Abstract
Many virulence-related, bacterial effector proteins are translocated directly into the cytosol of host cells by the type III secretion (TTS) system. Translocation of most TTS effectors requires binding by specific chaperones in the bacterial cytosol, although how chaperones promote translocation is unclear. To provide insight into the action of such chaperones, we studied the consequences of binding by the Yersinia chaperone SycE to the effector YopE by NMR. These studies examined the intact form of the effector, whereas prior studies have been limited to well ordered fragments. We found that YopE had the characteristics of a natively unfolded protein, with its N-terminal 100 residues, including its chaperone-binding (Cb) region, flexible and disordered in the absence of SycE. SycE binding caused a pronounced disorder-to-order transition in the Cb region of YopE. The effect of SycE was strictly localized to the Cb region, with other portions of YopE being unperturbed. These results provide stringent limits on models of chaperone action and are consistent with the chaperone promoting formation of a three-dimensional targeting signal in the Cb region of the effector. The target of this putative signal is unknown but appears to be a bacterial component other than the TTS ATPase YscN.
A large number of Gram-negative bacterial pathogens translocate virulence-related effector proteins directly into the cytosol of eukaryotic host cells via the type III secretion (TTS)2 system (1). Translocation of most effectors requires their prior association in the bacterial cytosol with specific dimeric chaperone proteins (2). A large family of such chaperones exists, with individual chaperones functioning in the translocation of only a single or just a few corresponding effectors. Although chaperones have limited sequence identity (≤ 20%), their folds and modes of effector binding are well conserved (3–13). Chaperone dimers provide rigid and globular surfaces, around which effectors wrap an ∼25–100-residue chaperone-binding (Cb) region in strikingly extended conformation (4, 6, 9, 12, 13). This conformation is generally similar among effectors despite lack of obvious sequence homology.
Two models have been proposed to explain the role of the extended conformation of the effector Cb region. In the first, the extended conformation is considered to maintain or prime unfolding of the effector (4) for transport through the narrow ∼20–30 Å diameter bore of the TTS needle (14). In the second, the extended conformation is suggested to form a discrete three-dimensional signal targeting the chaperone-effector complex to a TTS component required for translocation (6). Recent results demonstrating binding between a Salmonella chaperone-effector complex and the TTS ATPase as well as ATP hydrolysis-dependent unfolding of the effector by the ATPase are consistent with both unfolding and targeting models (15).
To provide further insight into mechanisms of chaperone action, we investigated the structural and dynamic changes an intact effector undergoes upon chaperone binding. Prior studies have been incomplete, being limited to crystallographic studies of structured portions of protein fragments. This has been due to the aggregation-prone nature of free intact effectors and the intractability of intact chaperone-effector complexes to crystallization. To circumvent these issues, we devised methods for obtaining free intact YopE, a well studied Yersinia pseudotuberculosis effector, and intact SycE-YopE chaperone-effector complexes at concentrations and conditions suitable for NMR spectroscopy.
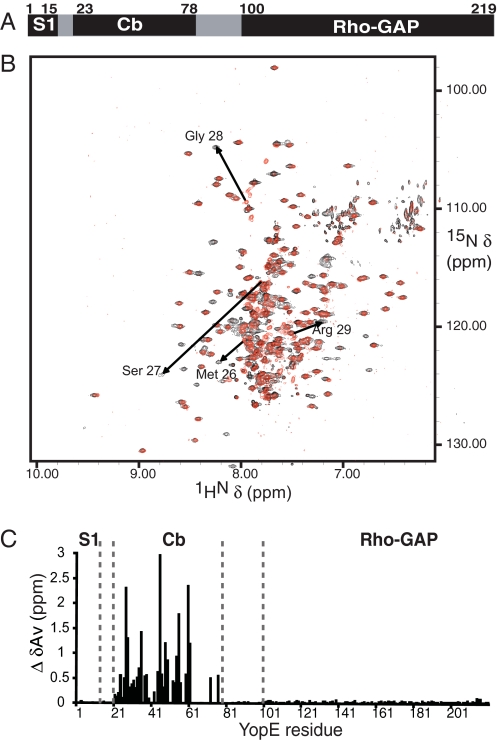
In common with other effectors, YopE requires not only the Cb region (residues 23–78) for translocation (16) but also a second discrete region, termed here signal 1 (S1) (Fig. 1A). Although one line of evidence indicates that S1 is composed of the N-terminal ∼15 amino acids of YopE (17), other results indicate it is composed of the first ∼15 mRNA codons of YopE (18). These possibilities are not exclusive, and indeed ribonucleic and proteinaceous signals may be important at different times after host cell contact (2). Following the S1 and Cb regions in effectors are host cell interaction domains, which in YopE consist of a single functionality. Residues 100–219 of YopE have RhoGAP activity (Fig. 1A) (19), which is required for disrupting host cell actin and antagonizing phagocytic uptake of Y. pseudotuberculosis by macrophages.
