Abstract
The p53 tumor-suppressor protein, a key regulator of cellular responses to genotoxic stress, is stabilized and activated after DNA damage. This process is associated with posttranslational modifications of p53, some of which are mediated by the ATM protein kinase. However, these modifications alone may not account in full for p53 stabilization. p53's stability and activity are negatively regulated by the oncoprotein MDM2, whose gene is activated by p53. Conceivably, p53 function may be modulated by modifications of MDM2 as well. We show here that after treatment of cells with ionizing radiation or a radiomimetic chemical, but not UV radiation, MDM2 is phosphorylated rapidly in an ATM-dependent manner. This phosphorylation is independent of p53 and the DNA-dependent protein kinase. Furthermore, MDM2 is directly phosphorylated by ATM in vitro. These findings suggest that in response to DNA strand breaks, ATM may promote p53 activity and stability by mediating simultaneous phosphorylation of both partners of the p53-MDM2 autoregulatory feedback loop.
Damage to cellular DNA activates repair mechanisms as well as signal transduction pathways that lead to cell cycle arrest or programmed cell death. Several of these responses are mediated by stabilization and activation of the p53 protein, a transcription factor encoded by a tumor-suppressor gene that controls a central junction of these pathways (1–3). The stabilization of p53 appears to be associated with posttranslational modifications of this protein invoked by DNA damage. These modifications include phosphorylation, dephosphorylation, and acetylation on various residues (3–8). The role of these modifications in p53 stabilization has become a major focus of interest.
Two protein kinases have been implicated directly in p53 modifications induced by ionizing radiation (IR) and radiomimetic chemicals: ATM, required for the initial phase of p53 accumulation in response to this damage (9–14), and ATR, involved in the later phase of this process (13, 15). In response to IR and radiomimetic treatment of cells, ATM is activated and mediates rapid phosphorylation of p53 on Ser15 (11–14), whereas ATR seems to be involved in the subsequent maintenance of this phosphorylation (15). Importantly, both of these kinases phosphorylate p53 in vitro on Ser15 (12–15). A third enzyme implicated in cellular responses to DNA damage, the DNA-dependent protein kinase (DNA-PK), is capable of phosphorylating p53 in vitro on Ser15 and Ser37 (16), but is probably not necessary for damage-induced p53 activation and accumulation (17–19).
ATM, ATR, and DNA-PK belong to a family of protein kinases with carboxyl-terminal domains displaying similarity to phosphoinositide 3 kinases. Members of this family are involved in controlling genome stability, cell cycle progression, and responses to DNA damage in various organisms (20–23). Lack of ATM in humans causes the genetic disorder ataxia-telangiectasia (A-T), characterized by neurodegeneration, immunodeficiency, genome instability, cancer predisposition, sensitivity to IR, and defective activation of cell cycle checkpoints by DNA damage (21, 24). DNA-PK deficiency in mice leads to severe combined immunodeficiency (scid), which shares many features with A-T (23, 25).
Despite the apparent association between Ser15 phosphorylation and p53 stabilization after DNA damage, we provide evidence that Ser15 phosphorylation might be dispensable for ATM-dependent p53 stabilization induced by DNA damage, suggesting that other ATM-mediated protein modifications may be more important to this process. A potential target of ATM activity could be the oncoprotein MDM2, a major negative regulator of p53 (26–28). MDM2 binds to the amino terminus of p53, represses p53's transactivation activity, and targets it to proteasome-mediated degradation (26–31). The destabilization of p53 requires both the amino- and carboxyl-terminal regions of MDM2 (30–33), as well as its nuclear export capability (34, 35). Recent observations suggest that MDM2 functions as an E3 ubiquitin-protein ligase in p53 degradation (36). The MDM2 gene is positively regulated by p53, thereby creating a negative feedback loop (26–28).
In this report we show that after exposure of cells to IR or a radiomimetic chemical, and before p53 accumulation, MDM2 is phosphorylated rapidly in an ATM-dependent, DNA-PK-independent manner. Moreover, ATM phosphorylates recombinant MDM2 in vitro on at least two sites, suggesting that MDM2 might be a direct in vivo target of ATM after DNA damage. These findings suggest a novel mechanism for ATM-dependent p53 stabilization.
