transfection were purified as described (20).
Human embryonic kidney 293 cells (2 × 107/dish) in DMEM containing 10% FBS were plated in twenty 15-cm dishes for 70–80% confluency after 24 h. Each plate was passaged 1:4 in 15-cm dishes, and 16 h later the medium was replaced with complete Iscove's modified Dulbecco's medium (Invitrogen) containing 10% FBS and incubated for 3 h before transfection. pXX6 helper plasmid (90 μg) was combined with 30 μg of control or AR-siRNA-scAAV vector plasmid, 30 μg of pXX2 helper plasmid, 0.25 m CaCl2, 0.25 m NaCl, 1.5 mm sodium phosphate, and 0.05 m Hepes, pH 7.2. Cells were incubated for 24 h, and medium was replaced with DMEM containing 2% FBS. Virus was collected 48 h post-transfection by lysing cells using 3 freeze-thaw cycles. scAAV vectors were purified by isopycnic centrifugation in CsCl (ρ = 1.4 g/ml) using an SW41 rotor (Beckman Instruments) at 40,000 rpm for 48 h at 10 °C. Fractions were collected and semiquantitated by slot-blot, and peak fractions were pooled and dialyzed in phosphate-buffered saline (PBS) overnight at 4 °C.
To test transduction efficiency of the scAAV serotype II in CWR-R1 cells, cells were infected with scAAV type II ptrs-U1a-green fluorescent protein virus in medium containing 2% FBS. Cells were harvested 1 and 9 days post-infection. Green fluorescent protein expression in transduced cells was analyzed by fluorescence-activated cell sorting using a FACScan1 cytometer (BD Biosciences). Forward and side scatter parameters were set according to cell size, and the setting for fluorochrome detection was adjusted so that fluorescence intensity of uninfected negative control cells was within the first decade of the four-scale log plot.
Cell Proliferation Assays—CWR-R1 prostate cancer cells (1.6 × 105/well) in 12-well plates were allowed to grow in prostate growth medium for 24 h. Cells were infected with 103 viral particles/cell in medium containing 2% FBS by rocking for 30 min at room temperature and incubated overnight at 37 °C in serum-containing medium. Cells were transferred to serum-free, phenol-red-free medium in the absence and presence of DHT and EGF. Duplicate wells were treated and assayed on days 1, 3, and 5 after infection. One-tenth volume of 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfo-phenyl)-2H-tetrazolium monosodium salt (WST-8, Dojindo Molecular Technologies, Gaithersburg, MD) was added to the wells and incubated for 2.5 h at 37 °C, and absorbance was determined at 450 nm using a plate reader. The effect of inhibiting PKC on cell proliferation was assayed using cells plated in medium containing 2% FBS. The next day cells were treated with calphostin (Calbiochem), a PKC inhibitor, in the absence and presence of EGF. Proliferation assays were performed 1, 3, 5, and 7 days after treatment.
Plasmids—Expression vectors pCMVhAR for full-length human AR (21), pCMVhAR-(1–660) for the AR NH2-terminal, DNA binding and hinge regions, and AR-(507–660) for part of the AR NH2-terminal domain, DNA binding domain, and hinge region were described (1, 22). Mutations in pCMVhAR were generated by PCR amplification and verified by sequencing. FLAG-AR-(507–660) was created by subcloning pCMVhAR mutants into EcoRI- and SalI-digested pCMV-FLAGb. Glutathione S-transferase (GST) fusion vector GST-AR-(1–660) was prepared as described (23). GST-AR-(1–660)-S578A was generated by site-directed mutagenesis using the QuikChange kit (Stratagene) and confirmed by sequencing. Prostate-specific antigen enhancer luciferase reporter vector (PSA-Enh-Luc) was provided by Michael Carey (University of California, Los Angeles) and contains the PSA upstream enhancer region (24). MMTV-Luc was provided by Stanley M. Hollenberg and Ron M. Evans (Salk Institute).
Immunochemistry—AR phospho-Ser-578 antipeptide antibody was raised in rabbits (21st Century Bio, Marlboro, MA) by immunizing with two bovine serum albumin-coupled peptides that were based on human AR sequence 572GALTCGSCKVFFKRA586. Peptide 1 was acetyl-Gly-Ser(P)-aminobutyrate-KVFFKRA-amino-hexanoic acid-C-amide. Peptide 2 was acetyl-C-aminohexanoic acid-GALT-aminobutyrate-Gly-Ser(P)-aminobutyrate-KVFFKRA-amide. Nonphosphopeptide acetyl-C-amino-hexanoic acid-GALT-amino-butyrate-GS-amino-butyrate-KVFFKRA-amide was used to establish binding specificity. Aminobutyrate replaced cysteine to avoid disulfide bonding, aminohexanoic acid was a six-carbon spacer, and the carboxyl-terminal cysteine was coupled to bovine serum albumin.
Immunoblotting was performed using monkey kidney COS-1 cells maintained in DMEM containing 2 mm l-glutamine, 10% bovine calf serum, penicillin, streptomycin, and 20 mm Hepes, pH 7.2. COS cells (2.5 × 106/10-cm dish) were transfected with 2 μg of DNA using DEAE-dextran (25). Cells were treated 24 h after transfection with and without 10 ng/ml EGF for 5 h. For MAP kinase inhibition studies, cells were treated for 1 h before transfection with and without U0126 (Promega). The next day cells were serum-starved in the absence and presence of U0126, and media were replaced 24 h later with and without 10 ng/ml EGF for 5 h. Cells were rinsed with PBS, scraped into 1.5 ml of cold PBS, and centrifuged at 12,000 × g for 2 min. The buffer was aspirated, and cells were resuspended and vortexed for 10 s in 50–100-μl buffer containing 1% Nonidet P-40, 0.15 m NaCl, 1% sodium deoxycholate, 0.1% SDS, 0.5 mm EDTA, 50 mm Tris-HCl, pH 7.5, 0.02 mg/ml pancreas extract, 1 mm phenylmethylsulfonyl fluoride, 0.005 mg/ml Pronase, 0.0005 mg/ml thermolysin, 0.003 mg/ml chymotrypsin, and 0.33 mg/ml papain (Roche Applied Science). After a 15-min incubation on ice, lysates were centrifuged for 15 min at 20,000 × g, and protein concentrations were determined using the Bio-Rad assay. Protein (25 μg) was incubated with and without 2.5 units λ-phosphatase (Sigma) in phosphatase buffer (Sigma) containing 2 mm MnCl2. To inhibit λ-phosphatase activity, lysates were incubated with 2.5 mm sodium vanadate, 10 mm sodium fluoride, and phosphatase Mixture Inhibitors 1/2 (Sigma) for 30 min at 25 °C.
To express FLAG-tagged constructs in CWR-R1 cells, 3 μg of DNA/5 × 106 cells/10-cm dish was transfected using Effectene (Qiagen). The next day CWR-R1 cells were treated with and without 10 ng/ml EGF for 5 h, harvested in cold PBS from 5 pooled dishes for each treatment group, and lysed in immunoprecipitation buffer containing 0.5% Nonidet P-40, 10% glycerol, 0.05 m sodium fluoride, 0.15 m NaCl, 50 mm Tris-HCl, pH 7.6, and phosphatase and protease inhibitors. For immunoprecipitation of full-length AR expressed in COS cells, 0.5 μm DHT was added to the lysis buffer. Approximately 1 mg of protein from two 10-cm COS cell dishes was precleared using Sepharose CL-4B (Sigma) and immunoprecipitated using anti-FLAG M2 affinity resin (Sigma). The final pellets were washed 3 times with immunoprecipitation buffer with and without 0.5 μm DHT and resuspended in 60 μl of 2× sample buffer containing 3.3% SDS, 10% glycerol, 0.2% 2-mercaptoethanol, and 20 mm Tris-HCl, pH 6.8, and analyzed by immunoblot.
For immunoblots of cell extracts, 4% total protein was separated on 10% acrylamide gels containing SDS. After electrophoresis, gel proteins were electroblotted to Immobilon-P membrane (Millipore Corp., Bedford, MA) overnight. Transfer blots were blocked overnight at 4 °C in 5% milk, 0.9% NaCl, 0.05% Tween 20, and 0.01 m Tris-HCl, pH 7.5. Blots were incubated for 1–2 h with 2.5 μg/ml rabbit polyclonal AR52 antibody targeting human AR NH2-terminal amino acid residues 544–558 (21), anti-FLAG M2 monoclonal antibody (1:2000 dilution, F-3165 Sigma), β-actin antibody AC-15 (1:5000 dilution, Abcam, Inc.), or Ku (p70) and Ku (p80) antibodies (0.5 μg/ml, MS-329 and MS-285, Lab Vision Corp., Fremont, CA). Anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary IgG antibodies (1:10,000 dilution, Amersham Biosciences) were incubated for 30 min at room temperature. Signals were detected using chemiluminescence (SuperSignal West Dura Extended Duration Substrate, Pierce).
