Abstract
We previously reported that glutamine was a major source of carbon for de novo fatty acid synthesis in a brown adipocyte cell line. The pathway for fatty acid synthesis from glutamine may follow either of two distinct pathways after it enters the citric acid cycle. The glutaminolysis pathway follows the citric acid cycle, whereas the reductive carboxylation pathway travels in reverse of the citric acid cycle from α-ketoglutarate to citrate. To quantify fluxes in these pathways we incubated brown adipocyte cells in [U-13C]glutamine or [5-13C]glutamine and analyzed the mass isotopomer distribution of key metabolites using models that fit the isotopomer distribution to fluxes. We also investigated inhibitors of NADP-dependent isocitrate dehydrogenase and mitochondrial citrate export. The results indicated that one third of glutamine entering the citric acid cycle travels to citrate via reductive carboxylation while the remainder is oxidized through succinate. The reductive carboxylation flux accounted for 90% of all flux of glutamine to lipid. The inhibitor studies were compatible with reductive carboxylation flux through mitochondrial isocitrate dehydrogenase. Total cell citrate and α-ketoglutarate were near isotopic equilibrium as expected if rapid cycling exists between these compounds involving the mitochondrial membrane NAD/NADP transhydrogenase. Taken together, these studies demonstrate a new role for glutamine as a lipogenic precursor and propose an alternative to the glutaminolysis pathway where flux of glutamine to lipogenic acetyl-CoA occurs via reductive carboxylation. These findings were enabled by a new modeling tool and software implementation (Metran) for global flux estimation.
Glutamine is utilized at a high rate by rapidly growing cells, including almost all cultured cell lines, where it is required at super-physiological concentrations of 2–4 mm for optimal growth (1). Recently, we evaluated the role of glutamine as a substrate for lipogenesis in a transformed wild type (WT)5 and IRS-1 knock-out brown adipose cell lines developed by Kahn and co-workers (2). Using isotopomer spectral analysis (ISA) we found that WT cells utilized glutamine for over 40% of their lipogenic acetyl-CoA (3). Glutamine was the largest precursor for lipogenic carbon, supplying more acetyl-CoA units than glucose or any other single source. This unexpected result led us to investigate glutamine metabolism in brown fat cell lines in more detail.
The pathway for glutamine utilization in rapidly dividing cell is generally described as “glutaminolysis” where glutamine enters the citric acid cycle (CAC) as α-ketoglutarate traversing the cycle to oxaloacetate and exits as pyruvate or aspartate (1, 4). Pyruvate then could be converted to acetyl-CoA and citrate in the lipogenic pathway. A major argument for glutaminolysis is the need for a large supply of anaplerotic substrates for rapidly growing cells, which explains the high rate of glutamine utilization (5). An alternative to glutaminolysis is the reductive carboxylation pathway where glutamine enters the CAC as α-ketoglutarate and is converted to citrate via isocitrate dehydrogenase (IDH) operating in reverse of the CAC direction. Evidence for reductive carboxylation in transformed cells is the finding that [5-14C]glutamine labels acetyl-CoA-derived carbons of lipids (6, 7). Additionally, reversal of IDH has been detected in perfused liver and heart as evidenced by the mass isotopomer distribution (MID) of [13C]citrate following perfusion with [13C]glutamine (8, 9). Although previous studies have detected label in citrate and lipids via the reductive carboxylation pathway, fluxes through the metabolic network connecting glutamine metabolism to lipids have not been quantified.
A complication in the analysis of the lipogenic pathway from glutamine is the role of the three mammalian IDH enzymes. The high mitochondrial NADH/NAD ratio will favor flux through the mitochondrial NAD-dependent isocitrate dehydrogenase toward α-ketoglutarate. Thus, the CAC enzyme is unlikely to be involved in lipogenesis from glutamine. Likewise, in rapidly growing cells the need for cytosolic NADPH for biosynthesis will favor flux of the cytosolic NADP-dependent enzyme toward α-ketoglutarate as shown recently in 3T3-L1 cells (10). This study also found that overexpression of cytosolic NADP-dependent enzyme in mice leads to increased lipogenesis in liver and fat, consistent with a reaction producing cytosolic NADPH. Thus, the candidate for flux in reverse of the CAC is the mitochondrial NADP-dependent enzyme (IDPm). Early studies established the feasibility of reductive carboxylation flux through this enzyme in hepatic models (11, 12). Flux of NADP-dependent enzyme toward α-ketoglutarate in vivo is supported by the affinity constant of the enzyme for CO2 (Km ∼ 1.6 mm), which is very close to physiological CO2 concentration (1.5 mm) and the affinity of the enzyme for NADPH, which is 100-fold higher than for NADP+ (13). Additionally, reductive carboxylation flux through IDPm has been hypothesized to play a key role in metabolic regulation of CAC cycle flux via a substrate cycle involving NAD/NADP transhydrogenase located at the inner mitochondrial membrane (14). However, there may be other roles for IDPm. Recent studies have provided evidence for flux through IDPm toward α-ketoglutarate generating NADPH in cellular defense against oxidative stress (15, 16). This role for IDPm may explain the high levels of the enzyme in non-lipogenic tissues, including heart and skeletal muscle. Although it is reasonable that the flux through the enzyme may differ in lipogenic and non-lipogenic tissues, it has been difficult to study such complicated pathways systematically. The major difficulty may be the absence of a comprehensive method for the analysis of metabolic fluxes through all of the relevant pathways.
