Abstract
The mitogen-activated protein kinase JNK1 suppresses interleukin-3 withdrawal-induced cell death through phosphorylation of the BH3-only pro-apoptotic Bcl-2 family protein Bad at Thr-201. It is unknown whether JNK1 regulates glycolysis, an important metabolic process that is involved in cell survival, and if so, whether the regulation depends on Thr-201 phosphorylation of Bad. Here we report that phosphorylation of Bad by JNK1 is required for glycolysis through activation of phosphofructokinase-1 (PFK-1), one of the key enzymes that catalyze glycolysis. Genetic disruption of Jnk1 alleles or silencing of Jnk1 by small interfering RNA abrogates glycolysis induced by growth/survival factors such as serum or interleukin-3. Proteomic analysis identifies PFK-1 as a novel Bad-associated protein. Although the interaction between PFK-1 and Bad is independent of JNK1, Thr-201 phosphorylation of Bad by JNK1 is required for PFK-1 activation. Thus, our results provide a novel molecular mechanism by which JNK1 promotes glycolysis for cell survival.
The c-Jun N-terminal protein kinase (JNK;2 also known as stress-activated protein kinase, SAPK) (1), a subfamily of the mitogen-activated protein kinase (MAPK) superfamily, has two ubiquitously expressed isoforms, JNK1 and JNK2, and a tissue-specific isoform JNK3 with different splicing variant (2–4). Between JNK1 and JNK2, JNK1 is the main c-Jun kinase (5–7). JNK is activated by a variety of extracellular stimuli, including growth factors, cytokines, oncogenes, and environmental stresses through a MAP kinase module, i.e. MAP3K → MAP2K → MAPK (4). Two MAP2Ks (JNKK1/MKK4/SEK1 and JNKK2/MKK7) for JNK have been identified (8–11). Several MAP3Ks, such as MEKK1 (12), MEKK2 (13), apoptosis signal-regulating kinase (ASK1) (14), mixed lineage kinase (MLK) (15), TGFβ-activated kinase (TAK1) (16), tumor repression locus-2 (Tpl-2) (17), dual leucine zipper-bearing kinase (DLK) (18), and thousand and one amino acid kinase (Tao) (19), have been reported to act as MAP3Ks for JNK. Activation of JNK is also regulated by scaffold proteins such as JIP, β-arrestin, JSAP1 (20–22), and protein phosphatases such as mitogen-activated protein phosphatases (23). In addition, the transcription factor NF-κB can negatively or positively regulate JNK activation, depending on the nature of the stimulus (7, 24–29). JNK plays a critical role in many cellular activities, from growth control to programmed cell death, and deregulation of JNK activity has been implicated in many human diseases and certain types of cancer (28, 30–33).
Bad is a BH3-only pro-apoptotic Bcl-2 family protein, having two splicing forms Bads and BadL (34). Bad plays a critical role in the cross-talk between the growth/survival factor signaling pathway and the intrinsic death machinery (35). The pro-apoptotic activity of Bad is regulated by extracellular stimuli via post-translational modifications including phosphorylation (35–38). In response to the stimulation of growth/survival factors such as interleukin-3 (IL-3) and insulin-like growth factor-I (IGF-I), Bad is phosphorylated at the “regulatory serines” (Ser-112, Ser-136, and Ser-155) by a group of protein kinases such as cAMP-dependent protein kinase, Akt, Rsk, and p70S6K (37–40). The hyperphosphorylated form of Bad is inactive in terms of its pro-apoptotic function as it is sequestrated in the cytoplasm via its interaction with 14-3-3, a group of cytoplasmic anchorage proteins that interact with proteins through specific phospho-serine/threonine motifs (35). Withdrawal of growth/survival factors leads to dephosphorylation of Bad, which translocates to the mitochondria, where it binds prosurvival Bcl-2 family proteins such as Bcl-XL and thereby allows the oligomerization of Bax and Bak (41). This results in increased mitochondria permeability, release of cytochrome c, caspase activation, and ultimately, apoptotic cell death. Recently, it has been shown that JNK suppresses IL-3 withdrawal-induced apoptosis in hematopoietic pro-B FL-5.12 cells via phosphorylation of Bad at Thr-201 (42). Phosphorylation of Bad by JNK at Thr-201 is also required for its interaction with 14-3-3 (42).
In addition to regulation of apoptosis, Bad is involved in regulation of glycolysis, a metabolic process that is required for cell survival (43). It has been shown that inactivation of Bad by phosphorylation at the regulatory serines (Ser-112, Ser-136, and Ser-155) is required for the activity of mitochondrial glucokinase, one of the key enzymes that are responsible for glycolysis and ATP production (43). Thus, inactivation of Bad may be critical for suppression of apoptosis and maintaining of glycolysis. Here we report that JNK1-mediated phosphorylation of Bad at Thr-201 is required for maintaining normal glycolysis for cell survival through activation of phosphofructokinase-1 (PFK-1), which is another key enzyme in glycolysis.
MATERIALS AND METHODS
Reagents—Antibodies against Bad (C-20), Bcl-XL (H5), PFK-1(K-15), and Xpress (M-21) were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against JNK (antibody 333 for immunoprecipitation and 666 for immunoblotting) were from Pharmingen. Anti-M2 antibody (F1804), triose phosphate isomerase, aldolase, α-glycerophosphate, dehydrogenase, and fructose-6-phosphate were from Sigma. The specific JNK inhibitor SP600125 was synthesized and purified (29). [γ-32P]ATP and [35S]methionine were from PerkinElmer Life Sciences. [5-3H]glucose was from GE Healthcare. Lipofectamine 2000 and the colloidal blue staining kit were from Invitrogen.