Reactivity of AR phospho-Ser-578 antisera was determined by immunoblot of bovine serum albumin (BSA), BSA-coupled AR-(572–586) non-phosphorylated peptide and AR-(572–586) phospho-Ser-578 peptides 1 and 2 immunogens, and wild-type FLAG-AR-(507–660) and the S578A mutant. COS cells (2.5 × 106/10-cm dish) were transfected with 2 μg of wild-type FLAG-AR-(507–660) and the S578A mutant, and 24 h later cells were placed in serum-free, phenol-red free media for 24 h and treated with and without 10 ng/ml EGF for 5 h. Cells were lysed in immunoprecipitation buffer containing phosphatase and protease inhibitors, immunoprecipitated using anti-FLAG M2 affinity resin (Sigma), and resolved on a 12% acrylamide gel containing SDS. Transfer blots were probed with AR phospho-Ser-578 antisera (1:100 dilution) and 2.5 μg/ml AR52 antibody at room temperature for 2 h and with secondary antibody as described above. After chemiluminescence detection, membranes were rinsed with distilled water and incubated for 10 min at room temperature with 0.2% Ponceau S in 0.1% glacial acetic acid.
Immunocytochemistry was performed in COS cells (1 × 105 cells/well) in 12-well plates with a coverglass (26). Cells were transfected using Effectene with 0.2 μg of wild-type FLAG-AR-(507–660) or the S578A mutant and serum-starved 24 h. Cells were fixed for 10 min at room temperature with 4% paraformaldehyde in PBS, permeabilized for 5 min at 4 °C with 0.2% Triton-X-100 in PBS, blocked for 1 h at room temperature with 0.5% bovine serum albumin in PBS (13), and incubated for 1 h in 0.5% bovine serum albumin containing AR52 antibody (2.5 μg/ml) and for 30 min at room temperature with fluorescein isothiocyanate-conjugated anti-rabbit secondary antibody (1:75 dilution, Jackson ImmunoResearch Laboratories, Inc.). Slides were viewed using an Olympus BX60 microscope with original magnification of 40×.
In Vitro Kinase Assays—GST-AR-(1–660) and GST-AR-(1–660)-S578A were expressed in BL21 Escherichia coli cells treated with 1 mm isopropyl-β-thiogalactoside for 24 h at 16 °C during log phase growth. Glutathione-agarose beads (Amersham Biosciences) were incubated for 1 h at 4 °C with sonicated bacterial supernatants containing GST-AR fusion proteins. Beads were washed 3 times with PBS containing 1% Triton-X-100 followed by 3 washes with kinase buffer containing 10 mm EGTA, 0.1 m MgCl2, and 0.4 m Mes, pH 6.0. Part of the sample eluted with sample buffer was analyzed on 8–12% acrylamide gradient gels containing SDS. A serial dilution of bovine serum albumin was analyzed in parallel to estimate protein recovery. Bound protein (10–15 μg) was assayed for PKC phosphorylation. GST beads were resuspended in 30 μl of kinase buffer containing 10 or 100 μCi of [γ-32P]adenosine triphosphate (3000 Ci/mmol) and 20 ng of purified PKC (catalytic subunit, rat brain, Calbiochem) in the absence and presence of 1 μm unlabeled adenosine triphosphate and either 2.5 μg of calf thymus histone H1 (Calbiochem) or 2.5 μg of wild-type or mutant GST-AR-(1–660) fusion protein. After 10 min at 30 °C, reaction products were resolved on 8–12% acrylamide gradient gels containing SDS, which were dried and analyzed by autoradiography. To verify equal loading, gels were rehydrated in PBS and stained with Coomassie Blue, or half of the input resin was resuspended in 2× SDS sample buffer and analyzed by immunoblotting using AR52 antibody. Autoradiographs were quantitated by densitometric scanning using Image-Pro Analyzer Software (Media Cybernetics, Inc., Bethesda, MD).
Nuclear Extraction—FLAG-AR fusion vectors were transfected into COS cells (2.5 × 106/10-cm dish) using DEAE dextran. The next day cells were serum-starved and 24 h later treated with 10 ng/ml EGF for 5 h. Nuclear extracts were prepared using the Nuclear Extract kit (Active Motif, Carlsbad, CA). Cells were washed with ice-cold PBS containing phosphatase inhibitors, scraped, and pelleted for 5 min at 2600 × g, resuspended in 550 μl of hypotonic buffer, and incubated for 15 min on ice. Detergent (25 μl) was added, and samples were vortexed for 10 s at the highest setting. Cell suspensions were centrifuged for 30 s at 14,000 × g, and cytoplasmic extracts were stored at -80 °C. Nuclear cell pellets were resuspended in 50 μl of complete lysis buffer containing 1 mm dithiothreitol and protease inhibitor mixture. Lysates were vortexed for 10 s, and suspensions were incubated for 30 min on ice on a rocking platform at 4 °C. Extracts were vortexed 30 s and centrifuged 10 min at 14,000 × g, and nuclear fractions were transferred to pre-chilled microcentrifuge tubes. Protein concentration was determined, and nuclear and cytoplasmic extracts were analyzed by immunoblotting using 2.5 μg/ml AR52, tubulin-α (2 μg/ml, Thermo Fisher Scientific, Fremont, CA) and Laminin B1 antibodies (2 μg/ml, Active Motif).
RESULTS
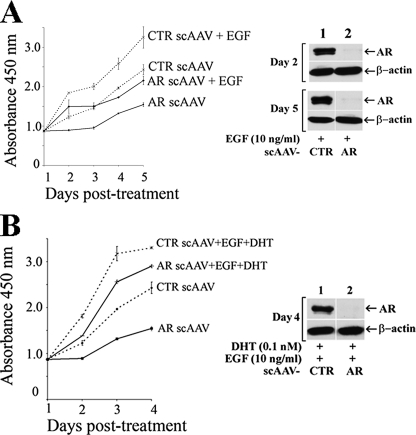
AR-dependent CWR-R1 Prostate Cancer Cell Proliferation— We investigated the requirement for AR in castration-recurrent prostate cancer cell growth using the CWR-R1 cell line that was derived from the castration-recurrent CWR22 xenograft of human prostate cancer (6). Growth studies were performed using CWR-R1 cells infected with control or AR-targeted siRNA-scAAV in the absence and presence of DHT and/or EGF. AR-siRNA-scAAV-infected cells grew more slowly than control cells in the absence and presence of 0.1 nm DHT (Fig. 1A). The >90% reduction in AR levels in CWR-R1 cells treated with and without DHT, assayed by immunoblot 2 and 5 days after AR-siRNA-scAAV infection (Fig. 1B), suggested that CWR-R1 cell growth was stimulated by AR both in the absence and presence of androgen.
FIGURE 1.
Inhibition of CWR-R1 cell growth by AR-siRNA-scAAV in the absence and presence of DHT. A, CWR-R1 cells were infected with control or AR-targeted siRNA-scAAV (103 virus particles/cell) and cultured with or without 0.1 nm DHT. Colorimetric assays at 450 nm using WST-8 reagent were performed in duplicate to measure cell proliferation daily up to 5 days. Medium was replaced every other day with or without 0.1 nm DHT as indicated. B, immunoblots of endogenous AR in CWR-R1 cells were performed 2 and 5 days after infecting cells with control siRNA scAAV (CTR) or AR-siRNA-scAAV in the absence (top panels) and presence of 0.1 nm DHT (bottom panels).
EGF also stimulated control CWR-R1 cells to grow faster than cells infected with AR-siRNA-scAAV (Fig. 2A, left panel). The reduction in AR levels determined by immunoblot of EGF-treated CWR-R1 cells 2 and 5 days after AR-siRNA-scAAV infection (Fig. 2A, right panels) provided evidence that EGF stimulation of CWR-R1 cell growth was mediated in part by AR. However, the attenuated but significant growth response of AR-siRNA-scAAV-infected cells to EGF suggested that EGF also stimulated cell proliferation through signaling mechanisms that were independent of AR. The stimulatory effect of EGF together with DHT on growth of control and AR-siRNA-scAAV-infected cells was greater than DHT or EGF alone and approached maximal levels within 3 days (Fig. 2B, left panel). Growth of AR-siRNA-scAAV-infected cells treated with DHT and EGF was less attenuated compared with control cells even though AR levels were reduced as shown by immunoblot (Fig. 2B, right panels). The results indicate that AR increases CWR-R1 prostate cancer cell growth in response to DHT or EGF and that EGF and DHT act synergistically through AR.
FIGURE 2.
Inhibition of EGF and EGF plus DHT-stimulated CWR-R1 cell growth using AR-siRNA-scAAV. A, CWR-R1 cells were infected with control or AR targeted siRNA-scAAV and cultured with or without 10 ng/ml EGF. Cell proliferation was assayed in the absence or presence of 10 ng/ml EGF as described in Fig. 1 (left panel). AR expression was determined by immunoblotting CWR-R1 cell lysates 2 and 5 days after infecting cells with control siRNA-scAAV or AR-siRNA-scAAV in the presence of EGF (right panels). B, CWR-R1 cells were infected with control or AR targeted siRNA-scAAV and treated with and without 10 ng/ml EGF and 0.1 nm DHT (left panel). Immunoblots of CWR-R1 cell AR were performed 4 days after infecting cells with control siRNA-scAAV or AR-siRNA-scAAV in the presence of 10 ng/ml EGF and 0.1 nm DHT (right panel).
Androgen-independent AR Transactivation—To pursue evidence that AR functions in castration-recurrent prostate cancer in the absence of androgen, we tested whether EGF can increase endogenous AR transcriptional activity in CWR-R1 cells using an MMTV-Luc reporter vector in the absence and presence of DHT. As expected, DHT increased AR transcriptional activity ∼100-fold, with a further 3-fold increase after the addition of EGF (Fig. 3A). A similar effect of DHT with and without EGF on AR transcriptional activity was seen after transfection of a control siRNA oligonucleotide that did not reduce AR levels (Figs. 3, A and B, lane 5). However, AR-targeted siRNA oligonucleotide-3 that reduced AR levels (Fig. 3B, lane 3) greatly reduced AR transactivation, with only a 2-fold increase in activity remaining in the presence of DHT and an ∼12-fold increase in response to DHT and EGF (Fig. 3A). In response to EGF alone, the ∼3-fold increase in AR transcriptional activity in the presence of control siRNA was abrogated by AR duplex siRNA oligonucleotide-3 (Fig. 3C).