Here, we have tested the hypothesis that flux of glutamine to lipogenic acetyl-CoA involves a reorganization of CAC metabolism including reductive carboxylation flux through IDPm. Studies were conducted with stable isotopes and mass spectrometry. The data analysis included our previously described ISA approach (3). However, we enhanced the analysis by employing a novel comprehensive modeling approach to quantify many additional fluxes from isotopic data. This powerful method provided quantitative flux estimates of the network describing glutamine metabolism in brown adipocytes. The details of this approach and its implementation with software (Metran) will be provided elsewhere.
EXPERIMENTAL PROCEDURES
Cell Culture and Adipocyte Differentiation—Brown adipocyte cells were cultured with the same procedure as in Yoo et al. (3). Briefly, WT brown preadipocytes were grown in 6-well plates to confluence in differentiation medium, DMEM containing 25 mm glucose, 4 mm glutamine, 20 nm insulin, 1 nm triiodothyronine, and 10% fetal bovine serum as well as 44 mm NaHCO3 (day 0). The medium was then replaced with fresh induction medium, which was differentiation medium plus 0.125 mm indomethacin, 0.25 mm isobutylmethylxanthine, and 5 μm dexamethasone. On days 2 and 4, the medium was replaced with fresh differentiation medium. To estimate metabolic fluxes, 13C-labeled glutamine and glucose were substituted for the unlabeled metabolites in the medium as described for specific experiments. To evaluate the effect of inhibitors of IDP, the differentiation medium was supplemented with oxalomalate or 2-methylisocitrate as indicated and incubated at 37 °C for 10 min, followed by addition of 13C-labeled substrates. After a 6-h incubation at 37 °C, the medium was removed and cells were extracted for organic/amino acids or lipids. Stock solution of 2-methylisocitrate was prepared from 2-methylisocitrate lactone (17) according to Dr. Wolfgang Buckel (Philipps-Universitaet, Marburg, Germany) as follows: threo-diastereoisomer of 2-methylisocitrate lactone (1:1 mixture of natural (2R,3S) and unnatural (2S,3R) diastereomers) in 1 m potassium hydroxide solution was incubated at 80 °C for 20 min and then neutralized to pH 7 with HCl. Biochemicals were obtained from Sigma. [5-13C]Glutamine, [U-13C5]glutamine, [U-13C]glucose, and [U-13C]aspartate were obtained from Cambridge Isotope Laboratories (Andover, MA). Tissue culture media were obtained from Invitrogen.
Isolation and Derivatization of Organic/Amino Acids and Lipids—For isolation of organic/amino acids, the procedure described in Fiehn et al. (18) was modified as follows. 0.7 ml of methanol and 25 μl of water were added to each well of the 6-well plate immediately after removal of the medium. After 15 min of incubation at room temperature, the methanol extract was mixed with 0.7 ml of water and 0.37 ml of chloroform in a 15-ml tube. Vigorous vortexing was followed by centrifugation at 3000 × g for 3 min. The chloroform layer was removed, and the methanol/water layer was centrifuged again at 3000 × g for 3 min. The methanol/water layer was then transferred to a glass vial and evaporated. The residue was dissolved in 70 μl of methoxyamine hydrochloride (20 mg/ml in pyridine) and vortexed. After 90 min of incubation at 37 °C, organic/amino acids were derivatized for gas chromatography/mass spectrometry analysis with 70 μl of N-methyl-N-tert-butyldimethylsilyl-trifluoroacetamide at 70 °C for 30 min. Isolation of lipids and derivatization of palmitate into methyl ester were performed as described previously (3).
Gas Chromatography/Mass Spectrometry Analysis—Samples labeled with [5-13C]glutamine and [U-13C]glutamine were analyzed for palmitate isotopomer distribution as previously reported (3). Labeling of palmitate was determined from relative intensities of ions at m/z 270–286. For the analysis of organic/amino acid derivatives, samples were injected into a Hewlett-Packard model 5890 Series II Gas Chromatograph equipped with a DB-XLB (60 mm × 0.25 mm id × 0.25 μm) capillary column (J&W Scientific, Folsom, CA), connected to an HP 5971 Mass Selective Detector operating under ionization by electron impact at 70 eV. Mass spectra were recorded in the range of 50–550 atomic mass units at 1.5 scans/s. The initial temperature of the column was 100 °C, held for 5 min, increased to 300 °C at 10 °C/min and held for 5 min. Labeling of the carbon atoms was determined from the mass isotopomer distributions of the following fragments: pyruvate (m/z 174, C1–C3), lactate (m/z 233, C2–C3; m/z 261, C1–C3), alanine (m/z 232, C2–C3; m/z 260, C1–C3), malate (m/z 233, C1–C4), aspartate (m/z 316, C2–C4; m/z 418, C1–C4), glutamate (m/z 330, C2–C5; m/z 432, C1–C5), glutamine (m/z 431, C1–C5), citrate (m/z 233, C1–C6). Measured intensities were corrected for baseline noise, and mass isotopomer distributions were obtained by integration (19). Mass isotopomer data are expressed as fractional abundances, i.e. for each fragment the sum of all mass isotopomers equals one.