Plasmids—pGEX-KG murine Bad was a gift from Dr. Stanley Korsmeyer. T201V mutation was introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), pCDNA3.1-Hygro (+) vectors encoding wild-type (WT) M2-mBad, and M2-mBad(T201V) were generated by the addition of a N-terminal M2 tag to mBad and confirmed by DNA sequencing. Murine PFK-1 clone (clone number: MGC-6544) was from ATCC (Manassas, VA) and subcloned into pCDNA3.1-HisB vector.
Cell Culture and Transfection—IL-3-dependent hematopoietic cell line FL5.12, FL5.12/Bcl-XL, FL5.12 Bcl-XL/M2-Bad were generated and cultured as described previously (42). In IL-3 deprivation experiments, FL5.12 Bcl-XL/Bad cells were washed with phosphate-buffered saline three times and recultured in media lacking IL-3. Jnk1–/– mouse embryonic fibroblasts(MEFs) were prepared from Jnk1 null mice, a gift from Dr. Michael Karin (44), and immortalized as described (26). Bad–/– fibroblasts (a gift from Dr. Stanley Korsmeyer) and COS-1 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, 100 units of penicillin/ml, and 100 μg of streptomycin/ml. Transfection was performed using ExGene 500 (MBI Fermentas), according to the manufacturer's protocol, or by electroporation with a Bio-Rad Gene Pulser at 960 microfarads/250 V.
Mass Spectrometry—Bad-associated proteins were immunoprecipitated from 8 mg of cytosolic extracts of FL5.12/Bcl-XL cells or FL5.12 Bcl-XL/Bad stable cells. Proteins were separated by SDS-PAGE. Gels were stained with colloidal Coomassie Blue, and protein bands of interest were excised and in-gel digested. The resultant tryptic peptides were identified as described previously (45).
Protein Kinase Assays and Immunoblotting—Immune complex kinase assays and glutathione S-transferase (GST)-c-Jun pull down kinase assays were performed and quantitated as described previously (8). Briefly, kinase assays were carried out for 60 min at 30 °C in 20 mm HEPES (pH 7.6), 20 mm MgCl2, 1 mm dithiothreitol, 10 μm nonradioactive ATP, and 10 μCi of [γ-32P]ATP. WT or mutant GST-Bad or GST-c-Jun (2–4 μg) was used as substrate. Reactions were terminated by the addition of 4× SDS sample buffer and heated at 95 °C for 5 min. The proteins were separated by SDS-PAGE and visualized by Coomassie Brilliant Blue staining. Phosphorylated proteins were detected and quantitated by PhosphorImager (GE Healthcare). Active JNK1 was immunoprecipitated from the lysates prepared from cells stimulated by UV (60 J/m2, 2 min), using anti-JNK1 antibody (antibody 333). Immunoblotting was performed as described previously (46).
PFK-1 Assay—PFK-1 activity was measured as described previously (47). Briefly, cells were washed by phosphate-buffered saline once and lysed with M2 buffer (20 mm Tris-HCl, 0.5% Nonidet P-40, 250 mm NaCl, 3 mm EDTA, 3 mm EGTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μm dithiothreitol, 1 mm Na3VO4, 10 mm p-nitrophenyl phosphate). Lysates were centrifuged at 13,000 rpm for 15 min at 4 °C. 20 μg of supernatants were mixed with 1 ml of reaction buffer (50 mm HEPES (pH 7.0), 100 mm KCl, 5 mm MgCl2, 1.5 mm ATP, 0.15 mm NADH, 5 mm Na2HPO4, 0.1 mm AMP, 1 mm NH4Cl, 5 units of triose phosphate isomerase/ml, 0.5 units of aldolase/ml, 0.5 units of α-glycerophosphate dehydrogenase/ml, and 5 mm fructose-6-phosphate). Absorbance at 340 nm was read at room temperature every 2 min for a period of 10 min in an Ultrospec 1100 pro-spectrophotometer (GE Healthcare). Data are expressed as the change in absorbance at 340 nm/min/mg of protein × 103.
Measurement of Glycolysis—Glycolysis was measured by monitoring the conversion of [5-3H]glucose to [3H]H2Oas described previously (48). Briefly, 106 cells were washed once in phosphate-buffered saline prior to resuspension in 1 ml of Krebs buffer followed by incubation for 30 min at 37 °C. Cells were pelleted, resuspended in 0.5 ml of Krebs buffer containing 10 mm glucose. The reaction was started by the addition of 10 mCi of [5-3H]glucose. After 1 h at 37 °C, triplicate 50-μl aliquots were transferred to uncapped PCR tubes containing 50 μl of 0.2 n HCl. Each tube was transferred to a scintillation vial containing 0.5 ml of H2O such that the water in the vial and the contents of the PCR tube were not mixed with each other. The vials were sealed, and diffusion was allowed to occur for 3 days. The amounts of diffused and undiffused 3H were determined by scintillation counting. Appropriate [3H]glucose-only and 3H2O-only controls were included for the calculation of 3H2O in each sample that reflects the rate of glycolysis.
In Vitro Translation—The pCDNA3.1-based PFK-1 and Bcl-XL were translated in vitro using the rabbit reticulocyte lysate TnT-coupled transcription/translation system (Promega, Madison, WI) following the manufacturer's instructions.
siRNA Transfection and Retroviral Infection—siRNA specific for murine JNK1 or green fluorescent protein was designed by using the SiDESIGN center (Dharmacon, Lafayette, CO). The target sequence for Jnk1 is 5′-TGATTCAGATGGAGTTAGA-3′, and for Gfp, the sequence is 5′-GAAGAAGATGGTGCGCTCC-3′. The siRNA was transfected into cells using Lipofectamine 2000 following the manufacturer's instructions. Retroviral particles were produced using the pSOS vector system, as described previously (49). The FL5.12 Bcl-XL/M2-Bad cells were infected with the retroviruses four consecutive times (with 6-h intervals). After 48 h, the infection procedure was repeated to enrich the frequency of infection. The bulk infected cells were collected for experiments 48–72 h after the second round of infection.