Materials and Methods
Protein Detection and Immunoprecipitation.
Cells were collected by centrifugation or scraping, and cellular pellets were washed with cold PBS and lysed for 1 hr at 4°C in 150 mM NaCl/10 mM Hepes, pH 7.2/0.5% Nonidet P-40/2 mM EDTA/2 mM EGTA and protease inhibitors. After centrifugation at 14,000 × g for 15 min at 4°C, the supernatant was collected for further experiments. For immunoblotting analysis, total cellular extracts were electrophoresed in 8% polyacrylamide gels and transferred overnight at 4°C and 230 mA onto nitrocellulose membranes (Nitrocell MFF; Pharmacia). For immunoprecipitation, cell lysates were precleared by constant mixing for 1.5 hr with protein A/G-Sepharose beads (Pharmacia). The beads were removed by centrifugation, and the supernatant was mixed constantly overnight with the appropriate antibody. Immune complexes were adsorbed onto protein A/G-Sepharose beads, boiled, and electrophoresed on 8% polyacrylamide gels. In some experiments, cell pellets were resuspended directly in protein sample buffer and subjected to gel electrophoresis. Conditions for immunodetection were adapted for each protein source and method of preparation.
Phosphatase Treatment of Immune Complexes.
Immunoprecipitates were washed three times with 150 mM NaCl/50 mM Tris, pH 8.9/5 mM MgCl2/1 mM DTT; 10 units of shrimp alkaline phosphatase (SAP; Amersham) were added, and the reaction was continued for 30 min at 37°C.
Recombinant Proteins.
The complete ORF of HDM2, HDM2 fragments spanning amino acids 82–312 and 330–491, and an MDM2 fragment containing amino acids 328–437, were obtained by PCR using the corresponding cDNA clones as templates and subcloned in the vectors pGEX6P or pGEX3X as glutathione S-transferase-fused proteins. These constructs were expressed in Escherichia coli XL1 and isolated by glutathione-agarose chromatography. An S-tagged fragment of HDM2 spanning amino acids 1–115 in the vector pET-29a was generously donated by A. Levine and D. Freedman (Princeton University, Princeton, NJ). A cytomegalovirus-driven expression construct encoding human p53(S15A) with Ser15 replaced by Ala (8) was kindly provided by T. Unger (The Weizmann Institute of Science, Rehovot, Israel).
Results
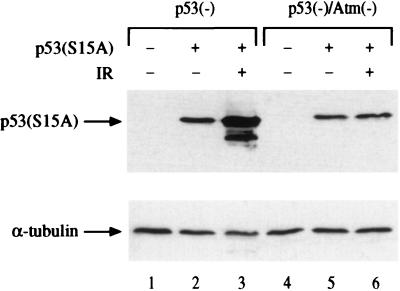
To examine the role of Ser15 phosphorylation in damage-induced stabilization of p53, we expressed mutant p53 with Ser15 substituted by Ala on p53-null and p53/Atm-null backgrounds. This substitution did not prevent effective IR-induced p53 stabilization, which remained ATM-dependent (Fig. 1). Hence, Ser15 phosphorylation alone cannot account for ATM-mediated p53 stabilization in this experimental system. This raised the possibility that modifications responsible for p53 stabilization may not be confined to the p53 molecule. We therefore sought to determine whether MDM2 also could be a target for DNA damage-induced modifications within cells.
Figure 1.
Radiation-induced accumulation of p53(S15A). A plasmid encoding a mutant p53 protein, in which Ser15 was replaced by Ala (8), was transfected into ap29 murine p53-null cells (37) or into ap24 double-mutant cells lacking both p53 and Atm (37). After 48 hr, some of the cultures were treated with 7.5 Gy of gamma radiation and harvested 75 min later. Total cellular extracts were subjected to SDS/PAGE and immunoblot analysis by using a mixture of the anti-p53 antibodies PAb1801 and DO-1. The faster-migrating band observed in lane 3 is a truncated form of p53 often observed when p53 is produced at elevated levels.