FIGURE 3.
Inhibition of DHT and/or EGF stimulated AR transcriptional activity using AR siRNA. CWR-R1 cells were transfected with 0.1 μg of MMTV-Luc with or without 10 nm AR siRNA-3 or control siRNA (CTR). The next day cells were cultured for 24 h with or without 0.1 nm DHT in the absence or presence of 10 ng/ml EGF (A) or in the absence and presence of EGF alone (C), and luciferase activity was determined. In B, COS cell extracts (30 μg of protein/lane) from cells transfected with pCMV5 empty vector (p5, lane 1) or pCMV-AR in the absence (lane 2) or presence of 10 nm AR-siRNA-3 (lane 3), AR-siRNA-4 (lane 4), or control siRNA (lane 5) were analyzed by immunoblot using AR52 (2.5 μg/ml) and β-actin antibodies (1:5000 dilution).
The results indicate that EGF can activate AR in the CWR-R1 prostate cancer cell line in the absence of androgen and that EGF and DHT act synergistically to increase AR transcriptional activity. These data together with the cell growth studies presented above support the hypothesis that AR activation by EGF is sufficient to drive prostate cancer cell growth.
EGF-dependent AR Phosphorylation—Sequence analysis using NetPhos 2.0 (27) indicated 15 consensus serine, threonine, or tyrosine phosphorylation sites between AR residues 507 and 660 that comprise part of the AR NH2-terminal region, the DNA binding domain, and hinge region (Fig. 4A). Immunoblots of wild-type FLAG-AR-(507–660) expressed in COS cells revealed a 21-kDa protein and, after treatment with EGF, an additional slower migrating 23-kDa form (Fig. 4B, lanes 1–3). The EGF-dependent slower migrating form was eliminated by treatment with λ-phosphatase in the absence but not in the presence of phosphatase inhibitors (Fig. 4B, lanes 3–8). The slower migrating 23-kDa band was also observed in response to EGF with FLAG-AR-(507–660)-C576A, which has a cysteine mutation in the first zinc module that eliminates DNA binding (data not shown). The appearance of an EGF-dependent and phosphatase-sensitive slower migrating form of FLAG-AR-(507–660) indicated that EGF induces phosphorylation at one or more sites between AR residues 507 and 660 independent of AR binding to DNA.
FIGURE 4.
EGF-dependent phosphorylation at AR Ser-515. A, schematic representation of 15 predicted phosphorylation sites in part of the AR NH2-terminal region (N-term), DNA binding domain (DBD), and hinge region that were mutated in FLAG-AR-(507–660) and tested for band shift on immunoblots. B, COS cells were transfected with 2 μg of FLAG-AR-(507–660), serum-depleted for 24 h, and treated for 5 h with and without 10 ng/ml EGF as indicated. Cells were harvested, and lysates were incubated with and without 2.5 units of λ-phosphatase for 30 min at 30 °C in the absence (lanes 1–4) and presence of phosphatase inhibitors (lanes 5–8). C, COS cells (upper panel) and CWR-R1 cells (lower panel) were transfected with 2 μg of wild-type (wt) FLAG-AR-(507–660) or the S515A and S578A mutants. Cells were serum-depleted for 24 h, treated with and without 10 ng/ml EGF for 5 h, collected, and lysed in the presence of phosphatase inhibitors for immunoprecipitation using FLAG affinity resin. AR52 antibody was used to detect wt and mutant FLAG-AR-(507–660) and an associated slower migrating band indicative of phosphorylation. Also indicated is the nonspecific IgG band. D, reduced AR Ser-515 phosphorylation by mitogen-activated protein kinase inhibitor, U0126. COS cells were treated in the absence (lanes 1–2) and presence of increasing concentrations of U0126 (lanes 3–8) for 1 h before transfection with 2 μg of FLAG-AR-(507–660). The next day cells were serum-starved for 24 h in the absence and presence of U0126. Cells were treated again for 5 h with and without U0126 in the absence (lanes 1, 3, 5, and 7) and presence of 10 ng/ml EGF (lanes 2, 4, 6, and 8). FLAG-AR-(507–660) was detected using AR52 antibody, and β-actin served as the loading control.
To identify the EGF-dependent AR phosphorylation site(s), single serine or threonine to alanine and tyrosine to phenylalanine mutations were introduced into FLAG-AR-(507–660) at the consensus phosphorylation sites highlighted in Fig. 4A. Immunoblots of cell extracts before and after treatment with EGF indicated that only the AR NH2-terminal MAP kinase consensus S515A mutation eliminated the slower migrating 23-kDa form of FLAG-AR-(507–660) when assayed in COS and CWR-R1 cells (Fig. 4C, lanes 1–4). This result provided evidence that AR is phosphorylated at Ser-515 in response to EGF. In addition, the PKC consensus site mutation S578A in the AR DNA binding domain increased the relative proportion of the slower migrating 23-kDa phospho-Ser-515 form in the presence of EGF (Fig. 4C, lanes 5 and 6). MAP kinase-dependent phosphorylation at AR Ser-515 was supported by the decrease in intensity of the slower migrating 23-kDa band after treatment with both EGF and increasing concentrations of the MAP kinase inhibitor, U0126 (Fig. 4D).
AR phosphorylation at Ser-578 was indicated by the reactivity of an AR phospho-Ser-578-specific antibody that recognized two AR-(572–586) phospho-Ser-578-conjugated peptides used as immunogens but not unphosphorylated AR-(572–586) (Fig. 5A, lanes 3–5). The AR phospho-Ser-578-specific antibody also recognized the faster migrating 21-kDa form of wild-type AR-(507–919) but not the S578A mutant (Fig. 5B, upper panel, lanes 2–5), whereas both forms were detected using the AR52 antibody (Fig. 5B, lower panel). Treatment with λ-phosphatase reduced the reactivity of the phospho-Ser-578 antibody reactivity with wild-type AR-(507–919) (data not shown). The results suggest that the EGF-dependent increase in AR transcriptional activity and CWR-R1 cell growth are associated with MAP kinase-dependent phosphorylation at AR Ser-515 in the NH2-terminal region and modulation by phosphorylation at Ser-578 in the DNA binding domain.
FIGURE 5.
Phosphorylation at AR Ser-578. A, immunoblots without protein (lane 1), 2.5 μg of bovine serum albumin (BSA, lane 2), bovine serum albumin-coupled nonphosphorylated AR-(572–586) peptide (lane 3), and AR-(572–586) phospho-Ser-578 peptides-1 and -2 (lanes 4 and 5) were separated on 10% acrylamide gels. Transfer blots were incubated with AR anti-phospho-Ser-578 antiserum (1:100 dilution) as described under “Experimental Procedures.” Equivalent loading of the conjugated AR peptides was confirmed by staining the transfer blot with 0.2% Ponceau S (lower panel). B, COS cells were transfected with 2 μg of pSG5 empty vector (-, lane 1), wild-type FLAG-AR-(507–660) (lanes 2 and 3), and FLAG-AR-(507–660)-S578A (lanes 4 and 5). The next day cells were transferred to serum-free medium and 24 h later treated for 5 h in the absence and presence of 10 ng/ml EGF as indicated. Cell extracts were immunoprecipitated using FLAG-M2 affinity resin and separated on a 12% acrylamide gel containing SDS, and transfer blots were incubated with AR anti-phospho-Ser-578 antisera (1:100 dilution, upper panel) and 2.5 μg/ml AR52 antibody (lower panel) as described under “Experimental Procedures”.
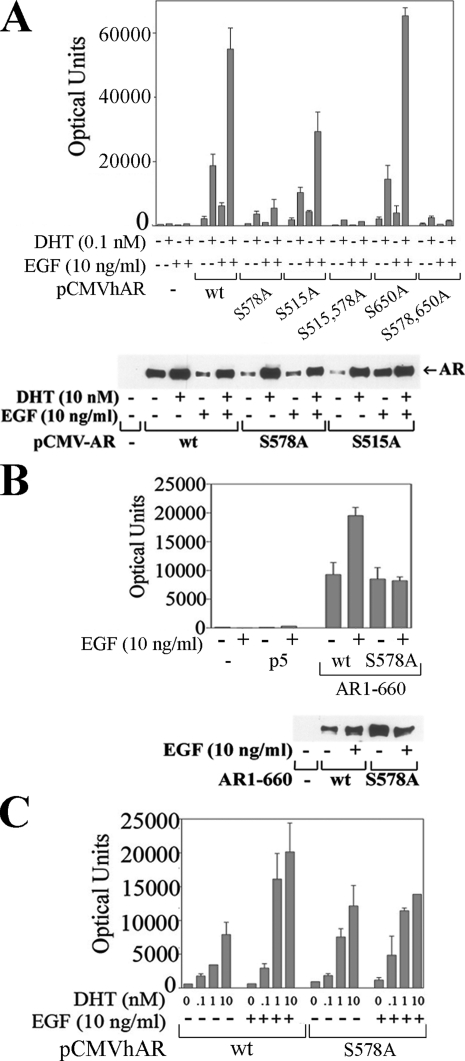
AR Phosphorylation Alters Transcriptional Activity—We investigated further the link between EGF-dependent AR phosphorylation and increased AR transcriptional activity using full-length wild-type AR and serine to alanine mutants expressed with a PSA-Enh-Luc reporter in CWR-R1 cells. AR transcriptional activity increased ∼3-fold in response to EGF in the absence and presence of DHT (Fig. 6A, upper panel). A similar response was seen with AR-S650A, which has a mutation in the previously reported Ser-650 phosphorylation site in the hinge region of AR (28, 29). AR-S515A transcriptional activity increased in response to EGF and DHT, but overall activity was less than wild type. In contrast, EGF did not increase transcriptional activity of AR-S578A in the absence or presence of DHT or when the S578A mutation was combined with the S515A or S650A mutation. When assayed by immunoblot (Fig. 6A, lower panel), expression of AR-S578A and AR-S515A was similar to wild-type AR, as was androgen-dependent AR stabilization that results from the AR N/C interaction (4, 23).