Statistic Analysis—All statistic analyses were performed by using the Student's t test. A probability level of 0.05 or less was considered as significant. Data are presented as mean values ± standard error.
RESULTS AND DISCUSSION
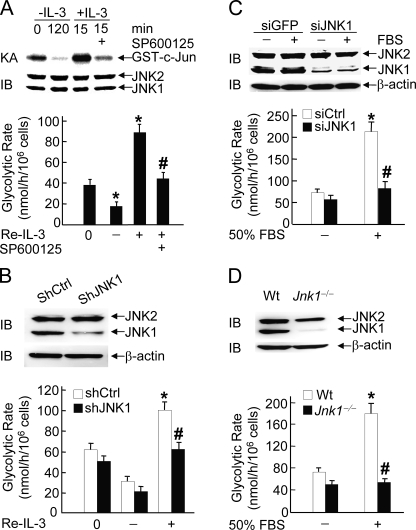
JNK1 Is Essential for IL-3- or Serum-induced Glycolysis—Maintenance of normal glycolysis is important for cell survival (50–52). The observations that phosphorylation of Bad at the regulatory serines (Ser-112, Ser-136, and Ser-155) is required for glycolysis (43), and JNK-mediated phosphorylation of Bad at Thr-201 for cell survival (42) prompted us to test whether JNK1 regulates glycolysis, using hematopoietic pro-B FL5.12 cells stably expressing Bcl-XL and Bad (for simplicity, referred to as FL-5.12 cells hereinafter unless indicated otherwise) (36, 42). Cells were subjected to IL-3 withdrawal for 2 h followed by the readdition of IL-3 for 15 min in the presence or absence of the specific JNK inhibitor SP600125. Under this condition, JNK1 activity was significantly reduced upon IL-3 withdrawal, stimulated by IL-3 readdition, and inhibited by SP600125 (Fig. 1A), consistent with the previous report (42). Metabolically labeling the cells with [5-3H]glucose revealed that the glycolytic rate was reduced following IL-3 withdrawal but became robust upon IL-3 readdition (Fig. 1A), consistent with the previous report that IL-3 up-regulates glycolysis in parental FL5.12 cells (53). The JNK inhibitor SP600125 significantly blocked IL-3 readdition-stimulated glycolysis (Fig. 1A). Similar results were obtained with FL-5.12 cells expressing specific retroviral shRNA of JNK1 but not control shRNA (Fig. 1B). The stimulation of glycolysis by IL-3 readdition was modest (Fig. 1B). This is most likely due to the high basal level of glycolysis that may be independent on IL-3.
FIGURE 1.
JNK1 is essential for IL-3- or serum-induced glycolysis. A, FL5.12 Bcl-XL/Bad (FL-5.12) cells were deprived of IL-3 for 2 h (–IL-3) followed by IL-3 readdition (+IL-3) for 15 min. The specific JNK inhibitor SP600125 (10 μm) was added 30 min prior to IL-3 readdition (Re-IL-3). The glycolytic rate (*, p < 0.01 when compared with 0 h; #, p < 0.01 when compared with IL-3 readdition for 15 min) was determined by measuring the conversion of [5-3H]glucose to [3H]H2O, as described under “Materials and Methods.” The results are presented as means ± S.E. and represent three individual experiments. The JNK activity was measured by immune complex kinase assays (KA) using GST-c-Jun as substrate, and the expression level of JNK was detected by immunoblotting (IB) using anti-JNK antibody (Pharmingen, 666.6). IP, immunoprecipitation. B, FL-5.12 cells were infected by retroviral vectors encoding the control shRNA (shCtrl) or shRNA of JNK1 (shJNK1). Cells were deprived of IL-3 for 2 h (–IL-3) followed by IL-3 readdition (+IL-3) for 15 min. The effect of shRNA of JNK1 on JNK1 expression was detected by immunoblotting and the glycolytic rate (*, p < 0.01 when compared with 0 h; #, p < 0.01 when compared with control shRNA cells treated with IL-3 readdition for 15 min) was determined, as described in panel A. C, immortalized WT MEFs were transfected with siCtrl or siJNK1 (200 nm each) followed by stimulation without or with 50% FBS for 15 min. The glycolytic rate (*, p < 0.01 when compared with unstimulated siCtrl cells; #, p < 0.01 when compared with siCtrl cells treated with 50% FBS stimulation for 15 min) was determined as described in panel A. D, WT or Jnk1–/– MEFs were stimulated without or with 50% FBS for 15 min, and the glycolytic rate (*, p < 0.01 when compared with unstimulated WT MEFs; #, p < 0.01 when compared with WT MEFs treated with 50% FBS stimulation for 15 min) was determined as described in panel A. ns, nonspecific.
This notion was further supported by the observation that in immortalized MEFs, silencing of JNK1 by siRNA or genetic disruption of Jnk1 alleles abolished fetal bovine serum (FBS)-induced glycolysis (Fig. 1, C and D). Thus, JNK1 is required for growth factor-induced glycolysis. Given the fact that up-regulation of glycolysis is a direct effect of IL-3, rather than a compensatory result of growth factor stimulation of cell growth and proliferation (53), IL-3-induced JNK1-dependent higher glycolytic rate may be beneficial to cell survival under stress conditions.