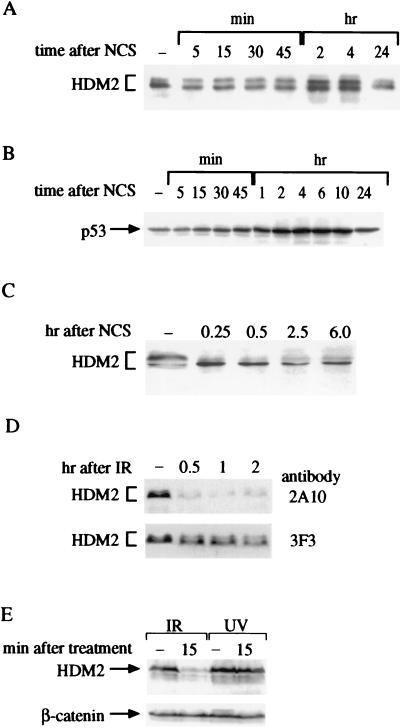
The electrophoretic mobility pattern of murine MDM2 and its human homolog, HDM2, was followed in cultured cells shortly after treatment with IR, UV radiation, or the radiomimetic chemical neocarzinostatin (NCS). Like IR, NCS induces DNA double-strand breaks by free radical attack and elicits the same hypersensitivity of A-T cells (38). HDM2 and MDM2 are found in several isoforms generated through alternative splicing and posttranslational modifications (ref. 39 and see below). A reproducible shift in the mobility of HDM2 toward a faster-migrating form was observed within minutes after treatment with NCS (Fig. 2A) or IR (not shown) well before the rise in p53 level (Fig. 2B). A similar phenomenon was observed in human p53-null Saos-2 cells (Fig. 2C), implying that this rapid, damage-induced alteration of HDM2 is not p53-dependent (see also Fig. 4B). It is of note that in most cases phosphorylation results in slower electrophoretic migration of the phosphorylated protein, although examples of faster migration after phosphorylation have been noted (40, 41).
Figure 2.
DNA damage-induced alterations of cellular HDM2. (A) HDM2 immunoprecipitated from cellular extracts of a human lymphoblastoid cell line (C3ABR) at various time points after addition of 80 ng/ml NCS. Immune complexes obtained by using the anti-HDM2 mAb IF2 were subjected to SDS/PAGE followed by immunoblotting with a mixture of mAbs 2A10, 1D6, 3G5, SMP14, and IF5. Note the rapid change toward accelerated electrophoretic mobility of HDM2 already apparent 5 min after treatment and the slower increase in its overall level that accompanies p53 accumulation (see Fig. 1B). (B) p53 levels in total cellular extracts of NCS-treated C3ABR lymphoblasts, detected by using the anti-p53 antibody DO-1. (C) HDM2 in total cellular extracts of p53-null Saos-2 cells. The blot was reacted with the same mixture of antibodies as in A. Note the shift of the major band toward a faster-migrating position. (D) Altered immunoreactivity of HDM2 after exposure to IR. Saos-2 cells were harvested at the indicated time points after treatment with 5 Gy of gamma radiation. Before their irradiation the cells were pretreated with 30 μM of the proteasome inhibitor MG132 for 2 hr to allow HDM2 accumulation. Total cellular extracts were immunoblotted with the two indicated anti-HDM2 mAbs. Note the differential decrease in immunoreactivity with 2A10 and the shift of the major (faster) band toward accelerated mobility observed by using 3F3. (E) Immunoreactivity of HDM2 with the 2A10 antibody decreases after exposure to ionizing but not UV radiation. Saos-2 cells were treated with either 10 Gy of gamma radiation or 30 J/m2 of UV radiation, and HDM2 was visualized as in Fig. 1D. In B–E, equal amounts of protein were loaded in all lanes in each. This is demonstrated in E by using a β-catenin antibody. In A, immunoprecipitates in each lane represent equal numbers of cells.