FIGURE 1.
Effect of SycE binding confined to the Cb region of YopE. A, functional YopE regions: S1, the Cb region, and the RhoGAP domain. B, 15N,1H TROSY NMR spectra of free (red) and SycE-bound (black) 15N,2H-labeled YopE. Arrows show examples of YopE cross-peaks differing between free (arrow tail) and SycE-bound forms (arrowhead). C, the difference between chemical shifts (ΔδAv) for the spectrum in B assigned to YopE residues.
The studies reported here revealed that the N-terminal 100 residues of YopE, including the S1 and Cb regions, were disordered and flexible in the absence of bound SycE, as is characteristic of natively unfolded proteins. SycE binding brought about a pronounced disorder-to-order transition in the Cb region but had no effect on other portions of YopE. These results provide support for a targeting model of chaperone action and are inconsistent with unfolding models. No association between SycE-YopE and the Yersinia TTS ATPase YscN was detected, suggesting that the target of the putative signal in the SycE-YopE complex is a Yersinia component other than YscN.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—His-tagged SycE-YopE complexes were prepared as described previously (6), except as follows. For production of 15N-labeled, 13C-labeled, or dually labeled SycE-YopE, bacteria were grown at 37 °C to mid-log phase in minimal medium containing 15N-labeled (NH4)2SO4 (1 g/liter), [13C] glucose (2 g/liter), or both; induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside; and grown for 18 h at 25 °C. For production of triple labeled (2H,15N,13C) SycE-YopE, bacteria were grown in 2X-YT medium at 37 °C to mid-log phase, at which point bacteria were harvested by centrifugation (10,000 × g, 20 min, 4 °C), washed twice in M9 salts, and resuspended in a 0.25 volume of minimal medium containing D2O, 15N-labeled (NH4)2SO4, and [13C] glucose. Bacteria were then grown for 20 h at 25 °C.
Bacteria were harvested by centrifugation and lysed by sonication in lysis buffer (500 mm NaCl, 50 mm sodium phosphate, pH 8.0, 5 mm imidazole, 5 mm MgCl2, and 10 mm β-mercaptoethanol supplemented with 1 mm phenylmethylsulfonyl fluoride and 20 μg/ml DNase). The lysate was clarified by centrifugation (30,000 × g, 20 min, 4 °C), and SycE-YopE was applied to a Ni2+ chelation column and eluted using a 5–500 mm imidazole gradient in lysis buffer. The eluate was concentrated by ultrafiltration (30-kDa molecular weight cut off), and further purified by size-exclusion chromatography (Superdex 75) in 500 mm NaCl, 50 mm sodium phosphate, pH 8.0, and 1 mm dithiothreitol.
The YopE(RhoGAP) fragment (residues 81–219), containing a C-terminal His tag (LEHHHHHH), was expressed using pET28b (Novagen) and produced with isotopic labeling and purified as above.
A biotinylated version of SycE-YopE, with YopE containing a C-terminal 21-residue biotinylation sequence (KLPAGGGLNDIFEAQKIEWHE), was expressed using pAC-6 in Escherichia coli AVB101 cells (Avidity). In vivo biotinylated SycE-YopE was purified as described previously (6), except that precipitation was carried out with 50% (NH4)2SO4, and the protein was dialyzed in Tris buffer, pH 8.