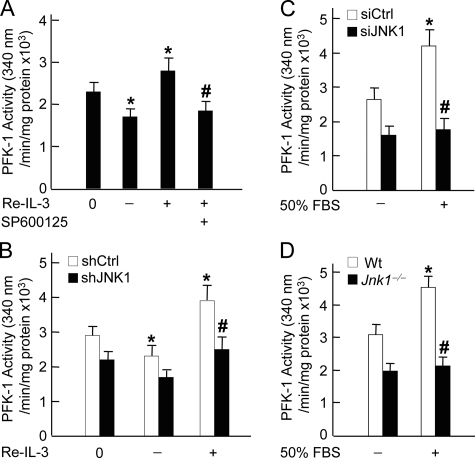
JNK Regulates Activation of PFK-1—Hexokinase and PFK-1 are two rate-limiting enzymes in the glycolytic process (54–56). Previous studies have shown that phosphorylation of Bad at Ser-112, Ser-136, and Ser-155 by several protein kinases (cAMP-dependent protein kinase, Akt, and Rsk) is required for activation of glucokinase, which is one of the hexokinase isoforms (57, 58), thereby regulating the glycolytic process (43). Since Bad is also phosphorylated at Thr-201 by JNK1, we were curious about whether JNK1 regulates glycolysis through modulation of the activity of PFK-1 and hexokinase. In IL-3-dependent FL-5.12 cells, PFK-1 had high basal activity, as measured by PFK-1 assay (Fig. 2A). The activity of PFK-1 was partially reduced by IL-3 withdrawal and further enhanced by IL-3 readdition (Fig. 2A). This suggests that PFK-1 activity is likely controlled by IL-3-dependent and -independent mechanisms. However, the augmented PFK-1 activity by IL-3 readdition was completely abrogated by the JNK inhibitor SP600125 (Fig. 2A) or retroviral shRNA of JNK1 (Fig. 2B). Thus, the stimulation of PFK-1 by IL-3 depends on JNK1 activity. Consistent with this conclusion, FBS induced significant increase of PFK-1 activity in MEFs but was unable to do so in MEFs expressing siRNA of JNK1 or in JNK1 null MEFs (Fig. 2, C and D). Similar results were obtained with hexokinase, although to a less extent (data not shown). Taken together, JNK1 may regulate IL-3- or FBS-stimulated glycolysis by, at least in part, modulating the activity of PFK-1.
FIGURE 2.
JNK1 regulates the activation of PFK-1. A, FL-5.12 cells were deprived of IL-3 for 2 h (–IL-3) followed by IL-3 readdition (Re-IL-3, +IL-3) for 15 min. The specific JNK inhibitor SP600125 (10 μm) was added 30 min prior to IL-3 readdition. Cell lysates were analyzed for PFK-1 activity (*, p < 0.05 when compared with 0 h; #, p < 0.01 when compared with IL-3 readdition for 15 min), as described under “Materials and Methods.” B, FL-5.12 cells were infected by control shRNA (shCtrl) or shRNA of JNK1 (shJNK1) retrovirus. Cells were deprived of IL-3 for 2 h (–IL-3) followed by IL-3 readdition (+IL-3) for 15 min. PFK-1 activity (*, p < 0.05 when compared with 0 h; #, p < 0.05 when compared with shCtrl cells treated with IL-3 readdition for 15 min) was measured as described in panel A. C, WT MEFs were transfected with siCtrl or siJNK1 (200 nm each) followed by stimulation without or with 50% FBS for 15 min. PFK-1 activity (*, p < 0.05 when compared with unstimulated siCtrl cells; #, p < 0.01 when compared with siCtrl cells treated with 50%FBS stimulation for 15 min) was determined as described in panel A. D, WT or Jnk1–/– MEFs were stimulated without or with 50% FBS for 15 min. PFK-1 activity (*, p < 0.01 when compared with unstimulated WT MEFs; #, p < 0.01 when compared with WT MEFs treated with 50%FBS stimulation for 15 min) was determined as described in panel A.
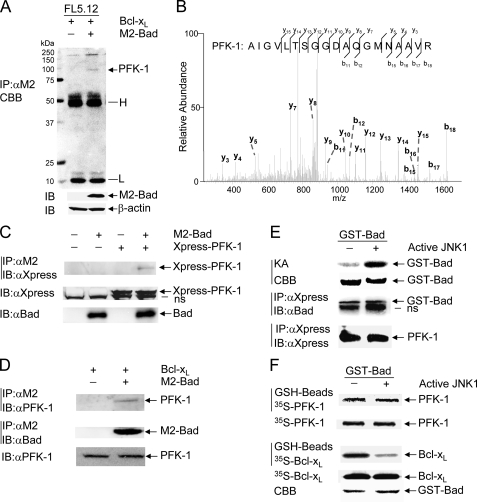
PFK-1 Is a Novel Bad-associated Protein—The observations that JNK1 inhibits IL-3 withdrawal-induced cell death through phosphorylation and inactivation of Bad (42) and that JNK1 regulates the rate of glycolysis and PFK-1 activity prompted us to examine the possible connection between PFK-1 and Bad. Using nano-HPLC electrospray ion trap mass spectrometry, we identified several proteins that were co-immunoprecipitated with Bad from FL-5.12 cells. One of Bad-associated proteins was PFK-1 (Fig. 3, A and B). Consistently, co-immunoprecipitation in combination of immunoblotting revealed that ectopically expressed Xpress-tagged PFK-1 or endogenous PFK-1 proteins were co-immunoprecipitated with M2-Bad proteins (Fig. 3, C and D). However, the interaction between Bad and PFK-1 was not affected by IL-3 (data not shown). To verify this observation, we employed an in vitro binding assay in which purified GST-Bad proteins were first phosphorylated by active JNK1 in the presence of either [γ-32P]ATP (Fig. 3E, top panel, positive control) or nonradioactive ATP. GST-Bad proteins phosphorylated with nonradioactive ATP were isolated by GSH beads and mixed with COS-1 cell extracts containing Xpress-PFK-1. Immunoprecipitation using anti-Xpress antibody in combination with immunoblotting using anti-Bad antibody revealed that both phosphorylated and nonphosphorylated GST-Bad proteins that were run at the position of 50 kDa in SDS-gel interacted with PFK-1 (Fig. 3E). In a parallel experiment, GST-Bad proteins phosphorylated with or without nonradioactive ATP were incubated with in vitro-translated 35S-labeled PFK-1 or the positive control [35S]-Bcl-XL. GST pull-down assays revealed that phosphorylation by active JNK1 significantly reduced the binding of GST-Bad to [35S]-Bcl-XL (Fig. 3F, low panel), consistent with the previous report (42). However, phosphorylation by active JNK1 had no detectable effect on the binding of GST-Bad to 35S-labeled PFK-1 (Fig. 3F, top panel). Thus, Bad directly interacts with the second rate-limiting enzyme of glycolysis, PFK-1, and the interaction is independent of its phosphorylation by JNK1. This is in analogy to the previous report that the interaction between Bad and glucokinase is also independent of Bad phosphorylation at the regulatory serines (43). It has been shown that Bad is involved in the assembly of the protein complex containing. The catalytic subunit of PP1 and cAMP-dependent protein kinase, glucokinase, and WAVE-1 (43). Future work is needed to determine whether PFK-1 is also a component of this complex.