Data Analysis—Fractional contribution of 13C-labeled carbon sources to fatty acid synthesis, D, and fractional new synthesis of fatty acids during time t, g(t), were estimated from the mass isotopomer distribution of palmitate based on the model of ISA as reported previously (3). Metabolic fluxes and their confidence intervals were determined by simultaneous fitting of external fluxes and mass isotopomer abundances of palmitate, pyruvate, lactate, alanine, malate, aspartate, glutamate, glutamine, and citrate to a detailed metabolic network model of adipocytes using the software Metran. Metran estimates fluxes by minimizing the difference between the observed and predicted measurements using an iterative least-squares regression procedure and isotopomer balancing (20). The objective of this routine is to evaluate a set of feasible fluxes that best account for the observed isotopomers and extracellular flux measurements. After metabolic fluxes were calculated, statistical analysis was automatically performed to obtain accurate standard deviations and 95% confidence intervals of fluxes by evaluating the sensitivity of the objective function with respect to fluxes as described by Antoniewicz et al. (21). Flux validation was accomplished by a statistical test for the goodness-of-fit based on the chi-square test for model adequacy. To ensure a global solution, flux estimation was repeated at least ten times starting with random initial values. All computations were performed with Matlab 6.5 and Matlab Optimization Toolbox (Mathworks Inc.).
To compare absolute fluxes in response to environmental perturbations, we determined, for each condition, the absolute palmitate synthesis flux. From ISA analysis we first obtained the fractional new synthesis of palmitate, i.e. g(6 h) value. Palmitate synthesis was then obtained as follows: palmitate synthesis flux (μmol/g-protein/h) = 25.7 × g(6 h). The factor 25.7 μmol/g-protein/h was determined from experiments where absolute uptake rates of glucose and glutamine were measured as reported previously (3).
RESULTS
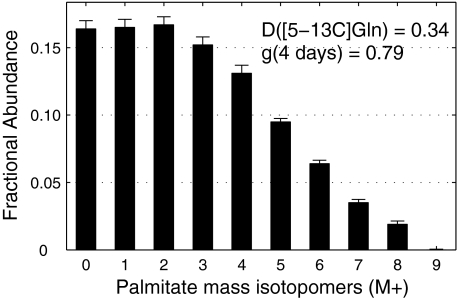
Quantifying the Reductive Carboxylation Pathway—When WT brown adipocyte cells were incubated from day 2 to day 6 of differentiation in medium containing 25 mm unlabeled glucose and 4 mm [5-13C]glutamine, the MID of palmitate was significantly labeled up to M8 (Fig. 1). ISA of palmitate indicated that glutamine provided 34% of the acetyl-CoA subunits for palmitate synthesis and that 79% of total palmitate was synthesized de novo over the 4-day period. The next issue was the relative flux through specific pathways from glutamine to lipogenic acetyl-CoA. De novo fatty acid synthesis, using glutamine as carbon source, may occur by either of two pathways: glutaminolysis, which converts C3 and C4 of glutamine to acetyl-CoA, and reductive carboxylation, which converts C4 and C5 of glutamine to acetyl-CoA. To evaluate the relative fluxes from glutamine to lipid via reductive carboxylation and glutaminolysis we compared the contribution of [5-13C]glutamine and [U-13C]glutamine to lipogenic acetyl-CoA using ISA. ISA estimated that [U-13C]glutamine contributed 38–40% of the lipogenic acetyl-CoA. The ISA D value for [U-13C]glutamine represents the sum of fluxes through the two pathways. Thus, we estimate that 85–90% of the glutamine flux to lipogenic acetyl-CoA was via reductive carboxylation in this cell model. These results indicate that one of the IDH is readily oxidizing NADPH or NADH to form citrate used for lipogenesis.
FIGURE 1.
Mass isotopomer distribution of palmitate isolated from WT brown adipocytes under incubation in medium containing 4 mm [5-13C]glutamine from day 2 to day 6 (mean ± S. E.; n = 3). Contribution of [5-13C]glutamine to palmitate synthesis (D value) and fractional new synthesis of palmitate (g value) determined by ISA.
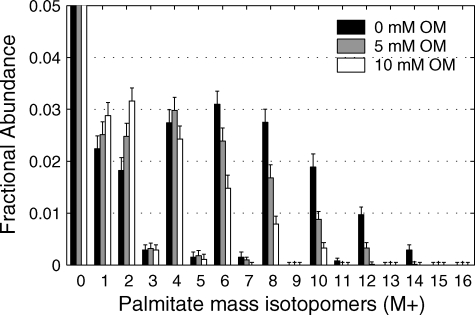
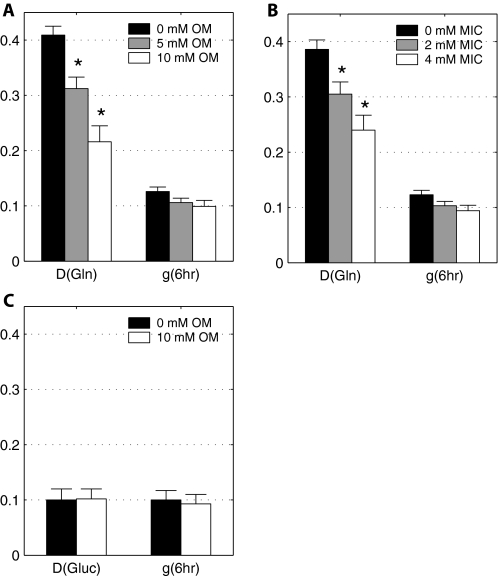
The Contribution of [U-13C]Glutamine to Palmitate Synthesis Affected by Inhibitors of NADP-isocitrate Dehydrogenase—Because of the large flux to lipid via the reductive carboxylation pathway, we quantified the effect of two reported inhibitors of NADP-dependent IDH, oxalomalate and 2-methylisocitrate, on lipogenesis. Cells were incubated in DMEM in the presence of 4 mm [U-13C]glutamine and either oxalomalate (5 or 10 mm) or 2-methylisocitrate (2 or 4 mm). The effect of oxalomalate on the MID of palmitate indicates that oxalomalate deceases flux of glutamine to lipid (Fig. 2). This is evident from the decrease in the higher mass isotopomers (M6-M12) with increasing oxalomalate. A similar result was observed with 2-methylisocitrate (supplemental Fig. S1). ISA analysis indicated that the major effect of these drugs was on the fractional contribution of glutamine to lipogenic acetyl-CoA (D value) rather than on the fractional synthesis rate, g(t) (Fig. 3, A and B). Systematic decrease in D values at higher concentration of oxalomalate or 2-methylisocitrate was evident, whereas g(6 h) values from conditions with three different concentrations of oxalomalate did not change significantly. In contrast, studies with [U-13C]glucose in DMEM demonstrated that these compounds did not affect the flux of glucose to lipogenic acetyl-CoA (Fig. 3C). Thus, the inhibitor studies supported a role for IDPm and/or cytosolic NADP-dependent enzyme in the reductive carboxylation reaction.