Denaturation and Renaturation—SycE-YopE was denatured in 8 m urea and 50 mm sodium phosphate, pH 8.0, and bound to nickel-nitrilotriacetic acid-agarose beads. Denatured YopE was eluted from these beads with 8 m urea, 50 mm sodium phosphate, pH 4.0, and renatured by dilution in 1 m arginine, 500 mm NaCl, 50 mm Tris, pH 8.0, 1 mm dithiothreitol, and 5 mm EDTA at a final concentration of 1 μm (determined by A280). For renaturation of SycE-YopE complexes, a 2-fold molar excess of denatured, unlabeled SycE was included with YopE. SycE was isolated from the unbound fraction of unlabeled, denatured SycE-YopE applied to nickel-nitrilotriacetic acid-agarose beads or purified in its native form as described previously (3). Renaturation solutions containing YopE alone or YopE mixed with SycE were stirred vigorously for a minimum of 1 h at 4 °C and then dialyzed in 500 mm NaCl, 50 mm Tris, pH 8.0, 1 mm dithiothreitol, and 5 mm EDTA. Renatured and aggregated proteins were separated by size-exclusion chromatography (Superdex 200) in 50 mm sodium phosphate, pH 6.1.
NMR Data Collection—NMR spectra for SycE-YopE, free YopE, and YopE(RhoGAP) were typically measured at protein concentrations of 15–60, 2–4, and 45 mg/ml, respectively. Samples for NMR analysis were prepared in 45 mm sodium phosphate buffer, pH 6.1 (95% H2O, 5% D2O), and all NMR spectra were collected at 298 K and 700-MHz 1H frequency on a Bruker Avance spectrometer with a cryoprobe. Sequential assignment was analyzed with the program CARA (20), and stretches of sequentially connected spin systems were mapped to the amino acid sequence of YopE with the program MAPPER (21). For free YopE, all but four of 191 resonances were assigned, and for SycE-YopE, all but 25 resonances were assigned, of which about half are from the RhoGAP domain. ΔδAv represents √((Δ1H)2 + 0.2(Δ15N)2)/2.
Binding Experiments with Biotinylated SycE-YopE and YopE— Biotinylated SycE-YopE (650 μg) was incubated with 100 μl of streptavidin-agarose beads for 1 h at 25 °C in 500 μl of binding buffer (150 mm NaCl, 50 mm sodium phosphate buffer, pH 8.0, 5% glycerol, 1% Triton X-100, and 10 mm β-mercaptoethanol), and beads were washed three times with 700 μl of binding buffer. A portion of the streptavidin-agarose beads containing SycE-YopE was washed 15 times with 700 μl of 10 mm glycine, pH 2.8, to remove SycE. YopE remaining on beads was renatured by washing five times with binding buffer. This procedure completely removed SycE and resulted in only slight loss of YopE, as assayed by Western blotting and silver staining.
Lysates were prepared from Y. pseudotuberculosis 126 and Y. pseudotuberculosis 126(ΔyopE). This latter strain was generated through standard allelic exchange procedures, with the coding region for yopE being replaced by the coding region for kanR.3 Wild-type and ΔyopE Y. pseudotuberculosis were grown in BHI medium at 37 °C until mid-log density, and type III secretion was induced by addition of sodium oxalate and MgCl2 to final concentrations of 20 mm. After 3 h, bacteria were harvested by centrifugation (10,000 × g, 20 min, 4 °C), resuspended in 25 ml of binding buffer (supplemented with 1 mm phenylmethylsulfonyl fluoride and 0.5 mm E-64) per liter of bacterial culture, and lysed by sonication. The lysate was clarified by centrifugation (30,000 × g, 20 min, 4 °C), and endogenous biotinylated proteins were removed by incubation with 400 μl of streptavidin-agarose beads (Pierce) for 1 h at 4 °C, followed by centrifugation (3,500 × g, 5 min, 4 °C).
Lysates (∼333 μl) from wild-type Y. pseudotuberculosis cultures (∼8 ml) were added to 100 μl of streptavidin-agarose beads containing bound SycE-YopE; lysates from the ΔyopE strain were added to streptavidin-agarose beads containing only bound YopE. Resulting slurries were rocked for 95 min at 4 °C and washed three times for wild-type lysate and 30 times for ΔyopE lysate with 700 μl of binding buffer. Bound proteins were removed from beads by boiling in 2× SDS-PAGE sample buffer, resolved by 12% SDS-PAGE, and transferred to a polyvinylidene difluoride membrane. Membranes were probed with anti-YscN (gift of O. Schneewind) (22) or anti-SycE rabbit polyclonal antibodies, for which binding was visualized using horse-radish peroxidase-conjugated goat anti-rabbit secondary antibodies (Santa Cruz Biotechnology) and ECL Plus (Amersham Biosciences). Anti-SycE antibodies were produced in rabbits by standard means using purified SycE as an antigen (Antibodies Inc.).