FIGURE 3.
PFK-1 is a novel Bad-associated protein. A, cell extracts of FL5.12/Bcl-XL and FL-5.12 were subjected to immunoprecipitation with anti-M2 antibody, followed by SDS-PAGE, and stained with colloidal Coomassie Brilliant Blue (CBB). Transfected M2-Bad was analyzed by immunoblotting using anti-M2 antibody. The arrow indicates the major Bad-associated protein, which was identified as PFK-1 (see panel B below). H, IgG heavy chains; L, IgG light chains. B, the major Bad-associated protein identified in panel A was excised and sequenced by nano-HPLC electrospray ion trap mass spectrometry, as described under “Materials and Methods.” C, COS-1 cells were transfected with mammalian expression vectors encoding PFK-1 or M2-Bad (2 μgof each), without or with empty vector (2 μg). After 40 h, M2-Bad was immunoprecipitated (IP) with anti-M2 antibody, and M2-Bad-associated PFK-1 was immunoblotted (IB) with anti-Xpress antibody. Total M2-Bad or Xpress-PFK-1 proteins in the cell extracts were analyzed by immunoblotting with anti-Bad or anti-Xpress antibody, respectively. D, M2-Bad was immunoprecipitated from stable FL5.12/Bcl-XL or FL-5.12 cells with anti-M2 antibody, and M2-Bad-associated PFK-1 was analyzed by immunoblotting using anti-PFK-1 antibody. The amounts of immunoprecipitated M2-Bad proteins and total PFK-1 proteins were examined by immunoblotting with anti-Bad and anti-PFK-1 antibody, respectively. E, purified GST-Bad proteins were incubated without or with active JNK1 in a kinase reaction buffer in the presence of [γ-32P] ATP (top two panels, KA for kinase assays) or 17 μm nonradioactive ATP (low two panels). GST-Bad proteins modified with nonradioactive ATP were mixed with cell extracts of COS-1 cells that have been transfected with mammalian expression vector encoding Xpress-PFK-1. Xpress-PFK-1 was immunoprecipitated with anti-Xpress antibody, and Xpress-PFK-1-associated GST-Bad proteins were analyzed by immunoblotting using anti-Bad antibody. The amount of Xpress-PFK-1 in the immune complex was analyzed by immunoblotting with anti-Xpress antibody. F, GST-Bad proteins modified with nonradioactive ATP (as described in panel E) were mixed with in vitro translated 35S-labeled PFK-1 or 35S-labeled Bcl-XL for 12 h and extensively washed. GST-Bad-associated 35S-PFK-1 or 35S-Bcl-XL was visualized by radioautography. One-tenth of the input 35S-PFK-1 or 35S-Bcl-XL was analyzed by radioautography. The amount of GST-Bad proteins was examined by Coomassie Brilliant Blue staining. ns, nonspecific.
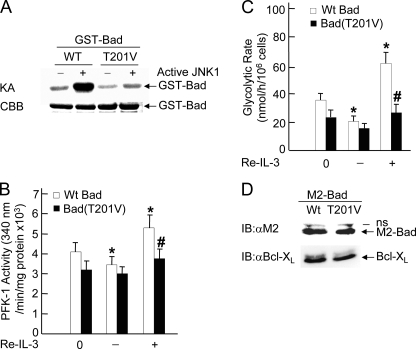
Phosphorylation of Bad at Thr-201 by JNK1 Is Required for PFK-1 Activation and Glycolysis—Although the interaction between Bad and PFK-1 is independent of Bad phosphorylation by JNK1, it is possible that the phosphorylation is required for Bad to regulate PFK-1 activity. To test this hypothesis, FL5.12/Bcl-XL cells were transiently transfected with WT M2-Bad or M2-Bad(T201V) in which threonine 201 has been replaced by valine. Although WT Bad was significantly phosphorylated by active JNK1, Bad(T201V) mutant was not, as measured by in vitro kinase assays (Fig. 4A). IL-3-withdrawal partially reduced PFK-1 activity, and its readdition significantly stimulated PFK-1 activity in cells expressing WT M2-Bad (Fig. 4B), consistent with the results obtained by using stable FL-5.12 cells (Fig. 2). In contrast, IL-3-withdrawal or readdition only slightly affected the activity of PFK-1 in cells expressing M2-Bad(T201V) mutant (Fig. 4B). Similarly, the effect of IL-3 on the rate of glycolysis was also greatly reduced in cells expressing M2-Bad(T201V) mutant when compared with that in cells expressing WT M2-Bad (Fig. 4C). The difference in IL-3-induced PFK-1 activity or glycolysis between FL5.12 Bcl-XL/Bad and FL5.12 Bcl-XL/Bad(T201V) cells was not the result of the difference in expression levels of M2-Bad and M2-Bad(T201V), as analyzed by immunoblotting using anti-M2 antibody (Fig. 4D). These results indicate that Thr-201 phosphorylation of Bad is required for the regulation of PFK-1 activity and glycolysis. This is in analogy to the previous report that the phosphorylation of Bad at the regulatory serines is required for activation of glucokinase (43). Future work is needed to determine how phosphorylation of Bad at Thr-201 by JNK1 or the regulatory serines regulates the activity of PFK-1 or glucokinase, respectively.