FIGURE 6.
AR Ser-578 required for EGF-induced AR transactivation in CWR-R1 and Ishikawa cells. A, CWR-R1 cells were transiently transfected with 0.1 μg of PSA-Enh-Luc and 10 ng wt pCMV-AR and the S578A, S515A, S515A-S578A, S650A, and S578A-S650A mutants and incubated for 24 h with and without 0.1 nm DHT and 10 ng/ml EGF as indicated, and luciferase activity was determined (upper panel). COS cells were transfected using DEAE dextran as described under “Experimental Procedures” with 2 μg of pCMV5 empty vector (-), wild-type pCMV-AR (wt), and the S578A and S515A mutants (lower panel). COS cells were incubated in the absence and presence of 10 nm DHT and 10 ng/ml EGF as indicated, and immunoblots were performed to confirm similar expression levels. B, CWR-R1 cells were transfected with 0.1 μg of PSA-Enh-Luc and 50 ng of pCMV-AR-(1–660) wt and S578A mutant and incubated for 24 h with and without 10 ng/ml EGF, and luciferase activity was determined (upper panel). Similar expression levels were determined by transfecting COS cells with 2 μg of wt pCMV-AR-(1–660) and the S578A mutant and incubating cells for 24 h in the absence and presence of 10 ng/ml EGF (lower panel). C, Ishikawa cells were transfected with 0.1 μg of PSA-Enh-Luc and 25 ng of wild-type pCMV-AR (wt) or S578A mutant and incubated for 24 h with increasing concentrations of DHT with and without 10 ng/ml EGF as indicated, and luciferase activity was determined.
The weaker transcriptional activity of the AR-S578A DNA binding domain mutant did not result from loss of DNA binding. This was evident from AR-(1–660)-S578A, a constitutively active AR NH2-terminal and DNA binding domain fragment that retained transcriptional activity of wild-type AR-(1–660) using the PSA-Enh-Luc reporter (Fig. 6B, upper panel). However, similar to results with full-length AR-S578A (Fig. 6A), transcriptional activity of AR-(1–660)-S578A was not increased by EGF when expression levels of AR-(1–660)-S578A were similar to wild-type AR-(1–660) (Fig. 6B, lower panel). AR-(1–660)-S578A also constitutively activated the MMTV-Luc reporter even though full-length AR-S578A was inactive with this promoter (data not shown).
To further establish a requirement for Ser-578 in the EGF-dependent increase in AR transcriptional activity, we performed transcription assays in human endometrial cancer Ishikawa cells using the PSA-Enh-Luc reporter. In the presence of increasing concentrations of DHT, AR-S578A transcriptional activity was similar to wild-type AR (Fig. 6C). This differed from CWR-R1 cells where AR-S578A transcriptional activity was less than wild-type AR (Fig. 6A). However, in agreement with results using the CWR-R1 cell line, the EGF-dependent increase in wild-type AR transcriptional activity in the presence of DHT was diminished by the AR S578A mutation in Ishikawa cells (Fig. 6C). The results suggest that phosphorylation at AR Ser-578 is required for the AR transcriptional response to EGF.
Functional Effects of AR Phosphorylation by PKC—Ser-578 is a predicted consensus phosphorylation site for PKC, a kinase that acts downstream of EGF signaling (30). We performed in vitro kinase assays using the PKC catalytic subunit with wild-type GST-AR-(1–660) and the S578A mutant. PKC-dependent phosphorylation of GST-AR-(1–660) was reduced 30–35% by the S578A mutation when equivalent amounts of protein were assayed by immunoblot (Fig. 7A, lanes 1 and 2), where histone H1 served as a PKC substrate control (Fig. 7A, lane 3). When averaged over multiple experiments, the S578A mutation decreased GST-AR-(1–660) phosphorylation by ∼50% (Fig. 7B). The EGF-dependent and independent increase in CWR-R1 cell growth was reduced by the PKC-specific inhibitor calphostin (Fig. 7C). The results suggest that EGF signaling through PKC-dependent phosphorylation of AR Ser-578 increases AR transcriptional activity and AR-mediated CWR-R1 cell growth.
FIGURE 7.
PKC-mediated phosphorylation at AR Ser-578 and EGF-dependent CWR-R1 cell growth. A, in vitro kinase assays were performed using GST-AR-(1–660) (2.5 μg, lane 1) and GST-AR-(1–660)-S578A (2.5 μg, lane 2) expressed in E. coli and purified by adsorption to glutathione beads. Histone H1 served as a PKC substrate control (2.5 μg, lane 3). Assays were performed as described under “Experimental Procedures” using the PKC catalytic subunit in the presence of 10 μCi of [γ-32P]adenosine triphosphate (upper panel). Parallel immunoblots were probed with AR52 antibody (lower panel). Samples were analyzed by autoradiography and band intensities measured by densitometry. NS designates a nonspecific phosphorylated band. B, data from four independent experiments described in A were averaged. C, inhibition of CWR-R1 cell proliferation by calphostin. CWR-R1 cells were plated and serum-starved the next day for 24 h and treated as described under “Experimental Procedures” in serum-free media with and without 10 ng/ml EGF alone or with 50 nm calphostin, a PKC inhibitor (day 0). Media and additives were replenished every other day over 7 days. Cell proliferation indexes were measured using WST-8 reagent on days 3, 5, and 7 after seeding.
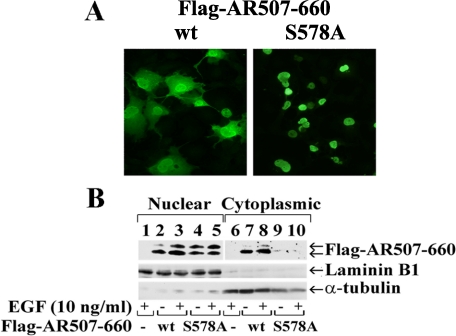
AR Ser-578 Phosphorylation Influences Nuclear-Cytoplasmic Shuttling—The effect of the S578A mutation on AR subcellular localization was investigated using wild-type and mutant FLAG-AR-(507–660). Immunostaining showed that wild-type FLAG-AR-(507–660) distributed between the nucleus and cytoplasm of transfected COS cells, indicative of nuclear-cytoplasmic shuttling (Fig. 8A, left panel). The phosphomimetic FLAG-AR-(507–660)-S578D distributed similarly between the nucleus and cytoplasm (data not shown), whereas immunostaining of FLAG-AR-(507–660)-S578A was exclusively nuclear (Fig. 8A, right panel).
FIGURE 8.
Increased nuclear localization of the AR S578A mutant. A, COS cells were transfected with 0.2 μg of wild-type (wt) FLAG-AR-(507–660) or the S578A mutant and serum-starved the next day for 24 h. FLAG-AR-(507–660) and the S578A mutant are represented by green fluorescence detected using AR52 antibody and fluorescein isothiocyanate-conjugated secondary anti-rabbit antibody. Original magnification 40×. B, nuclear and cytoplasmic fractions were prepared as described under “Experimental Procedures” and analyzed by immunoblotting using AR52 antibody that recognizes FLAG-AR-(507–660) wt and the S578A mutant. Laminin-B1 and α-tubulin served as nuclear and cytoplasmic extract controls, respectively.
AR compartmentalization was also investigated by comparing nuclear and cytoplasmic extracts of cells expressing FLAG-AR-(507–660) and the S578A mutant before and after treatment with EGF. In agreement with the immunostaining results, wild-type FLAG-AR-(507–660) was distributed in both the nuclear and cytoplasmic fractions (Fig. 8B, lanes 2, 3, 7, and 8). The slower migrating 23-kDa phospho-Ser-515 form was more prominent in the nuclear fraction in response to EGF. FLAG-AR-(507–660)-S515A lacked the slower migrating 23-kDa form (Figs. 4 and 5) and distributed in both nuclear and cytoplasmic extracts similar to wild type (data not shown). However, in agreement with the immunostaining results, FLAG-AR-(507–660)-S578A was almost entirely nuclear, and the proportion of the phospho-Ser-515 form was increased (Fig. 8B, lanes 4, 5, 9, and 10). Parallel immunoblotting of nuclear laminin-B1 and cytoplasmic α-tubulin substantiated the subcellular fractionation results. Cell extracts contained similar amounts of wild-type and mutant FLAG-AR-(507–660) (data not shown), suggesting that the smaller amount of the S578A mutant in the cytoplasmic fraction did not result from degradation. The results suggest that phosphorylation at AR Ser-578 limits nuclear phosphorylation at Ser-515 and modulates AR nuclear-cytoplasmic shuttling.