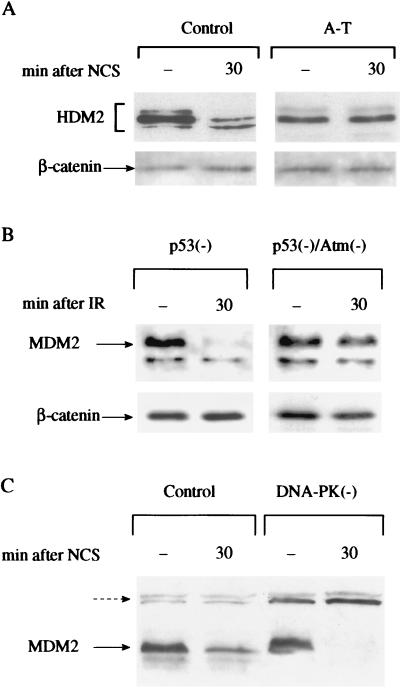
Figure 4.
Damage-induced modifications of HDM2 or MDM2 are dependent on ATM but not DNA-PK. (A) Immunoblot analysis of HDM2 in total cellular extracts of control (L-39) and A-T (L-6) lymphoblasts before and after NCS treatment. The blots were reacted with the same antibody mixture as in Fig. 2A. (B) 2A10 immunoreactivity of MDM2 in total cellular extracts of murine ap29 cells lacking p53 and ap24 cells lacking both p53 and Atm (37) before and after treatment with 7.5 Gy of ionizing radiation. (C) 2A10 immunoreactivity of MDM2 in total cellular extracts of A9 murine control cells and 494 cells derived from the “slip” mice lacking DNA-PK activity (25) after NCS treatment. Note the complete loss of 2A10 immunoreactivity in DNA-PK-deficient cells. Cross-reacting bands (broken arrow) indicate relative protein amounts in different lanes.
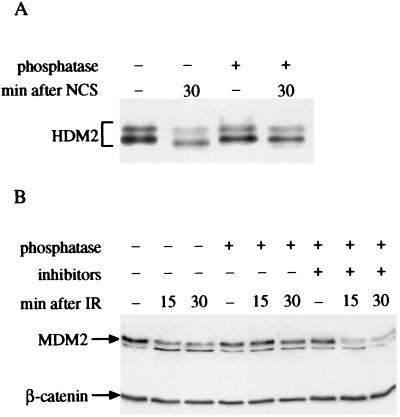
The structural modification of HDM2 also was manifested as a marked reduction in its immunoreactivity with the mAb 2A10, whereas reactivity with another mAb, 3F3, was retained (Fig. 2D). The 2A10 and 3F3 mAbs are directed against epitopes within the carboxyl-terminal half and the extreme amino-terminal portion of HDM2, respectively (42). This effect was not observed after treatment with UV radiation (Fig. 2E). Significantly, both types of alterations in HDM2 or MDM2 were reversed by treatment with phosphatase, but not in the presence of phosphatase inhibitors (Fig. 3), indicating that they arose through phosphorylation. Thus, rapid phosphorylation of HDM2/MDM2 followed treatment of cells with DNA breaking agents.
Figure 3.
Alkaline phosphatase (AP) reverses damage-induced HDM2 or MDM2 modifications. (A) Immunoblot analysis of HDM2 immunoprecipitated from a human lymphoblastoid cell line (NL553) treated with 80 ng/ml NCS. NCS-treated cells were lysed 30 min after addition of the drug to the cultures. Cellular extracts were immunoprecipitated with the IF2 antibody, and the immune complexes were treated with shrimp alkaline phosphatase as described in Materials and Methods and subjected to SDS/PAGE and immunoblotting with the same antibody mixture as in Fig. 2A. Note the reversion of HDM2's mobility shift by AP treatment. (B) Immunoblot analysis of murine MDM2 using the 2A10 antibody. p53-null mouse fibroblasts (ap29) infected with a recombinant retrovirus encoding mouse MDM2 were treated with 7.5 Gy of ionizing radiation and harvested at various time points after irradiation. Cellular extracts were incubated for 30 min at 30°C, without (lanes 1–3) or with (lanes 4–6) calf intestine alkaline phosphatase (CIAP), or with CIAP and phosphatase inhibitors (50 mM NaF, 10 mM NaVO4; lanes 7–9). The band migrating slightly ahead of MDM2 is a nonspecific-background band also observed in MDM2-null cells.