FIGURE 4.
JNK1-phosphorylated Bad regulates PFK-1 activation and glycolysis. A, purified WT GST-Bad and GST-Bad(T201V) mutant proteins were incubated with active JNK1 for 60 min, and phosphorylation of GST-Bad proteins was measured by in vitro kinase assays (KA) in the presence of [γ-32P]ATP. The amount of GST-Bad was examined by Coomassie Brilliant Blue (CBB) staining. B and C, FL5.12/Bcl-XL cells were transfected with WT M2-Bad or M2-Bad(T201V) (20 μg of each). 48 h later, cells were deprived of IL-3 for 2 h (–IL-3) followed by IL-3 readdition (Re-IL-3, +IL-3) for 15 min. PFK-1 activity (B, *, p < 0.05 when compared with cells transfected with WT M2-Bad at 0 h; #, p < 0.05 when compared with cells transfected with WT M2-Bad treated with IL-3 readdition for 15 min) and the glycolytic rate (C, *, p < 0.01 when compared with cells transfected with WT M2-Bad at 0 h; #, p < 0.01 when compared with cells transfected with WT M2-Bad treated with IL-3 readdition for 15 min) were measured as described in the legends for Figs. 1 and 2. D, the expression levels of Bad and Bcl-XL in transfected FL5.12/Bcl-XL cells were analyzed by immunoblotting using anti-M2 and anti-Bcl-XL antibody, respectively. IB, immunoblotting; IP, immunoprecipitation.
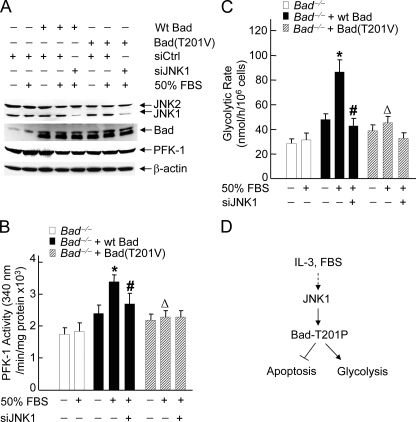
It is possible that JNK1 may regulate PFK-1 activity and glycolysis in Bad-dependent and -independent mechanisms. We reasoned that if JNK1 regulates PFK-1 activity and glycolysis only through phosphorylation of Bad at Thr-201, inactivation of JNK1 should not further affect PFK-1 activity and glycolysis in cells expressing M2-Bad(T201V) mutant. To test this hypothesis, Bad null MEFs were transiently transfected with WT M2-Bad, M2-Bad(T201V), or empty vector, with or without siRNA of JNK1 or control siRNA. Immunoblotting analysis revealed that expression of JNK1 was significantly silenced by its specific siRNA but not the control siRNA (Fig. 5A), whereas the expression of M2-Bad and M2-Bad(T201V) were at similar levels (Fig. 5A). PFK-1 assay showed that in Bad null cells expressing WT M2-Bad, FBS significantly stimulated the activity of PFK-1, and the stimulation was reduced by siRNA of JNK1 (Fig. 5B). However, in Bad null cells expressing M2-Bad(T201V) mutant, FBS only slightly stimulated PFK-1 activity, and more importantly, siRNA of JNK1 had no detectable effect on PFK-1 activity (Fig. 5B). Similar results were obtained when the glycolytic rate was analyzed (Fig. 5C). Thus, JNK1 regulates PFK-1 activation and glycolysis through phosphorylation of Bad at Thr-201 in response to IL-3 or FBS.
FIGURE 5.
JNK1-mediated activation of PFK-1 and glycolysis depends on its phosphorylation of Bad at Thr-201. A, Bad–/– cells were transfected with mammalian expressing vector encoding WT M2-Bad, M2-Bad(T201V), or empty vector (2μg of each), without or with siCtrl or siJNK1 (200 nm each). The expression levels of JNK, Bad, and PFK-1 were analyzed by immunoblotting with the antibodies against JNK, Bad, or PFK-1, respectively. B and C, after 48 h, cells were treated without or with 50% FBS for 15 min. The activity of PFK-1 (B, *, p < 0.01 when compared with untreated cells transfected with WT M2-Bad; #, p < 0.01 and ▵, p < 0.05 when compared with cells transfected with WT M2-Bad treated with 50% FBS stimulation for 15 min, respectively) and the glycolytic rate (C, *, p < 0.01 when compared with untreated cells transfected with WT M2-Bad; # and ▵, p < 0.01 when compared with cells transfected with WT M2-Bad treated with 50% FBS stimulation for 15 min) were measured as described in the legends for Figs. 1 and 2. D, the schematic presentation of how JNK1 regulates PFK-1 activity via phosphorylation of Bad at Thr-201, thereby contributing to glycolysis.
In summary, our results suggest that nonphosphorylated Bad is ready to interact with PFK-1, but only JNK1-phosphorylated Bad is able to regulate PKF-1 activity. Thus, in addition to inhibition of the pro-apoptotic function of Bad, JNK1-mediated phosphorylation of Bad at Thr-201 is required for regulating the activity of PFK-1, which in turn promotes glycolysis for cell survival (Fig. 5D).
Acknowledgments
We are grateful to Dr. Stanley Korsmeyer for reagents that make this work possible.
This work was supported, in whole or in part, by National Institutes of Health Grants CA100460 and GM071409 (to A. L.) and CA090516 (to J. X.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: JNK, c-Jun N-terminal protein kinase; SAPK, stress-activated protein kinase; MAP, mitogen-activated protein; MAPK, MAP kinase; MAP3K, MAPK kinase kinase; MEKK, MAPK and ERK kinase; ERK, extracellular signal-regulated kinase; TGF, transforming growth factor; PFK-1, phosphofructokinase-1; IL-3, interleukin-3; Bad, bcl-associated death protein; FBS, fetal bovine serum; GST, glutathione S-transferase; MEF, mouse embryo fibroblast; siRNA, small interfering RNA; siCtrl, control siRNA; siJNK1, siRNA of JNK1; shRNA, short hairpin RNA; WT, wild type; nano-HPLC, nano-high pressure liquid chromatography.