FIGURE 2.
Effect of oxalomalate on palmitate synthesis from [U-13C]glutamine. Shown is the mass isotopomer distribution of palmitate from WT brown adipocytes on day 4 under 6 h of incubation in medium containing 4 mm [U-13C]glutamine and 0, 5, or 10 mm oxalomalate (Error bars, mean ± S.E.; n = 3).
FIGURE 3.
Effect of increasing concentration of oxalomalate (A) and 2-methylisocitrate (B) on D(Gln) and g(6 h) values of palmitate synthesis from [U-13C]glutamine and from [U-13C]glucose (C). Asterisk (*) indicates significant difference compared with the control condition without inhibitor (p < 0.005). Error bars, mean ± S.E.
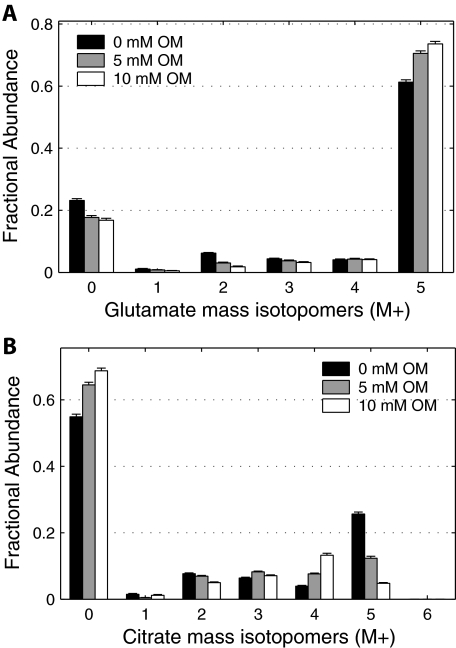
The effect of oxalomalate and 2-methylisocitrate on the contribution of glutamine to fatty acid synthesis was further investigated by analyzing the MID of other metabolites in the pathways of glutamine metabolism. The effect of oxalomalate was most pronounced in isotopomer distributions of glutamate and citrate, which are intermediates in the route of glutamine to fatty acid synthesis (Fig. 4). The M5 mass isotopomer of glutamate increased in fractional abundance upon treatment with higher concentration of oxalomalate (Figs. 4A). The relative intensity of M5 isotopomer of citrate, which can only originate from M5 isotopomer of α-ketoglutarate via reductive carboxylation, decreased systematically upon increase in oxalomalate concentration from 0 to 5 and 10 mm (Fig. 4B). The mass isotopomer abundances of malate, aspartate, pyruvate, and lactate from the same condition as above in the presence of 0, 5, or 10 mm oxalomalate are summarized in supplemental Fig. S2. These data indicate that the inhibitor has altered the labeling patterns of metabolites. The next step is to move beyond these data indicating changes in MID to estimates of fluxes in the metabolic network.
FIGURE 4.
Effect of increasing concentration of oxalomalate on labeling patterns of key intermediates of CAC. Mass isotopomer distributions of glutamate (A) and citrate (B) from WT brown adipocytes on day 4 under 6 h of incubation in medium containing 4 mm [U-13C]glutamine and 0, 5, or 10 mm oxalomalate. Data were corrected for natural isotope enrichments. Error bars, mean ± S.E.
To determine whether reductive carboxylation of α-ketoglutarate to citrate is a cytosolic or mitochondria process, the effects of benzenetricarboxylate (BTC), an inhibitor of the mitochondrial tricarboxylic acid transporter, were investigated (22). Brown adipocytes were incubated with BTC (5 mm) in DMEM containing [U-13C]glutamine for 6 honday 4. Flux of glutamine to lipogenic acetyl-CoA was estimated from the MID of palmitate with ISA. We found that BTC reduced the fractional contribution of glutamine to lipogenic acetyl-CoA (D value) from 0.36 ± 0.01 to 0.28 ± 0.02. Additionally, BTC reduced the fractional new synthesis, g(6 h) value, from 0.18 ± 0.01 to 0.09 ± 0.01. This corresponds to an overall 39% reduction in total flux of glutamine to lipid. Thus, these findings implicate the mitochondrial synthesis of citrate and reductive carboxylation flux through IDPm in de novo lipogenesis.