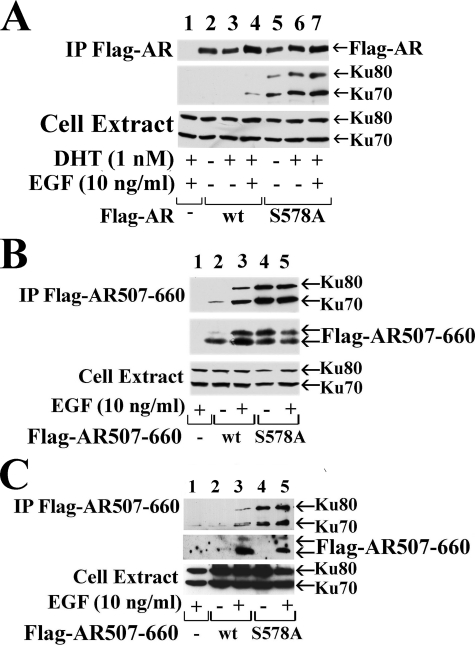
Phosphorylation at AR Ser-578 Modulates AR Interaction with Ku-70/80—The Ku-70/80 regulatory subunits of DNA-PK were shown to interact with the progesterone receptor DNA binding domain (31), which shares sequence similarity with the AR DNA binding domain (32). Ku-70/80 subunits were also implicated in AR transcriptional recycling (33). To address the influence of Ser-578 phosphorylation on AR interaction with Ku-70/80, we performed coimmunoprecipitation studies in COS cells using full-length wild-type FLAG-AR, the S578A mutant, and endogenous Ku-70/80. We found that Ku-70 and Ku-80 coimmunoprecipitated with FLAG-AR-S578A but only weakly with wild-type AR (Fig. 9A, upper two panels).
FIGURE 9.
AR Ser-578 mediates the interaction with Ku-70/80. COS cells (A and B) and CWR-R1 (C) cells were transfected with 2 μg of wild-type (wt), FLAG-AR, FLAG-AR-(507–660) and the S578A mutants as indicated. Cells were serum-starved the next day for 24 h and treated with and without 1 nm DHT and/or 10 ng/ml EGF for 5 h as indicated. Protein lysates were immunoprecipitated (IP) using anti-FLAG resin and analyzed by immunoblotting. AR-52 antibody was used to detect immunoprecipitated FLAG-AR and Ku-70/80-specific antibodies for coimmunoprecipitated proteins. Protein lysates (4% of total) were analyzed for endogenous Ku-70/80 (lower panels).
In similar experiments using FLAG-AR-(507–660), we found that the S578A mutant interacted with endogenous Ku-70 and Ku-80 to a greater extent than wild-type FLAG-AR-(507–660) in both COS (Fig. 9B) and CWR-R1 cells (Fig. 9C). Also, as seen with full-length FLAG-tagged AR, the interaction between wild-type FLAG-AR-(507–660) and Ku-70/80 increased in the response to EGF in both cell lines. A DNA binding mutant, FLAG-AR-(507–919)-C576A, interacted with Ku-70/80 to a similar extent as wild type (data not shown). The results suggest that EGF-dependent phosphorylation at AR Ser-578 modulates phosphorylation at Ser-515 and regulates AR interaction with Ku-70/80.
DISCUSSION
EGF Regulation of AR Transcriptional Activity—EGF signaling has been indirectly linked to increased AR transcriptional activity through the post-translational modification of AR coregulatory proteins. EGF increases phosphorylation of transcriptional intermediary factor 2, an SRC/p160 coactivator (17), and phosphorylation and multiple monoubiquitinylation of MAGE-11 of the melanoma antigen gene family (16). These changes stabilize the coregulator interaction with AR to increase AR transcriptional activity. In this report we provide evidence that EGF increases AR transcriptional activity through the coordinate phosphorylation of serine residues in the AR NH2-terminal and DNA binding domains.
We have shown that EGF increases CWR-R1 prostate cancer cell growth in an AR-dependent manner in the absence and presence of androgen. EGF acts synergistically with DHT to stimulate AR transcriptional activity and cell growth. EGF-dependent CWR-R1 prostate cancer cell proliferation was greatest in the presence of AR and DHT and was reduced by an inhibitor of PKC. One interpretation suggests that AR signaling in the absence and presence of DHT establishes a basal proliferation rate that is enhanced by EGF through mechanisms that include AR but are also independent of AR. EGF-dependent AR activation in the absence and presence of androgen is mediated by PKC-dependent phosphorylation at Ser-578 in the AR DNA binding domain and by MAP kinase-dependent phosphorylation at Ser-515 in the AR NH2-terminal region. The downstream functional effects of AR phosphorylation at these sites alter AR nuclear-cytoplasmic shuttling through interactions with Ku-70/80.
EGF-dependent AR Phosphorylation—Earlier mutagenesis studies demonstrated AR phosphorylation at Ser-81 and Ser-94 in the NH2-terminal region and at Ser-650 in the hinge region between the DNA and ligand binding domains (28). AR Ser-81 was suggested to be a substrate for the HER2-regulated kinase pathway (34), and AR phosphorylation at Ser-650 was linked to stress kinase modulation of AR transcriptional activity (29). However, mutations at these sites have relatively little effect on AR transcriptional activity (28, 35). Mass spectrometry confirmed AR phosphorylation at Ser-81, -94, and -650 and identified Ser-16, -256, -308, and -424 as phosphorylation sites in the AR NH2-terminal region (14). AR NH2-terminal Ser-213 was phosphorylated in nonproliferating prostate epithelial cells (36), and Akt-mediated phosphorylation at AR Ser-213 and Ser-791 was linked to anti-apoptotic and proliferative effects in prostate cancer cells (37, 38). However, the synergistic effects of Akt and AR on neoplastic proliferation of murine prostatic epithelium were independent of phosphorylation at these sites (39). More recently, prostate cancer growth was linked to Src and Ack1-mediated phosphorylation of AR tyrosine residues 267 and 363 in the AR NH2-terminal region (40).
Our current studies focused on the AR DNA binding domain and flanking NH2-terminal and hinge regions and confirmed that a mutation at Ser-650 has relatively little effect on AR transcriptional activity (28, 35). We also found that mutations at other previously reported phosphorylation sites in the region, including a recently reported Src-dependent tyrosine phosphorylation site Tyr-534 in the AR NH2-terminal region close to the DNA binding domain (41), did not diminish AR transcriptional activity in CWR-R1 cells in response to DHT with or without EGF. The relative lack of functional effects of mutations at most of the previously reported AR phosphorylation sites left unanswered the question of the significance of AR phosphorylation, particularly in prostate cancer cells where growth factor and mitogen signaling are increased.
Our studies indicate direct effects of EGF signaling on AR transcriptional activity through Ser-515 in the NH2-terminal region and Ser-578 in the DNA binding domain. MAP kinase phosphorylation site Ser-515 is positioned near the DNA binding domain (Fig. 10A) carboxyl-terminal to activation function-1 transactivation domain residues 142–337 required for androgen-dependent AR transcriptional activity (1, 42). AR Ser-515 is closer to tau-5 residues 360–485 (43), a transcriptional activation region whose activity is apparent in the AR NH2-terminal and DNA binding domain fragment that lacks the ligand binding domain (1, 42).
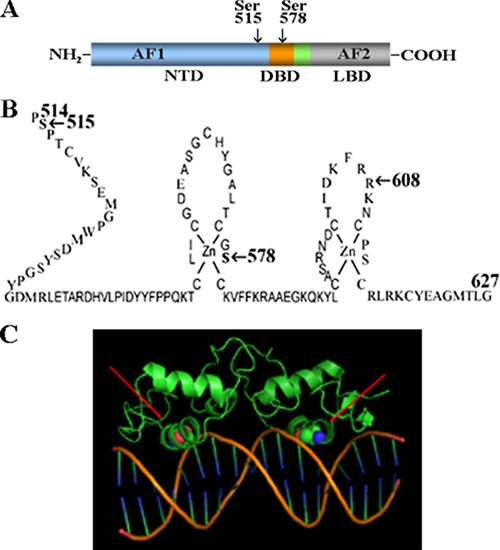
FIGURE 10.
AR schematic diagrams. A, full-length human AR amino acid residues 1–919 contain the NH2-terminal domain (NTD) with activation function-1 (AF1) and Ser-515, DNA binding domain (DBD) with Ser-578, and ligand binding domain (LBD) with activation domain-2 (AF2). B, ARNH2-terminal and DNA binding domain residues 514–627 showing the two zinc modules with highlighted AR phosphorylation sites Ser-515 and Ser-578. C, structure of the AR DNA binding domain dimer (green) bound to androgen response element DNA (orange and blue) (52) with space-filled Ser-578 indicated with red arrows.
MAP kinase signaling was previously implicated in androgen-dependent AR transcriptional activity (13, 17, 44). In vitro MAP kinase assays suggested phosphorylation of AR-36–643, an AR NH2-terminal and DNA binding domain fragment, in response to HER2/neu signaling (11). Although an AR S515A mutation reduced the signaling effects of HER2/neu, additional evidence for phosphorylation at this site was not reported. Here we show that EGF signaling slowed the gel electrophoretic migration of an AR NH2-terminal-DNA binding domain-hinge region fragment, an effect that was eliminated by treatment with λ-phosphatase. The EGF-dependent increase in AR transcriptional activity was linked to phosphorylation at Ser-515 and Ser-578. MAP kinase consensus site mutation S515A reduced AR transcriptional activity slightly but did not eliminate the AR transcriptional response to EGF in the absence and presence of androgen. Inhibition of Ser-578 phosphorylation by an S578A mutation increased phosphorylation at Ser-515, which provided evidence that AR phosphorylation at Ser-515 is linked to phosphorylation at Ser-578.