RESULTS
Renaturation of Free and SycE-bound YopE for NMR Studies— To carry out NMR characterization of the 52-kDa SycE-YopE complex, selectively labeled SycE-YopE complexes were prepared by a denaturation-renaturation protocol. Such complexes contained unlabeled SycE and isotopically labeled (with 15N, 13C, 2H, or a combination of these) YopE. For this procedure, biosynthetically labeled SycE-YopE complexes were recombinantly expressed and purified. Labeled YopE (23 kDa) was dissociated from labeled dimeric SycE (29 kDa/dimer) by urea-induced denaturation and then renatured with unlabeled SycE in a non-chaotropic buffer to produce selectively labeled SycE-YopE. Free YopE was generated similarly, except that SycE was omitted during renaturation.
Renatured SycE-YopE was verified to have circular dichroism spectra and gel filtration chromatography profiles indistinguishable from those of catalytically active complexes isolated in its native form (i.e. without denaturation; data not shown) (6). However, NMR analysis provided the most precise verification for renatured SycE-YopE and free YopE. 15N-1H transverse relaxation optimized spectroscopy (TROSY) spectra of renatured YopE and SycE-YopE (supplemental Fig. S1) were acquired and compared with that of YopE(RhoGAP), a truncated form of YopE. This truncated form contains only the RhoGAP domain (residues 81–219) and was isolated in its native form (i.e. without denaturation steps). A similar fragment (residues 90–219) was crystallized and demonstrated to have RhoGAP catalytic activity (19, 23). Without exception, all 147 cross-peaks in the 15N-1H TROSY spectrum of YopE(RhoGAP) superimposed with cross-peaks in the spectrum of renatured YopE as well as that of SycE-YopE. This complete superimposition indicated that renatured YopE and SycE-YopE had properly folded conformations in their RhoGAP domains and were suitable for structural analysis.
No Effect of SycE Binding on the YopE RhoGAP Domain—15N-1H TROSY spectra of YopE and SycE-YopE were compared to evaluate the effects of SycE binding on YopE. We observed 191 cross-peaks for the 15N-1H moieties of free YopE and 244 for SycE-bound YopE (Fig. 1B). The 244 YopE resonances in SycE-YopE were in excess of the 214 resonances expected (including a His tag, see “Experimental Procedures”), suggesting the existence of multiple conformations and possibly the incomplete suppression of side chain resonances. More importantly, the great majority of free YopE resonances (151 of 191) overlapped (<0.1 ppm change) with SycE-YopE resonances (Fig. 1B), which indicated that SycE binding had a limited effect on YopE. The 40 YopE resonances that had no counterparts in the SycE-YopE spectrum (i.e. those shifted upon SycE binding) were also notably absent in the spectrum of the YopE(RhoGAP) fragment. This indicated that SycE exerted its effect within the N-terminal 80 residues of YopE and had no effect on the RhoGAP domain.
Localized Effect of SycE on YopE—To characterize the effects of SycE on YopE at the resolution of individual residues, we assigned cross-peaks in 15N-1H TROSY spectra to corresponding residues. We acquired sequential backbone assignments for 2H,13C,15N-labeled YopE in its free or SycE-bound states using three-dimensional TROSY-HNCA, TROSY-HNCACB, and 15N-resolved [1H,1H] NOE spectroscopy (with a 100-ms mixing time) for the collection of 1HN-1HN NOEs (24–27).