Isotopic Steady State Confirmed—To perform more detailed metabolic studies of brown adipocytes estimating CAC-related fluxes required verification that the entire pathway was in isotopic steady state after 6 h of isotope incubation. An implicit assumption in flux determination from labeling data is that intracellular metabolite pools are in metabolic and isotopic steady state, i.e. that the 13C-labeling patterns of intermediary metabolites do not change in time. Previous work has estimated that substrate utilization was linear over time, consistent with metabolic steady state (3). However, these studies focused only on palmitate synthesis and were carried our for long incubations times, up to 72 h. To carry the modeling study forward we evaluated CAC intermediates and related amino acids within 6 h of incubation in 4 mm [U-13C]glutamine. The isotopomer distribution of eight metabolites was measured at 2, 4, and 6 h. The MID of eight key metabolites, including citrate and malate, was constant at each time point (supplemental Fig. S3). Therefore, the metabolic system was found to be at isotopic and metabolic steady state. Next, the metabolic flux of glutamine carbon to fatty acid synthesis was examined in detail with a metabolic network model constructed from metabolic reactions of glycolysis, fatty acid synthesis, malic enzyme, pyruvate carboxylase, and the CAC. The model included reversible reactions between α-ketoglutarate and citrate; malate and fumarate; malate and oxaloacetate; and glutamine, glutamate, and α-ketoglutarate. The model was implemented with a software tool, Metran, based on the concepts described elsewhere (19, 20).
Flux Estimation and Assessment of Goodness-of-fit—A steady state metabolic network consisting of 17 independent metabolic fluxes was constructed for WT cells. The network was analyzed with data on day 4 of differentiation in DMEM with 4 mm [U-13C]glutamine and 25 mm unlabeled glucose in medium supplemented with 0, 5, 10 mm oxalomalate or 0, 2, 4 mm 2-methylisocitrate. 17 fluxes for the six conditions were estimated by fitting 60 independent mass isotopomer abundances, hence there were 43 (= 60–17) redundant measurements for each condition. In all cases the minimized sum of squared residuals was lower than the threshold value of 64 (chi-square test for model adequacy at 95% confidence level), indicating that all flux models were statistically acceptable. Comprehensive data sets of the estimated fluxes in the presence of either 0, 5, 10 mm oxalomalate or 0, 2, 4 mm 2-methylisocitrate are summarized in Table 1.
TABLE 1.
Effect of oxalomalate and 2-methylisocitrate on the flux phenotype in WT brown adipocytes
Metabolic fluxes and their standard deviations were quantified by fitting MID of palmitate, pyruvate, lactate, alanine, malate, aspartate, glutamine, glutamate, and citrate to the metabolic network model. Shown are the estimated net fluxes of selected reactions (μmol/g-protein/h), fractional equilibrium of IDH and fumarase, and ISA model parameters (best fit ± S.D.).
| Reactions | Oxalomalate | 2-Methylisocitrate | |||||
|---|---|---|---|---|---|---|---|
| Pathway | 0 mm | 5 mm | 10 mm | 0 mm | 2 mm | 4 mm | |
| Glutamine uptake | Glutamine → AKG | 13.0 ± 0.8 | 10.1 ± 0.9 | 9.6 ± 2.1 | 12.2 ± 0.8 | 10.6 ± 1.0 | 8.0 ± 1.0 |
| Reductive carboxylationa | AKG + CO2 → Cit | 4.4 ± 0.8 | 0.9 ± 0.6 | 0.3 ± 0.5 | 3.6 ± 0.9 | 0.1 ± 1.0 | -2.1 ± 1.0 |
| α-Ketoglutarate to malate flux | AKG → Suc → Mal | 8.6 ± 1.1 | 9.1 ± 1.0 | 9.3 ± 1.6 | 8.5 ± 1.2 | 10.5 ± 1.6 | 10.2 ± 1.5 |
| Malic enzyme | Mal → Pyr + CO2 | 59.7 ± 9.2 | 28.4 ± 4.0 | 14.2 ± 2.5 | 52.9 ± 8.1 | 52.2 ± 13.1 | 31.1 ± 7.9 |
| Pyruvate carboxylase | Pyr + CO2 → OAC | 46.5 ± 9.1 | 18.3 ± 4.0 | 6.0 ± 3.1 | 40.5 ± 8.2 | 41.7 ± 12.6 | 22.8 ± 7.7 |
| Pyruvate dehydr. and citrate synth. | Pyr → AcCoA (+OAC) → Cit | 21.6 ± 1.1 | 20.9 ± 0.9 | 20.8 ± 0.9 | 21.8 ± 1.1 | 21.3 ± 1.3 | 21.4 ± 1.3 |
| Citrate lyase | Cit → OAC + AcCoA | 25.9 ± 0.8 | 21.8 ± 0.8 | 20.4 ± 0.8 | 25.4 ± 0.8 | 21.2 ± 0.8 | 19.4 ± 0.8 |
| Palmitate synthesis | 8 AcCoA → palmitate | 3.2 ± 0.1 | 2.7 ± 0.1 | 2.6 ± 0.1 | 3.2 ± 0.1 | 2.7 ± 0.1 | 2.4 ± 0.1 |
| Fractional equilibrium IDHb | Cit ↔ AKG + CO2 | 0.93 ± 0.04 | 0.92 ± 0.04 | 0.94 ± 0.03 | 0.93 ± 0.04 | 1.00 ± 0.01 | 0.92 ± 0.04 |
| Fractional equilibrium fumaraseb | Fum ↔ Mal | 0.97 ± 0.02 | 0.89 ± 0.05 | 1.00 ± 0.01 | 0.97 ± 0.02 | 1.00 ± 0.01 | 0.87 ± 0.07 |
| Fractional contribution glutamine | D(Gln) from ISA model | 0.41 ± 0.02 | 0.31 ± 0.02 | 0.22 ± 0.03 | 0.39 ± 0.02 | 0.31 ± 0.02 | 0.24 ± 0.03 |
| Fractional new synthesis palmitate | g(6h) from ISA model | 0.13 ± 0.01 | 0.11 ± 0.01 | 0.10 ± 0.01 | 0.12 ± 0.01 | 0.10 ± 0.01 | 0.09 ± 0.01 |
Negative flux of reductive carboxylation indicates a net flux in the direction from citrate to α-ketoglutarate.