Indeed, EGF-dependent phosphorylation at AR Ser-578 appears to be required for EGF stimulation of AR transcriptional activity. Ser-578 is positioned in the first zinc module of the AR DNA binding domain (Fig. 10B) within the P-box. Response element DNA binding specificity distinguished by the glucocorticoid receptor (GR) and estrogen receptor is associated with three P-box residues, GSXXV (45, 46). Human AR P-box 577GSCKV581 contains Ser-578 and is common to GR and the progesterone and mineralocorticoid receptors, each of which recognizes the GGTACANNNTGTTCT (N is any nucleoside) consensus response element (47–49). P-box sequence within this subfamily distinguishes TGTTCT half-site sequence from TGACCT estrogen response element consensus sequence (46). Within the AR receptor subgroup, DNA response element binding specificity was attributed to residues in the second zinc module and the carboxyl-terminal extension in the hinge region (50, 51).
Crystal structures of the AR DNA binding domain-androgen response element DNA complex indicate that human AR Ser-578 lies within the first α-helix that directly contacts the major groove of the DNA (Fig. 10C) (52). Human AR Ser-578 corresponds to human GR Ser-440. When GR Ser-440 was changed to glycine to mimic Gly-204 in human ER or to alanine, GR DNA binding specificity was reduced. Based on these studies, the AR Ser-578 hydroxyl group is predicted to modulate DNA binding specificity through a hydrogen bond network extending to Arg-608 in the second zinc module loop containing AR D-box 596ASRND600 (Fig. 10B) (53). Unlike the common P-box sequence shared by AR, GR, progesterone, and mineralocorticoid receptors, the AR D-box, thought to be involved in receptor dimerization and half site spacing recognition, differs from the AGRND D-box common to other members of this subgroup of steroid receptors (45, 54). AR Arg-608 corresponds to Arg-470 in human GR, for which a lysine substitution was predicted to alter DNA binding specificity by changing the hydrogen bonding pattern (53). The importance of AR Arg-608, which contacts the DNA phosphate backbone and is conserved in the steroid receptor family, is supported by the naturally occurring R608K mutation that causes partial androgen insensitivity.
A naturally occurring AR S578T mutation also causes grade 5 partial androgen insensitivity, where complete androgen insensitivity (grade 6/7) is associated with an external female phenotype in a genetic male (55, 56). The S578T mutation demonstrates the in vivo importance of Ser-578 in AR function. The partial response of recombinant AR-S578T to elevated androgen levels in a COS cell transcriptional assay (55) indicates some retention of DNA binding activity. The extent of phosphorylation at residue 578 may be diminished by threonine at this position and phospho-Ser-578 or phospho-Thr-578 might alter AR interaction with DNA (Fig. 10C) or associated proteins such as Ku-70/80. Rapid release and rebinding of steroid receptors to DNA in a dynamic hit-and-run model has revealed the transient nature of steroid receptor DNA binding required for activation of transcription (57, 58). Similar transient interactions were reported for AR binding to androgen response element DNA (59). EGF-dependent phosphorylation at AR Ser-578 may regulate AR association and dissociation from DNA and the magnitude of the transcriptional response.
We have provided evidence for PKC phosphorylation at AR Ser-578. Our results support previous evidence that phosphorylation by PKC in the DNA binding domain modulates nuclear receptor localization. PKC was shown to phosphorylate highly conserved Ser-78 between the two zinc modules in the DNA binding domain of hepatocyte nuclear factor-4α and at a corresponding position in retinoic acid receptor-α, retinoid X receptor-α, and thyroid hormone receptor-β (60). Hepatocyte nuclear factor-4α Ser-78 is conserved through most of the nuclear receptor superfamily, including Ser-212 in human estrogen receptor-α. However, the corresponding residue in the AR subgroup of steroid receptors is alanine and is Ala-586 in human AR. PKC phosphorylation at Ser-78 in the DNA binding domain of hepatocyte nuclear factor-4α and other nuclear receptors regulates nuclear localization (60). Hepatocyte nuclear factor-4α-S78A bound DNA, but the S78D phosphomimetic did not. The AR phosphomimetic S578D retained greater transcriptional activity than AR-S578A but less than wild-type AR (data not shown), and like wild-type AR, the S578D mutant was distributed in both the cytoplasm and nucleus. AR-S578D might be expected to bind less Ku-70/80, which could interfere with the dynamic model of steroid receptor binding to DNA.
The results raise the possibility that phosphorylation by PKC at conserved serine residues in the first zinc module of the DNA binding domain regulates DNA binding for the entire nuclear receptor family. Phosphorylation within the DNA binding domain may be a common regulatory mechanism that controls nuclear-cytoplasmic shuttling and DNA binding required for differential gene regulation.
AR Nuclear-Cytoplasmic Shuttling and DNA Binding Specificity—EGF-dependent AR transcriptional activity mediated by PKC-dependent phosphorylation at AR Ser-578 is linked to AR nuclear-cytoplasmic shuttling. Based on results with an AR NH2-terminal, DNA binding, and hinge region fragment, where the S578A mutant was exclusively nuclear, EGF-dependent AR phosphorylation at Ser-515 and Ser-578 modulates AR nuclear retention. In addition, AR coimmunoprecipitates with Ku-70/80, a DNA binding protein complex that regulates DNA-PK activity and other transcription factors (61). The greater nuclear retention of AR-(507–660)-S578A compared with wild type was associated with increased interaction with Ku-70/80. PKC-mediated AR phosphorylation at Ser-578 appears to regulate the AR interaction with Ku-70/80 and modulate AR nuclear-cytoplasmic shuttling and DNA binding. Our studies are in agreement with previous evidence that steroid receptor phosphorylation is linked to nuclear-cytoplasmic shuttling (62).
In agreement with evidence that AR-S578A interacts to a greater extent with Ku-70/80, full-length AR-S578A did not activate MMTV-Luc in Ishikawa cells (data not shown) even though AR-S578A transcriptional activity was similar to wild type in Ishikawa cells using the PSA-Enh-Luc reporter, which lacks a negative response element (NRE) sequence present in the MMTV promoter-enhancer. Ku-80 represses GR transactivation through high affinity, sequence-specific binding to double-stranded negative response element 1 at -394 to -381 in the MMTV long terminal repeat (63, 64). Repression of GR transactivation of the MMTV promoter correlated with recruitment of Ku-70/80 to negative response element 1 (64, 65, 66).
Loss of AR-S578A transactivation of MMTV-Luc did not result from inhibition of AR binding to DNA. In the context of the AR NH2-terminal and DNA binding domain constitutively active fragment, AR-S578A retains wild-type activity with MMTV-Luc (data not shown). This is consistent with a previous study suggesting that Ku-70/80 interacts preferentially with the AR ligand binding domain (33) and provides further evidence that the effect of the S578A mutation is to increase AR interaction with Ku-70/80. Our studies suggest that a stronger interaction between AR and Ku-70/80 is associated with loss of AR activation of MMTV. In addition, the results support the concept that EGF-dependent phosphorylation at Ser-578 modulates AR transactivation through its interaction with Ku-70/80.
EGF-dependent phosphorylation sites Ser-515 and Ser-578 in AR differ from the GR DNA-PK phosphorylation site that was linked to GR association with Ku-70/80. Human GR hinge region DNA-PK phosphorylation site Ser-508 implicated in GR nuclear retention (63, 66, 67) is positioned close to human GR nuclear targeting residues 479–498 and is displaced in position by two residues to the corresponding human AR hinge residue Ser-650 near AR nuclear targeting residues 617–633 (22, 68). We and others have shown that a mutation at Ser-650 has relatively little effect on AR transcriptional activity (28, 35). However, human AR has a DNA-PK consensus phosphorylation site 656TQ657 in the hinge region, and inhibition of DNA-PK was reported to reduce AR phosphorylation and nuclear export (69). Given that AR interacts with Ku-70/80, an interaction that is enhanced by the S578A mutation, and that Ku-70/80 has additional functions independent of DNA-PK (65), Ku-70/80 may regulate AR independent of DNA-PK. Ku-70/80 is reported to have DNA helicase activity (70), which may more directly influence AR transcriptional activity.
AR Function in Castration-recurrent Prostate Cancer—AR is an important transcriptional activator in castration-recurrent prostate cancer growth that follows a period of remission in response to androgen deprivation therapy. The AR gene is amplified in approximately one-third of prostate cancers (71). AR somatic gene mutations in prostate cancer can increase AR transactivation by androgens and other ligands. However, although AR mutations are relatively common in established prostate cancer cell lines, they are infrequent in prostate cancer tissue specimens and cannot account for the high incidence of castration-recurrent prostate cancer growth after androgen deprivation therapy. AR somatic mutations in prostate cancer, such as AR-H874Y in the CWR22 human prostate xenograft and the derived CWR-R1 prostate cancer cell line used in the present study (6, 15), tend to increase AR sensitivity to ligand-dependent transactivation through more efficient recruitment of SRC/p160 coactivators by activation domain-2 in the ligand binding domain (72). In contrast, ligand-independent AR activation may be independent of activation domain-2 in the ligand binding domain (73), mediated instead by growth factor and mitogen downstream signaling mechanisms (74). This is supported by the inability of flutamide to inhibit castration-recurrent prostate cancer cell growth (75, 76).