References
- 1.Hibi, M., Lin, A., Smeal, T., Minden, A., and Karin, M. (1993) Genes Dev. 7 2135–2148 [DOI] [PubMed] [Google Scholar]
- 2.Chang, L., and Karin, M. (2001) Nature 410 37–40 [DOI] [PubMed] [Google Scholar]
- 3.Davis, R. J. (2000) Cell 103 239–252 [DOI] [PubMed] [Google Scholar]
- 4.Lin, A. (2003) BioEssays 25 17–24 [DOI] [PubMed] [Google Scholar]
- 5.Liu, J., Minemoto, Y., and Lin, A. (2004) Mol. Cell Biol. 24 10844–10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabapathy, K., Hochedlinger, K., Nam, S. Y., Bauer, A., Karin, M., and Wagner, E. F. (2004) Mol. Cell 15 713–725 [DOI] [PubMed] [Google Scholar]
- 7.Liu, J., Yang, D., Minemoto, Y., Leitges, M., Rosner, M. R., and Lin, A. (2006) Mol. Cell 21 467–480 [DOI] [PubMed] [Google Scholar]
- 8.Lin, A., Minden, A., Martinetto, H., Claret, F. X., Lange-Carter, C., Mercurio, F., Johnson, G. L., and Karin, M. (1995) Science 268 286–290 [DOI] [PubMed] [Google Scholar]
- 9.Derijard, B., Raingeaud, J., Barrett, T., Wu, I. H., Han, J., Ulevitch, R. J., and Davis, R. J. (1995) Science 267 682–685 [DOI] [PubMed] [Google Scholar]
- 10.Lu, X., Nemoto, S., and Lin, A. (1997) J. Biol. Chem. 272 24751–24754 [DOI] [PubMed] [Google Scholar]
- 11.Tournier, C., Whitmarsh, A. J., Cavanagh, J., Barrett, T., and Davis, R. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7337–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minden, A., Lin, A., McMahon, M., Lange-Carter, C., Derijard, B., Davis, R. J., Johnson, G. L., and Karin, M. (1994) Science 266 1719–1723 [DOI] [PubMed] [Google Scholar]
- 13.Hammaker, D. R., Boyle, D. L., Chabaud-Riou, M., and Firestein, G. S. (2004) J. Immunol. 172 1612–1618 [DOI] [PubMed] [Google Scholar]
- 14.Ichijo, H., Nishida, E., Irie, K., ten Dijke, P., Saitoh, M., Moriguchi, T., Takagi, M., Matsumoto, K., Miyazono, K., and Gotoh, Y. (1997) Science 275 90–94 [DOI] [PubMed] [Google Scholar]
- 15.Tibbles, L. A., Ing, Y. L., Kiefer, F., Chan, J., Iscove, N., Woodgett, J. R., and Lassam, N. J. (1996) EMBO J. 15 7026–7035 [PMC free article] [PubMed] [Google Scholar]
- 16.Shibuya, H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E., and Matsumoto, K. (1996) Science 272 1179–1182 [DOI] [PubMed] [Google Scholar]
- 17.Salmeron, A., Ahmad, T. B., Carlile, G. W., Pappin, D., Narsimhan, R. P., and Ley, S. C. (1996) EMBO J. 15 817–826 [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, G., Merritt, S. E., Kortenjann, M., Shaw, P. E., and Holzman, L. B. (1996) J. Biol. Chem. 271 24788–24793 [DOI] [PubMed] [Google Scholar]
- 19.Chen, Z., Hutchison, M., and Cobb, M. H. (1999) J. Biol. Chem. 274 28803–28807 [DOI] [PubMed] [Google Scholar]
- 20.Ito, M., Yoshioka, K., Akechi, M., Yamashita, S., Takamatsu, N., Sugiyama, K., Hibi, M., Nakabeppu, Y., Shiba, T., and Yamamoto, K. I. (1999) Mol. Cell Biol. 19 7539–7548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald, P. H., Chow, C. W., Miller, W. E., Laporte, S. A., Field, M. E., Lin, F. T., Davis, R. J., and Lefkowitz, R. J. (2000) Science 290 1574–1577 [DOI] [PubMed] [Google Scholar]
- 22.Whitmarsh, A. J., Cavanagh, J., Tournier, C., Yasuda, J., and Davis, R. J. (1998) Science 281 1671–1674 [DOI] [PubMed] [Google Scholar]
- 23.Karin, M. (1995) J. Biol. Chem. 270 16483–16486 [DOI] [PubMed] [Google Scholar]
- 24.Karin, M., and Lin, A. (2002) Nat. Immunol. 3 221–227 [DOI] [PubMed] [Google Scholar]
- 25.Lin, A., and Karin, M. (2003) Semin. Cancer Biol. 13 107–114 [DOI] [PubMed] [Google Scholar]
- 26.Tang, G., Minemoto, Y., Dibling, B., Purcell, N. H., Li, Z., Karin, M., and Lin, A. (2001) Nature 414 313–317 [DOI] [PubMed] [Google Scholar]
- 27.Lin, A. (2006) Dev. Cell 10 277–278 [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., and Lin, A. (2007) Oncogene 26 3267–3278 [DOI] [PubMed] [Google Scholar]
- 29.Tang, F., Tang, G., Xiang, J., Dai, Q., Rosner, M. R., and Lin, A. (2002) Mol. Cell Biol. 22 8571–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonyak, M. A., Kenyon, L. C., Godwin, A. K., James, D. C., Emlet, D. R., Okamoto, I., Tnani, M., Holgado-Madruga, M., Moscatello, D. K., and Wong, A. J. (2002) Oncogene 21 5038–5046 [DOI] [PubMed] [Google Scholar]