Fractional equilibrium of reversible reactions is defined as feq = vexch/(|vnet|+vexch).
Comparison of Individual Carbon Fluxes—Flux of glutamine to α-ketoglutarate was based on glutamine uptake. In WT cells under the control condition without inhibitors, this flux of 13.0 μmol/g-protein/h was then divided into net flux of 8.6 ± 1.1 going to malate and a net flux of 4.4 ± 0.8 going to citrate via reductive carboxylation. In addition to the estimated net fluxes, mass isotopomer data also allowed quantification of the exchange of carbon between α-ketoglutarate and citrate. We observed almost complete equilibrium of the two pools (93 ± 4%). We also observed a large anaplerotic flux of glutamine into CAC at α-ketoglutarate. We estimated relative anaplerosis as the ratio of this flux to the sum of citrate synthase and glutamine to α-ketoglutarate. Using data for control conditions (Table 1), we estimated relative anaplerosis at 37% of CAC flux. This is comparable with a 38–40% estimate of glutamine anaplerosis obtained from ISA. Thus, we have confirmed the ISA results with the more complete Metran model.
In the presence of 5 and 10 mm oxalomalate, the net flux of reductive carboxylation was linearly reduced to 0.9 ± 0.6 and –0.3 ± 0.5 (i.e. negative flux corresponding to net flux from citrate to α-ketoglutarate), respectively, consistent with specific inhibition of IDP. The net flux of α-ketoglutarate to malate did not change significantly, while glutamine uptake decreased only slightly to 10.1 ± 0.9 and 9.6 ± 2.1 for 5 and 10 mm oxalomalate, respectively. Furthermore, the flux from malate to pyruvate catalyzed by malic enzyme was linearly decreased from 60 ± 9 to 28 ± 4 and 14 ± 3 upon increasing concentration of oxalomalate. The inhibitors did not affect citrate synthase flux. Treatment of WT cells with 0, 2, or 4 mm 2-methylisocitrate gave rise to parallel patterns for the fluxes of α-ketoglutarate to citrate as in the experiments with oxalomalate. The decrease in glutamine flux was similar (from 12.2 ± 0.8 to 10.6 ± 1.0 and 8.0 ± 1.0), resulting in a slight increase in the net flux from α-ketoglutarate to malate from 8.5 ± 1.2 to 10.5 ± 1.6 and 10.2 ± 1.5. In contrast with the oxalomalate, addition of 2-methylisocitrate had no effect on pyruvate cycling fluxes, i.e. malic enzyme and pyruvate carboxylation. Taken together, the results shown in Table 1 demonstrate decreased flux from α-ketoglutarate to citrate with the inhibitors. In addition, they detected an active pyruvate cycle between malate/oxaloacetate and pyruvate.
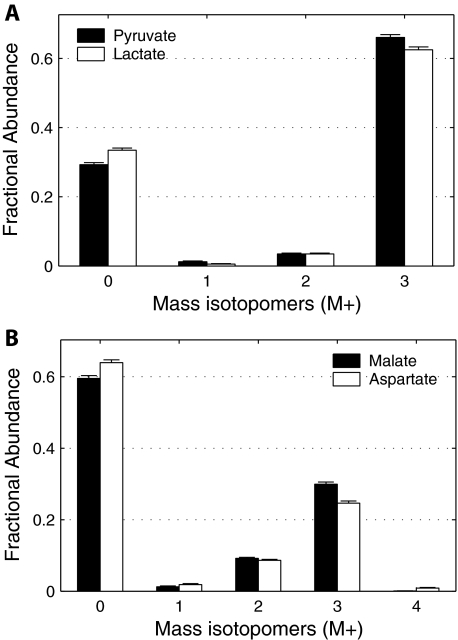
Independent Validation of Pyruvate Cycling—To provide additional support for the observed pyruvate cycling, a parallel experiment with 25 mm [U-13C]glucose and 4 mm unlabeled glutamine was performed on day 4. The MID of pyruvate, lactate (Fig. 5A), and malate, aspartate (Fig. 5B), showed conspicuous peaks of M3 isotopomers, consistent with large glycolytic flux and high level of pyruvate carboxylation, via either pyruvate carboxylase or malic enzyme. An alternative form of the pyruvate cycle involving phosphoenolpyruvate was rejected because there was no enrichment of phosphoenolpyruvate from [U-13C]glutamine. Thus, the [U-13C]glucose experiment confirmed the pyruvate carboxylation flux predicted by the Metran model of [U-13C]glutamine metabolism (Table 1). The [U-13C]glucose experiment provided further insight into the fluxes of the network. The relatively large abundance of pyruvate M0 indicated that glucose from the medium was not the sole source of pyruvate, consistent with rapid pyruvate cycling. The low abundance of M1 and M2 mass isotopomers in pyruvate indicated that flux through the oxidative branch of the pentose phosphate pathway was low relative to the high glycolytic rate of these transformed cells.