Chromatin studies support AR function in cell proliferation in castration-recurrent prostate cancer in the absence of androgen (77–81). Adeno-associated viral siRNA knockdown of AR diminished both DHT and EGF-stimulated CWR-R1 prostate cancer cell growth. However, residual levels of AR in the AR-siRNA-scAAV-treated cells appear to be sufficient to mediate prostate cancer cell growth in response to the synergistic effects of DHT and EGF, a finding relevant to clinical prostate cancer since androgens are present in castration-recurrent prostate cancer tissue (8). In addition, EGF was shown to induce site-specific phosphorylation and monoubiquitinylation of AR coregulator MAGE-11 required to interact with AR and increase transcriptional activity in the absence and presence of androgen (16). Thus, growth factor and mitogen signaling through multiple mechanisms that involve AR may account for the relapse of prostate cancer growth.
Acknowledgments
We thank Brian J. Kennerley, John T. Minges, Andrew T. Hnat, and K. Michelle Cobb for excellent technical assistance and Ying Wang for helpful suggestions and the control pSilencer 1.0-U6 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants HD16910 (NICHD, to United States Public Health Service) and P01-CA77739 (NCI). This work was also supported by United States Department of Defense Grant PC060628. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AR, androgen receptor; EGF, epidermal growth factor; DHT, dihydrotestosterone; MAP, mitogen-activated protein; DNA-PK, DNA-dependent protein kinase; MMTV-Luc, mouse mammary tumor virus luciferase reporter vector; DMEM, Dulbecco's modified Eagle medium; FBS, fetal bovine serum; scAAV, self-complementary adeno-associated virus; CTR, control; PBS, phosphate-buffered saline; PKC, protein kinase C; GST, glutathione S-transferase; PSA, prostate-specific antigen; GR, glucocorticoid receptor; Mes, 2-(N-morpholino)ethanesulfonic acid; wt, wild type; siRNA, small interfering RNA.
References
- 1.Simental, J. A., Sar, M., Lane, M. V., French, F. S., and Wilson, E. M. (1991) J. Biol. Chem. 266 510-518 [PubMed] [Google Scholar]
- 2.Chawla, A., Repa, J. J., Evans, R. M., and Mangelsdorf, D. J. (2001) Science 294 1866-1870 [DOI] [PubMed] [Google Scholar]
- 3.Wilson, E. M., and French, F. S. (1976) J. Biol. Chem. 251 5620-5629 [PubMed] [Google Scholar]
- 4.Kemppainen, J. A., Lane, M. V., Sar, M., and Wilson, E. M. (1992) J. Biol. Chem. 267 968-974 [PubMed] [Google Scholar]
- 5.Scher, H. I., and Sawyers, C. L. (2005) J. Clin. Oncol. 23 8253-8261 [DOI] [PubMed] [Google Scholar]
- 6.Gregory, C. W., Johnson, R. T., Mohler, J. L., French, F. S., and Wilson, E. M. (2001) Cancer Res. 61 2892-2898 [PubMed] [Google Scholar]
- 7.Gregory, C. W., Hamil, K. G., Kim, D., Hall, S. H., Pretlow, T. G., Mohler, J. L., and French, F. S. (1998) Cancer Res. 58 5718-5724 [PubMed] [Google Scholar]
- 8.Mohler, J. L., Gregory, C. W., Ford, O. H., Kim, D., Weaver, C. M., Petrusz, P., Wilson, E. M., and French, F. S. (2004) Clin. Cancer Res. 10 440-448 [DOI] [PubMed] [Google Scholar]
- 9.Titus, M. A., Schell, M. J., Lih, F. B., Tomer, K. B., and Mohler, J. L. (2005) Clin. Cancer Res. 11 4653-4657 [DOI] [PubMed] [Google Scholar]
- 10.Culig, Z., Hobisch, A., Cronauer, M. V., Radmayr, C., Trapman, J., Hittmair, A., Bartsch, G., and Klocker, H. (1994) Cancer Res. 54 5474-5478 [PubMed] [Google Scholar]
- 11.Yeh, S., Lin, H. K., Kang, H. Y., Thin, T. H., Lin, M. F., and Chang, C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5458-5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda, T., Mawji, N. R., Bruchovsky, N., and Sadar, M. D. (2002) J. Biol. Chem. 277 38087-38094 [DOI] [PubMed] [Google Scholar]
- 13.Gregory, C. W., Whang, Y. E., McCall, W., Fei, X., Liu, Y., Ponguta, L. A., French, F. S., Wilson, E. M., and Earp, H. S. (2005) Clin. Cancer Res. 11 1704-1712 [DOI] [PubMed] [Google Scholar]
- 14.Gioeli, D., Ficarro, S. B., Kwiek, J. J., Aaronson, D., Hancock, M., Catling, A. D., White, F. M., Christian, R. E., Settlage, R. E., Shabanowitz, J., Hunt, D. F., and Weber, M. J. (2002) J. Biol. Chem. 277 29304-29314 [DOI] [PubMed] [Google Scholar]
- 15.Tan, J. A., Sharief, Y., Hamil, K. G., Gregory, C. W., Zang, D. Y., Sar, M., Gumerlock, P. H., DeVere White, R. W., Pretlow, T. G., Harris, S. E., Wilson, E. M., Mohler, J. L., and French, F. S. (1997) Mol. Endocrinol. 11 450-459 [DOI] [PubMed] [Google Scholar]
- 16.Bai, S., and Wilson, E. M. (2008) Mol. Cell. Biol. 28 1947-1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory, C. W., Fei, X., Ponguta, L. A., He, B., Bill, H. M., French, F. S., and Wilson, E. M. (2004) J. Biol. Chem. 279 7119-7130 [DOI] [PubMed] [Google Scholar]
- 18.Myslinski, E., Amé, J. C., Krol, A., and Carbon, P. (2001) Nucleic Acids Res. 29 2502-2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel, G. R., and Pederson, T. (1989) Nucleic Acids Res. 17 7371-7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao, X., Li, J., and Samulski, R. J. (1998) J. Virol. 72 2224-2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubahn, D. B., Joseph, D. R., Sar, M., Tan, J. A., Higgs, H. N., Larson, R. E., French, F. S., and Wilson, E. M. (1988) Mol. Endocrinol. 2 1265-1275 [DOI] [PubMed] [Google Scholar]
- 22.Zhou, Z. X., Sar, M., Simental, J. A., Lane, M. V., and Wilson, E. M. (1994) J. Biol. Chem. 269 13115-13123 [PubMed] [Google Scholar]
- 23.He, B., Kemppainen, J. A., and Wilson, E. M. (2000) J. Biol. Chem. 275 22986-22994 [DOI] [PubMed] [Google Scholar]
- 24.Huang, W., Shostak, Y., Tarr, P., Sawyers, C., and Carey, M. (1999) J. Biol. Chem. 274 25756-25768 [DOI] [PubMed] [Google Scholar]
- 25.He, B., Bowen, N. T., Minges, J. T., and Wilson, E. M. (2001) J. Biol. Chem. 276 42293-42301 [DOI] [PubMed] [Google Scholar]
- 26.He, B., Bai, S., Hnat, A. T., Kalman, R. I., Minges, J. T., Patterson, C., and Wilson, E. M. (2004) J. Biol. Chem. 279 30643-30653 [DOI] [PubMed] [Google Scholar]
- 27.Blom, N., Gammeltoft, S., and Brunak, S. (1999) J. Mol. Biol. 294 1351-1362 [DOI] [PubMed] [Google Scholar]
- 28.Zhou, Z. X., Kemppainen, J. A., and Wilson, E. M. (1995) Mol. Endocrinol. 9 605-615 [DOI] [PubMed] [Google Scholar]
- 29.Gioeli, D., Black, B. E., Gordon, V., Spencer, A., Kesler, C. T., Eblen, S. T., Paschal, B. M., and Weber, M. J. (2006) Mol. Endocrinol. 20 503-515 [DOI] [PubMed] [Google Scholar]
- 30.Carpenter, G., and Cohen, S. (1990) J. Biol. Chem. 265 7709-7712 [PubMed] [Google Scholar]
- 31.Sartorius, C. A., Takimoto, G. S., Richer, J. K., Tung, L., and Horwitz, K. B. (2000) J. Mol. Endocrinol. 24 165-182 [DOI] [PubMed] [Google Scholar]
- 32.Marschke, K. B., Tan, J. A., Kupfer, S. R., Wilson, E. M., and French, F. S. (1995) Endocrine 3 819-825 [DOI] [PubMed] [Google Scholar]
- 33.Mayeur, G. L., Kung, W. J., Martinez, A., Izumiya, C., Chen, D. J., and Kung, H. J. (2005) J. Biol. Chem. 280 10827-10833 [DOI] [PubMed] [Google Scholar]
- 34.Mellinghoff, I. K., Vivanco, I., Kwon, A., Tran, C., Wongvipat, J., and Sawyers, C. L. (2004) Cancer Cell 6 517-527 [DOI] [PubMed] [Google Scholar]
- 35.Wong, H. Y., Burghoorn, J. A., Van Leeuwen, M., De Ruiter, P. E., Schippers, E., Blok, L. J., Li, K. W., Dekker, H. L., De Jong, L., Trapman, J., Grootegoed, J. A., and Brinkmann, A. O. (2004) Biochem. J. 383 267-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taneja, S. S., Ha, S., Swenson, N. K., Huang, H. Y., Lee, P., Melamed, J., Shapiro, E., Garabedian, M. J., and Logan, S. K. (2005) J. Biol. Chem. 280 40916-40924 [DOI] [PubMed] [Google Scholar]