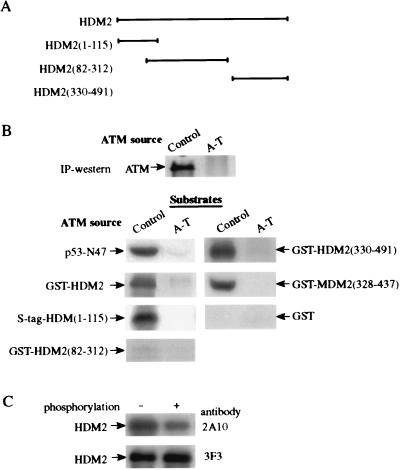
This modification of HDM2 was found to be ATM-dependent: it was observed in all six control human cell lines, but in none of six independent A-T cells lines (e.g., Fig. 4A) nor in murine cells lacking Atm (Fig. 4B). In contrast, cells derived from the “slip” mice that completely lack DNA-PK activity (25) maintained the ability to strongly reduce 2A10 immunoreactivity in response to IR (Fig. 4C), demonstrating that this phosphorylation was not dependent on DNA-PK.
Because these findings could be explained by HDM2 or MDM2 being direct targets of ATM's kinase activity, we tested whether these proteins can serve as ATM substrates in vitro. ATM immunoprecipitated from human lymphoblastoid cells (12) indeed did phosphorylate recombinant, full-length HDM2, as well as various fragments of HDM2 or MDM2 obtained as recombinant fusion proteins (Fig. 5 A and B). Importantly, this phosphorylation was accompanied by a reduction in HDM2 immunoreactivity with the 2A10 mAb (Fig. 5C). Although the degree of this reduction indicates that the efficiency of the phosphorylation in vitro is not 100%, it points to a similarity between the phosphorylation site(s) responsible for this phenomenon in vitro and in vivo. Phosphorylation was detected in the amino-terminal end of HDM2 and in a carboxyl-terminal region that, in MDM2, spans residues 328–437; an HDM2 fragment corresponding to positions 82–312 showed barely detectable phosphorylation (Fig. 5). These results indicate that ATM can phosphorylate HDM2 in vitro on at least two sites.
Figure 5.
In vitro phosphorylation of recombinant proteins by ATM. ATM was immunoprecipitated from control (L-40) and A-T (L-6) lymphoblastoid lines, and an immunoprecipitation-kinase reaction was performed for 15 min as described before (12). (A) Diagram showing various HDM2 fragments used as substrates (see Materials and Methods). The numbers in brackets denote the residues contained in each fragment. HDM2(1–115) was fused to an S tag, and all other fragments were produced as glutathione S-transferase (GST)-fusions. (B) In vitro phosphorylation of HDM2 and MDM2 fragments by ATM. Immunoprecipitated ATM was visualized by using immunoblotting. p53-N47, a polypeptide containing residues 1–47 of human p53 fused to the POU domain of the human transcription factor Oct-1 serving as a positive control substrate (12). Nonfused GST served as a negative control. (C) Reduction in immunoreactivity of recombinant HMD2 with the 2A10 mAb after in vitro phosphorylation by ATM. Full-length HDM2 was phosphorylated for 15 min by ATM and subsequently detected by immunoblotting with the antibodies 2A10 and 3F3.
Discussion
It is widely believed that posttranslational modifications play a key role in p53 stabilization and activation in response to DNA damage (3). Previous attempts to elucidate these mechanisms were focused on modifications of the p53 molecule and on their possible effect on the ability of p53 to bind to MDM2. Indeed, Shieh et al. (43) demonstrated that phosphorylation of p53 in vitro by DNA-PK on serines 15 and 37 reduces its affinity for MDM2. However, the dissociation obtained in the present study between Ser15 phosphorylation and damage-induced p53 stabilization (Fig. 1) suggests that protein modifications other than Ser15 phosphorylation may be involved in this process. DNA damage, indeed, does induce additional modifications throughout the p53 molecule (4–8). These modifications, particularly phosphorylation of Ser20 (7, 8), may contribute to abrogation of the p53-MDM2 association in some, though perhaps not all, situations (44). p53 modifications may also contribute to its transactivation capacity through enhanced DNA binding and expedited recruitment of specific p53 coactivators (5, 6, 45–47). The emerging picture is that of a complex activation–stabilization process associated with multiple protein modifications, each with an incremental contribution to the final outcome of this process.