FIGURE 5.
Flux from [U-13C]glucose to key intermediates of glycolysis and CAC. Mass isotopomer distributions of pyruvate and lactate (A) and malate and aspartate (B) from WT brown adipocytes on day 4 under 6 h of incubation in medium containing 25 mm [U-13C]glucose and 4 mm unlabeled glutamine. Data were corrected for natural isotope enrichments. Error bars, mean ± S.E.
DISCUSSION
Our previous report demonstrated that the contribution of glutamine to lipogenic acetyl-CoA was greater than that of glucose for wild-type brown adipocytes (3). This result was unexpected because glutamine is not generally viewed as a lipogenic precursor. Glutamine metabolism in transformed cells has been described as glutaminolysis, an oxidative pathway for energy production in the CAC and for generating NADPH through subsequent activity of malic enzyme (23). The pyruvate produced by glutaminolysis could be either converted to lactate and exported or converted to acetyl-CoA for utilization in fatty acid synthesis. In brown adipocytes, conversion of glutamine to lipids is supported by findings of high activity of glutaminase in vitro (24) and in vivo (25). Also, in vivo activity of NADP-dependent IDH was shown to be significantly higher in brown adipose tissue than in white adipose tissue (26). The studies cited above supporting glutaminolysis did not employ isotopes to estimate fluxes. Thus, the studies performed here provide a more detailed quantitative analysis of glutamine metabolism in brown adipocytes. We found that most of the glutamine carbon entering the CAC follows the oxidative path as predicted by glutaminolysis. However, approximately one third of the glutamine molecules travel to citrate via reductive carboxylation through IDH in reverse of the CAC direction. This reductive carboxylation flux is not simply isotopic exchange. It contributes 30–40% of the acetyl units for de novo lipogenesis. Thus, we found significant flux via an alternative to the glutaminolysis pathway in which glutamine supplies lipogenic carbon via reductive carboxylation. These results were obtained at 4.0 mm extracellular glutamine, which is greater than physiological plasma glutamine (∼0.6 mm). It is possible that the extracellular glutamine concentration influences the extent of reductive carboxylation. Thus, our findings are most relevant for understanding the metabolic characteristics of cells requiring high levels of glutamine for optimal growth.
The metabolic fluxes quantified in this study used both ISA- and Metran-based models. ISA was used to quantify lipogenesis from the two glutamine tracers over 48 h, and Metran was used for detailed fluxes of CAC and related pathways. Both ISA and Metran models use nonlinear regression methods to fit the data and found similar values for reductive carboxylation flux of glutamine to lipogenic acetyl-CoA. However, there are important differences. The ISA model used here requires only the MID of a single fatty acid, such as palmitate, as input whereas Metran models employ the MID of all of the metabolic intermediates (and selected fragments) detected in the relevant pathways. The ISA model is limited to estimating only two parameters, D and g(t), related to lipogenesis. In contrast, Metran models are flexible and estimate a large number of parameters. In a practical sense, the Metran provides much more information about a metabolic network from an isotopic study simply by collecting MID data on additional compounds present in the cell or tissue extract. This added information holds much promise as biochemists seek to integrate genomic, transcriptional, and proteomic data with network metabolic flux data.
A major finding in this study is the role of reductive carboxylation flux in lipogenesis. This flux was accompanied by near equilibration between total cellular α-ketoglutarate and citrate (Table 1). Thus, the reductive carboxylation reaction carries a high flux relative to the CAC flux. This near equilibrium of citrate and α-ketoglutarate has implications for the control of flux in this pathway. Studies with inhibitors supported a role for IDPm in reductive carboxylation of α-ketoglutarate to isocitrate. To permit IDPm to function as proposed here requires a continuous source of mitochondrial NADPH. The rapid cycling between α-ketoglutarate and citrate is consistent with proposed actions of a substrate cycle involving IDPm and the NAD/NADP transhydrogenase located at the inner mitochondrial membrane (14). The cycle begins at the inner mitochondrial membrane where H+-transhydrogenase mediates the transfer of reducing equivalents from NADH to NADPH. NADPH would next be consumed by reductive carboxylation where IDPm yields isocitrate and NADP. Citrate is then formed via the near equilibrium aconitase reactions. Mitochondrial citrate, derived from the sum of this reductive carboxylation path and citrate synthase, has three possible fates (1). It may be oxidized via mitochondrial NAD-dependent isocitrate dehydrogenase (2). It may exit the mitochondria on the citrate carrier and be converted to α-ketoglutarate via cytosolic NADP-dependent enzyme with generation of cytosolic NADPH (3). It may exit the mitochondria, encounter ATP citrate lyase in the cytosol, and form lipogenic acetyl-CoA plus oxaloacetate. Paths 1 and 2 contribute to the observed near equilibration between total cellular α-ketoglutarate and citrate (Table 1). Path 3 yields the large contribution of glutamine carbon to lipogenic acetyl-CoA (Fig. 3). This cycle is favored by conditions likely to occur in transformed cells, high demand for NADPH-requiring biosynthesis and low requirement for mitochondrial ATP synthesis. These conditions apply because differentiating brown adipocytes require NADPH for de novo lipogenesis and because the transformed cells obtain much of their ATP from cytosolic glycolysis. In summary, the tracer-based analyses conducted here provide a novel view of metabolic fluxes in transformed brown adipocytes that includes a unique role of glutamine as a precursor for lipogenesis.