- 37.Lin, H. K., Yeh, S., Kang, H. Y., and Chang, C. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7200-7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, L., Xie, S., Jamaluddin, M. S., Altuwaijri, S., Ni, J., Kim, E., Chen, Y. T., Hu, Y. C., Wang, L., Chuang, K. H., Wu, C. T., and Chang, C. (2005) J. Biol. Chem. 280 33558-33565 [DOI] [PubMed] [Google Scholar]
- 39.Xin, L., Teitell, M. A., Lawson, D. A., Kwon, A., Mellinghoff, I. K., and Witte, O. N. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7789-7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan, N. P., Liu, Y., Majumder, S., Warren, M. R., Parker, C. E., Mohler, J. L., Earp, H. S., and Whang, Y. E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8438-8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo, Z., Dai, B., Jiang, T., Xu, K., Xie, Y., Kim, O., Nesheiwat, I., Kong, X., Melamed, J., Handratta, V. D., Njar, V. C., Brodie, A. M., Yu, L. R., Veenstra, T. D., Chen, H., and Qiu, Y. (2006) Cancer Cell 10 309-319 [DOI] [PubMed] [Google Scholar]
- 42.Jenster, G., van der Korput, H. A., Trapman, J., and Brinkmann, A. O. (1995) J. Biol. Chem. 270 7341-7346 [DOI] [PubMed] [Google Scholar]
- 43.Berrevoets, C. A., Doesburg, P., Steketee, K., Trapman, J., and Brinkmann, A. O. (1998) Mol. Endocrinol. 12 1172-1183 [DOI] [PubMed] [Google Scholar]
- 44.Abreu-Martin, M. T., Chari, A., Palladino, A. A., Craft, N. A., and Sawyers C. L. (1999) Mol. Cell. Biol. 19 5143-5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umesono, K., and Evans, R. M. (1989) Cell 57 1139-1146 [DOI] [PubMed] [Google Scholar]
- 46.Mader, S., Kumar, V., de Verneuil, H., and Chambon, P. (1989) Nature 338 271-274 [DOI] [PubMed] [Google Scholar]
- 47.Zilliacus, J., Carlstedt-Duke, J., Gustafsson, J. A., and Wright, A. P. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 4175-4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freedman, L. P. (1992) Endocr. Rev. 13 129-145 [DOI] [PubMed] [Google Scholar]
- 49.Zilliacus, J., Wright, A. P., Carlstedt-Duke, J., and Gustafsson, J. A. (1995) Mol. Endocrinol. 9 389-400 [DOI] [PubMed] [Google Scholar]
- 50.Schoenmakers, E., Alen, P., Verrijdt, G., Peeters, B., Verhoeven, G., Rombauts, W., and Claessens, F. (1999) Biochem. J. 341 515-521 [PMC free article] [PubMed] [Google Scholar]
- 51.Schauwaers, K., De Gendt, K., Saunders, P. T., Atanassova, N., Haelens, A., Callewaert, L., Moehren, U., Swinnen, J. V., Verhoeven, G., Verrijdt, G., and Claessens, F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4961-4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaffer, P. L., Jivan, A., Dollins, D. E., Claessens, F., and Gewirth, D. T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4758-4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luisi, B. F., Xu, W. X., Otwinowski, Z., Freedman, L. P., Yamamoto, K. R., and Sigler, P. B. (1991) Nature 352 497-505 [DOI] [PubMed] [Google Scholar]
- 54.Glass, C. K. (1994) Endocr. Rev. 15 391-407 [DOI] [PubMed] [Google Scholar]
- 55.Lundberg Giwercman, Y., Nikoshkov, A., Lindsten, K., Byström, B., Pousette, A., Knudtzon, J., Alm, J., and Wedell, A. (2000) Horm. Res. 53 83-88 [DOI] [PubMed] [Google Scholar]
- 56.Quigley, C. A., De Bellis, A., Marschke, K. B., El-Awady, M. K., Wilson, E. M., and French, F. S. (1995) Endocr. Rev. 16 271-321 [DOI] [PubMed] [Google Scholar]
- 57.McNally, J. G., Müller, W. G., Walker, D., Wolford, R., and Hager, G. L. (2000) Science 287 1262-1265 [DOI] [PubMed] [Google Scholar]
- 58.Stavreva, D. A., Müller, W. G., Hager, G. L., Smith, C. L., and McNally, J. G. (2004) Mol. Cell. Biol. 24 2682-2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klokk, T. I., Kurys, P., Elbi, C., Nagaich, A. K., Hendarwanto, A., Slagsvold, T., Chang, C. Y., Hager, G. L., and Saatcioglu, F. (2007) Mol. Cell. Biol. 27 1823-1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, K., Montana, V., Chellappa, K., Brelivet, Y., Moras, D., Maeda, Y., Parpura, V., Paschal, B. M., and Sladek, F. M. (2007) Mol. Endocrinol. 21 1297-1311 [DOI] [PubMed] [Google Scholar]
- 61.Shaw, P. E., Schröter, H., and Nordheim, A. (1989) Cell 56 563-572 [DOI] [PubMed] [Google Scholar]
- 62.Galigniana, M. D., Housley, P. R., DeFranco, D. B., and Pratt, W. B. (1999) J. Biol. Chem. 274 16222-16227 [DOI] [PubMed] [Google Scholar]
- 63.Gottlieb, T. M., and Jackson, S. P. (1993) Cell 72 131-142 [DOI] [PubMed] [Google Scholar]
- 64.Giffin, W., Torrance, H., Rodda, D. J., Préfontaine, G. G., Pope, L., and Hache, R. J. (1996) Nature 380 265-268 [DOI] [PubMed] [Google Scholar]
- 65.Giffin, W., Gong, W., Schild-Poulter, C., and Haché, R. J. (1999) Mol. Cell. Biol. 19 4065-4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giffin, W., Kwast-Welfeld, J., Rodda, D. J., Préfontaine, G. G., Traykova-Andonova, M., Zhang, Y., Weigel, N. L., Lefebvre, Y. A., and Haché, R. J. (1997) J. Biol. Chem. 272 5647-5658 [DOI] [PubMed] [Google Scholar]
- 67.Carrigan, A., Walther, R. F., Salem, H. A., Wu, D., Atlas, E., Lefebvre, Y. A., and Haché, R. J. (2007) J. Biol. Chem. 282 10963-10971 [DOI] [PubMed] [Google Scholar]
- 68.Picard, D., and Yamamoto, K. R. (1987) EMBO J. 6 3333-3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shank, L. C., Kelley, J. B., Gioeli, D., Yang, C. S., Spencer, A., Allison, L. A., and Paschal, B. M. (2008) J. Biol. Chem. 283 10568-10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tuteja, N., Tuteja, R., Ochem, A., Taneja, P., Huang, N. W., Simoncsits, A., Susic, S., Rahman, K., Marusic, L., Chen, J., Zhang, J., Wang, S., Pongor, S., and Falaschi, A. (1994) EMBO J. 13 4991-5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linja, M. J., Savinainen, K. J., Saramäki, O. R., Tammela, T. L., Vessella, R. L., and Visakorpi, T. (2001) Cancer Res. 61 3550-3555 [PubMed] [Google Scholar]
- 72.Askew, E. B., Gampe, R. T., Stanley, T. B., Faggart, J. L., and Wilson, E. M. (2007) J. Biol. Chem. 282 25801-25816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dehm, S. M., and Tindall, D. J. (2006) J. Biol. Chem. 281 27882-27893 [DOI] [PubMed] [Google Scholar]
- 74.Kaarbø, M., Klokk, T. I., and Saatcioglu, F. (2007) BioEssays 29 1227-1238 [DOI] [PubMed] [Google Scholar]
- 75.Scher, H. I., and Kelly, W. K. (1993) J. Clin. Oncol. 11 1566-1572 [DOI] [PubMed] [Google Scholar]
- 76.Ilagan, R., Zhang, L. J., Pottratz, J., Le, K., Salas, S., Iyer, M., Wu, L., Gambhir, S. S., and Carey, M. (2005) Mol. Cancer Ther. 4 1662-1669 [DOI] [PubMed] [Google Scholar]
- 77.Zhang, L., Johnson, M., Le, K. H., Sato, M., Ilagan, R., Iyer, M., Gambhir, S. S., Wu, L., and Carey, M. (2003) Cancer Res. 63 4552-4560 [PubMed] [Google Scholar]
- 78.Furutani, T., Takeyama, K., Koutoku, H., Ito, S., Taniguchi, N., Suzuki, E., Kudoh, M., Shibasaki, M., Shikama, H., and Kato, S. (2005) Biosci. Biotechnol. Biochem. 69 2236-2239 [DOI] [PubMed] [Google Scholar]
- 79.Li, T. H., Zhao, H., Peng, Y., Beliakoff, J., Brooks, J. D., and Sun, Z. (2007) Nucleic Acids Res. 35 2767-2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan, X., Li, T., Wang, H., Zhang, T., Barua, M., Borgesi, R. A., Bubley, G. J., Lu, M. L., and Balk, S. P. (2006) Am. J. Pathol. 169 682-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamy, F., Brondani, V., Spoerri, R., Rigo, S., Stamm, C., and Klimkait, T. (2003) Prostate Cancer Prostatic. Dis. 6 27-33 [DOI] [PubMed] [Google Scholar]