Our findings presented herein disclose a new target of ATM-dependent protein modifications induced by DNA damage: the MDM2 protein. ATM-mediated phosphorylation of HDM2 precedes the initiation of damage-induced p53 accumulation (Fig. 2B), which is similar to the phosphorylation of Ser15 on p53 (11). The rapid phosphorylation of these two ATM targets correlates with the immediate enhancement of ATM's kinase activity after IR or radiomimetic treatment (12, 13). Thus, activated ATM may exert its effect on p53's activity and stability by mediating simultaneous phosphorylation of both partners of the p53-MDM2 autoregulatory feedback loop. It is noteworthy that this rapid phosphorylation differs significantly from MDM2's response to high-dose UV irradiation, which mainly compromises the rate of MDM2 gene expression (48).
DNA-PK also phosphorylates MDM2 in vitro (49); however, our data (Fig. 4C) imply that in cells it is dispensable for the type of MDM2 phosphorylation described in this study. This result correlates with recent evidence that damage-induced phosphorylation of Ser18, the equivalent of human Ser15 in murine p53, is also unaffected by lack of DNA-PK (18, 19), underscoring the distinction between in vitro and in vivo targets of protein kinases.
ATM phosphorylates in vitro residues within the amino-terminal end of HDM2 and a carboxyl-terminal region of HDM2/MDM2 (Fig. 5). Although fine mapping of these phosphorylation sites is in progress, it still is not clear whether the same sites are phosphorylated in vivo. Phosphorylation of the amino-terminal region may affect p53-MDM2 binding after radiation damage, because a nearby portion of MDM2 is directly involved in p53 binding (31, 32). On the other hand, modification of the MDM2 carboxyl-terminal domain, which is essential for targeting p53 for degradation (31, 33), may compromise the ability of MDM2 to destabilize p53, possibly interfering with its activity in p53 ubiquitination (36). Interestingly, Kubbutat et al. (33) showed recently that MDM2 portions that are critical for its degradative function overlap at positions 339–435, a region shown here to contain a major in vitro phosphorylation site of ATM (Fig. 5). Experiments are underway to determine the effects of ATM-mediated phosphorylation of MDM2 on its ability to bind p53 and mediate its degradation.
Our data provide a possible answer to recently raised speculations about the mechanism of damage-induced up-regulation of p53 (50), highlighting the major role of ATM in this process. Although ATM may contribute to p53 stabilization through modification of p53, we would like to propose that ATM-mediated phosphorylation of MDM2 also may play a significant role in this stabilization. These findings further elucidate the intricate web of signaling pathways that guard our genome.
Acknowledgments
We are indebted to Y. Ziv for valuable experimental advice, Y. Reiss for useful discussions, and Quark Biotech, Inc. (QBI) for their cooperation in this work. We thank A. Levine and D. Freedman for hybridoma cell lines and the S-tagged(1–115)HDM2 fragment, C. Westphal for the ap24 and ap29 cells, C. Jhappan for the 494 cells, G. Lozano for the p53-deficient and p53/MDM2-deficient cells, M. Lavin for the C3ABR cell line, and T. Unger for the CMV-p53(S15A) construct. This work was supported by research grants from the A-T Medical Research Foundation, the A-T Children's Project, the National Institutes of Health (RO1 NS31763), the Thomas Appeal (A-T Medical Research Trust), and the A-T Appeal (to Y.S.), as well as the National Institutes of Health (RO1 CA40099) and the Centers of Excellence Program administered by the Israeli Academy of Sciences and Humanities (to M.O.). T.G. is a recipient of a postdoctoral fellowship from the Israel Cancer Research Fund and the Feinberg Graduate School. This work was carried out in partial fulfillment of the requirements for the M.Sc. degree by R.K.