Supplementary Material
Acknowledgments
The brown preadipocyte cell line was generously provided by Dr. C. R. Kahn (Joslin Diabetes Center, Boston, MA). 2-Methylisocitrate lactone was a gift from Dr. Wolfgang Buckel (Philipps-Universitaet, Marburg, Germany).
This work was supported, in whole or in part, by National Institutes of Health Grant DK075850. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: WT, wild type; ISA, isotopomer spectral analysis; CAC, citric acid cycle; IDH, isocitrate dehydrogenase; MID, mass isotopomer distribution; IDPm, mitochondrial NADP-dependent isocitrate dehydrogenase; DMEM, Dulbecco's modified Eagle's medium.
References
- 1.Newsholme, E. A., Crabtree, B., and Ardawi, M. S. (1985) Biosci. Rep. 5 393–400 [DOI] [PubMed] [Google Scholar]
- 2.Fasshauer, M., Klein, J., Kriauciunas, K. M., Ueki, K., Benito, M., and Kahn, C. R. (2001) Mol. Cell. Biol. 21 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoo, H., Stephanopoulos, G., and Kelleher, J. K. (2004) J. Lipid Res. 45 1324–1332 [DOI] [PubMed] [Google Scholar]
- 4.Mazurek, S., and Eigenbrodt, E. (2003) Anticancer Res. 23 1149–1154 [PubMed] [Google Scholar]
- 5.Board, M., Humm, S., and Newsholme, E. A. (1990) Biochem. J. 265 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holleran, A. L., Briscoe, D. A., Fiskum, G., and Kelleher, J. K. (1995) Mol. Cell. Biochem. 152 95–101 [DOI] [PubMed] [Google Scholar]
- 7.D'Adamo, A. F., and Haft, D. E. (1989) J. Biol. Chem. 240 613–617 [PubMed] [Google Scholar]
- 8.Des Rosiers, C., Di Donato, L., Comte, B., Laplante, A., Marcoux, C., David, F., Fernandez, C. A., and Brunengraber, H. (1995) J. Biol. Chem. 270 10027–10036 [DOI] [PubMed] [Google Scholar]
- 9.Comte, B., Vincent, G., Bouchard, B., Benderdour, M., and Des Rosiers, C. (2002) Am. J. Physiol. 283 H1505–H1514 [DOI] [PubMed] [Google Scholar]
- 10.Koh, H. J., Lee, S. M., Son, B. G., Lee, S. H., Ryoo, Z. Y., Chang, K. T., Park, J. W., Park, D. C., Song, B. J., Veech, R. L., Song, H., and Huh, T. L. (2004) J. Biol. Chem. 279 39968–39974 [DOI] [PubMed] [Google Scholar]
- 11.Dalziel, K., and Londesborough, J. C. (1968) Biochem. J. 110 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wanders, R. J., van Doorn, H. E., and Tager, J. M. (1981) Eur. J. Biochem. 116 609–614 [DOI] [PubMed] [Google Scholar]
- 13.Reynolds, C. H., Kuchel, P. W., and Dalziel, K. (1978) Biochem. J. 171 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sazanov, L. A., and Jackson, J. B. (1994) FEBS Lett. 344 109–116 [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. J., Kang, B. S., and Park, J. W. (2005) Free Radic. Res. 39 441–448 [DOI] [PubMed] [Google Scholar]
- 16.Jo, S. H., Son, M. K., Koh, H. J., Lee, S. M., Song, I. H., Kim, Y. O., Lee, Y. S., Jeong, K. S., Kim, W. B., Park, J. W., Song, B. J., and Huh, T. L. (2001) J. Biol. Chem. 276 16168–16176 [DOI] [PubMed] [Google Scholar]
- 17.Brock, M., Maerker, C., Schutz, A., Volker, U., and Buckel, W. (2002) Eur. J. Biochem. 269 6184–6194 [DOI] [PubMed] [Google Scholar]
- 18.Fiehn, O., Kopka, J., Trethewey, R. N., and Willmitzer, L. (2000) Anal. Chem. 72 3573–3580 [DOI] [PubMed] [Google Scholar]
- 19.Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2007) Anal. Chem. 79 7554–7559 [DOI] [PubMed] [Google Scholar]
- 20.Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2007) Metab. Eng. 9 68–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoniewicz, M. R., Kelleher, J. K., and Stephanopoulos, G. (2006) Metab. Eng. 8 324–337 [DOI] [PubMed] [Google Scholar]
- 22.Rao, S., and Coleman, P. S. (1989) Exp. Cell Res. 180 341–352 [DOI] [PubMed] [Google Scholar]
- 23.McKeehan, W. L. (1982) Cell Biol. Int. Rep. 6 635–650 [DOI] [PubMed] [Google Scholar]
- 24.Kowalchuk, J. M., Curi, R., and Newsholme, E. A. (1988) Biochem. J. 249 705–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Almeida, A. F., Curi, R., Newsholme, P., and Newsholme, E. A. (1989) Int. J. Biochem. 21 937–940 [DOI] [PubMed] [Google Scholar]
- 26.Kochan, Z., Bukato, G., and Swierczynski, J. (1993) Biochem. Pharmacol. 46 1501–1506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.