- 31.Sadoshima, J., Montagne, O., Wang, Q., Yang, G., Warden, J., Liu, J., Takagi, G., Karoor, V., Hong, C., Johnson, G. L., Vatner, D. E., and Vatner, S. F. (2002) J. Clin. Investig. 110 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uehara, T., Bennett, B., Sakata, S. T., Satoh, Y., Bilter, G. K., Westwick, J. K., and Brenner, D. A. (2005) J. Hepatol. 42 850–859 [DOI] [PubMed] [Google Scholar]
- 33.Xia, X. G., Harding, T., Weller, M., Bieneman, A., Uney, J. B., and Schulz, J. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10433–10438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, E., Zha, J., Jockel, J., Boise, L. H., Thompson, C. B., and Korsmeyer, S. J. (1995) Cell 80 285–291 [DOI] [PubMed] [Google Scholar]
- 35.Gross, A., McDonnell, J. M., and Korsmeyer, S. J. (1999) Genes Dev. 13 1899–1911 [DOI] [PubMed] [Google Scholar]
- 36.Zha, J., Harada, H., Yang, E., Jockel, J., and Korsmeyer, S. J. (1996) Cell 87 619–628 [DOI] [PubMed] [Google Scholar]
- 37.Datta, S. R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y., and Greenberg, M. E. (1997) Cell 91 231–241 [DOI] [PubMed] [Google Scholar]
- 38.Harada, H., Becknell, B., Wilm, M., Mann, M., Huang, L. J., Taylor, S. S., Scott, J. D., and Korsmeyer, S. J. (1999) Mol. Cell 3 413–422 [DOI] [PubMed] [Google Scholar]
- 39.Harada, H., Andersen, J. S., Mann, M., Terada, N., and Korsmeyer, S. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9666–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H. G., Rapp, U. R., and Reed, J. C. (1996) Cell 87 629–638 [DOI] [PubMed] [Google Scholar]
- 41.Cheng, E. H., Wei, M. C., Weiler, S., Flavell, R. A., Mak, T. W., Lindsten, T., and Korsmeyer, S. J. (2001) Mol. Cell 8 705–711 [DOI] [PubMed] [Google Scholar]
- 42.Yu, C., Minemoto, Y., Zhang, J., Liu, J., Tang, F., Bui, T. N., Xiang, J., and Lin, A. (2004) Mol. Cell 13 329–340 [DOI] [PubMed] [Google Scholar]
- 43.Danial, N. N., Gramm, C. F., Scorrano, L., Zhang, C. Y., Krauss, S., Ranger, A. M., Datta, S. R., Greenberg, M. E., Licklider, L. J., Lowell, B. B., Gygi, S. P., and Korsmeyer, S. J. (2003) Nature 424 952–956 [DOI] [PubMed] [Google Scholar]
- 44.Sabapathy, K., Jochum, W., Hochedlinger, K., Chang, L., Karin, M., and Wagner, E. F. (1999) Mech. Dev. 89 115–124 [DOI] [PubMed] [Google Scholar]
- 45.Zhao, Y., Zhang, W., Kho, Y., and Zhao, Y. (2004) Anal. Chem. 76 1817–1823 [DOI] [PubMed] [Google Scholar]
- 46.Tang, G., Yang, J., Minemoto, Y., and Lin, A. (2001) Mol. Cell 8 1005–1016 [DOI] [PubMed] [Google Scholar]
- 47.Sapico, V., and Anderson, R. L. (1970) J. Biol. Chem. 245 3252–3256 [PubMed] [Google Scholar]
- 48.Liang, Y., Buettger, C., Berner, D. K., and Matschinsky, F. M. (1997) Diabetologia 40 1018–1027 [DOI] [PubMed] [Google Scholar]
- 49.Luo, Q., Kang, Q., Song, W. X., Luu, H. H., Luo, X., An, N., Luo, J., Deng, Z. L., Jiang, W., Yin, H., Chen, J., Sharff, K. A., Tang, N., Bennett, E., Haydon, R. C., and He, T. C. (2007) Gene (Amst.) 395 160–169 [DOI] [PubMed] [Google Scholar]
- 50.Raff, M. C. (1992) Nature 356 397–400 [DOI] [PubMed] [Google Scholar]
- 51.Vander Heiden, M. G., Plas, D. R., Rathmell, J. C., Fox, C. J., Harris, M. H., and Thompson, C. B. (2001) Mol. Cell Biol. 21 5899–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gottlob, K., Majewski, N., Kennedy, S., Kandel, E., Robey, R. B., and Hay, N. (2001) Genes Dev. 15 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauer, D. E., Harris, M. H., Plas, D. R., Lum, J. J., Hammerman, P. S., Rathmell, J. C., Riley, J. L., and Thompson, C. B. (2004) FASEB J. 18 1303–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, J. E. (1995) Rev. Physiol. Biochem. Pharmacol. 126 65–198 [DOI] [PubMed] [Google Scholar]
- 55.Rempel, A., Mathupala, S. P., Griffin, C. A., Hawkins, A. L., and Pedersen, P. L. (1996) Cancer Res. 56 2468–2471 [PubMed] [Google Scholar]
- 56.Layzer, R. B., Rowland, L. P., and Bank, W. J. (1969) J. Biol. Chem. 244 3823–3831 [PubMed] [Google Scholar]
- 57.Pelicano, H., Martin, D. S., Xu, R. H., and Huang, P. (2006) Oncogene 25 4633–4646 [DOI] [PubMed] [Google Scholar]
- 58.Cardenas, M. L., Cornish-Bowden, A., and Ureta, T. (1998) Biochim. Biophys. Acta 1401 242–264 [DOI] [PubMed] [Google Scholar]