Abbreviations
- DNA-PK
DNA-dependent protein kinase
- IR
ionizing radiation
- A-T
ataxia-telangiectasia
- NCS
neocarzinostatin
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Gottlieb T M, Oren M. Biochem Biophys Acta Rev Cancer. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal M L, Taylor W R, Chemov M V, Chemova O B, Stark G R. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Giaccia A J, Kastan M B. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 5.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Anderson C W, Appella E. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterman M J F, Stavridi E S, Waterman J L F, Halazonetis T D. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 7.Shieh S-Y, Taya Y, Prives C. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastan M B, Zhang Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 10.Khanna K K, Lavin M F. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 11.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1999;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 13.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 14.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, et al. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 15.Tibbetts R S, Brunbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S-Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burma S, Kurimasa A, Xie G, Taya Y, Araki R, Abe M, Crissman H A, Quyang H, Li G C, Chen D J. J Biol Chem. 1999;274:17139–17143. doi: 10.1074/jbc.274.24.17139. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez G S, Bryntesson F, Torres-Arzayus M I, Preisley A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo P A, Taccioli G E, et al. Nature (London) 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- 19.Araki R, Fukumura R, Fujimori A, Taya Y, Shiloh Y, Kurimasa A, Burma S, Li G C, Chen D J, Sato K, et al. Cancer Res. 1999;59:3543–3546. [PubMed] [Google Scholar]
- 20.Hoekstra M F. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- 21.Rotman G, Shiloh Y. Hum Mol Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 22.Rotman G, Shiloh Y. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- 23.Smith G C, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 24.Shiloh Y. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 25.Jhappan C, Morse H C, III, Fleischmann R D, Gottesman M M, Merlino G. Nat Genet. 1997;17:483–486. doi: 10.1038/ng1297-483. [DOI] [PubMed] [Google Scholar]
- 26.Prives C. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 27.Juven-Gershon T, Oren M. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- 28.Freedman D A, Wu L, Levine A J. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Midgley C A, Lane D P. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 30.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 31.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 32.Bottger A, Bottger V, Garcia-Echeverria C, Chene P, Hochkeppel H K, Sampson W, Ang K, Howard S F, Picksley S M, Lane D P. J Mol Biol. 1997;269:744–756. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- 33.Kubbutat M H, Ludwig R L, Levine A J, Vousden K H. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- 34.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao W, Levine A J. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westphal C H, Schmaltz C, Rowan S, Elson A, Fisher D E, Leder P. Cancer Res. 1997;57:1664–1667. [PubMed] [Google Scholar]
- 38.Goldberg I H, Kappen L S. In: Enediyne Antibiotics as Antitumor Agents. Borders D B, Doyle T W, editors. New York: Dekker; 1995. pp. 327–362. [Google Scholar]
- 39.Olson D C, Marechal V, Momand J, Chen J, Romocki C, Levine A J. Oncogene. 1993;8:2353–2360. [PubMed] [Google Scholar]
- 40.Dulic V, Lees E, Reed S I. Science. 1992;257:1958–1960. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 41.Dell'Angelica E C, Ooi C E, Bonifacino J S. J Biol Chem. 1997;272:15078–15084. doi: 10.1074/jbc.272.24.15078. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shieh S-Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 44.Ashcroft M, Kubbutat M H G, Vousden K H. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hupp T R, Meek D W, Midgley C A, Lane D. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 46.Gu W, Roeder R G. Cell. 1996;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 47.Lambert P F, Kashanchi F, Radonovich M F, Shiekhatter R, Brady J N. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 48.Wu L, Levine A. Mol Med. 1997;3:441–451. [PMC free article] [PubMed] [Google Scholar]
- 49.Mayo L D, Turchi J J, Berberich S J. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 50.Lane D. Nature (London) 1998;394:616–617. doi: 10.1038/29166. [DOI] [PubMed] [Google Scholar]