For free YopE, 91% of possible assignments were made. Residues lacking assignments were Met1 and Lys2 in S1; Leu55–Arg58, Arg62–Ile71, Phe73, Ile74, and Phe78 in Cb; and Gln149 in the RhoGAP domain. These gaps in the Cb region were likely due to overlapping resonances. For SycE-bound YopE, 95% of possible assignments were made. Residues lacking assignments were Met1 and Lys2 in S1; Ser36, Asn41, Leu43, Ser64, Ser69, Phe73, and Arg76 in the Cb region; and Gln149 in the RhoGAP domain. The assignment process for SycE-YopE was complicated by the fact that ∼25% of residues in the Cb region showed duplicated chemical shifts, and several residues in this region showed very weak peak intensities, in particular the segment comprising residues 67–78. Duplicated resonances likely arose from local conformational exchange induced by proline cis,trans-isomerization, conformational heterogeneity in regions with poorly defined secondary structure, or both. These characteristics are consistent with high temperature factors for the Cb region in the crystal structure of SycE-YopE(Cb) (6), especially in the α-helical region formed by residues 67–78.
Thirty-five of the 40 15N,1H TROSY cross-peaks of free YopE that were shifted upon SycE binding were assigned in both free and chaperone-bound states. Notably, these shifted and dually assigned cross-peaks all mapped to residues in the Cb region (Fig. 1C). The magnitude of these shifts was large, indicating that a major structural transition had been brought about by SycE binding. Four residues within the Cb region had resonances that were unshifted: Gln51, Ser65, Val66, and Ala67. The crystal structure of the SycE-YopE(Cb) fragment shows that these residues are not contacted by SycE and lack regular secondary structure (6). Comparison of dually assigned residues also revealed that SycE binding had no effect on the S1 region and likewise no effect on residues 81–100 connecting the Cb region to the RhoGAP domain. These results provide direct evidence for the effect of SycE binding being strictly localized to the Cb region of YopE and not extending outside the Cb region.
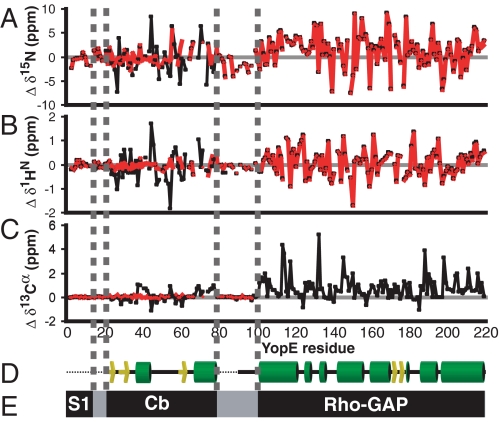
Random Coil in the Cb Region of Free YopE—We next addressed the issue of whether the Cb region of free YopE was in a folded state. Backbone chemical shifts of free YopE were compared with those expected for a random coil state. The deviations from random coil values of secondary chemical shifts (Δδ) in 15N, 1H, and 13C dimensions of the Cb region of free YopE were found to be close to zero, indicating that this region was in a random coil state (Fig. 2). In contrast, when SycE was bound, these same residues of YopE had values significantly different from those typical for a random coil state (Fig. 2) (Δδ ≠ 0). The deviation from random coil values of 13C-α chemical shifts in SycE-bound YopE suggested an α-helical secondary structure for Asp37–Leu43 and Ala67–Ser79 and a β-strand secondary structure for Gly24–Gln34, Glu48–Gly52, and Arg58–Leu63, agreeing almost exactly with the crystal structure of the SycE-YopE(Cb) complex (6). The only exception was Glu48–Gly52, which lacks β-strand characteristics in the crystal structure. These results provide evidence for SycE promoting a transition in the Cb region from an unstructured state to a structured conformation.
FIGURE 2.
Random coil conformation of the N-terminal half of free YopE. Chemical shift deviations from those expected for random coil conformation (Δδ, secondary chemical shifts) of 15N(A), 1H(B), and 13C-α (C) nuclei for free (red) or SycE-bound (black) 15N,2H,13C-labeled YopE after application of sequence-dependent corrections (36). 13C-α secondary chemical shifts were not observed for residues of the RhoGAP domain of free YopE, because of low sensitivity of the experiment, low sample concentration, and the highly structured nature of these residues. D, the secondary structure of YopE from crystal structures of SycE-YopE(Cb) (6) and the RhoGAP (23) domain aligned with secondary chemical shifts. Yellow arrows, β-strands; green cylinders, α-helices; black lines, loops. Regions not present in crystal structures are depicted as dotted lines. E, functional regions of YopE aligned with secondary chemical shifts and secondary structure.
As expected, there were large deviations from random coil values in secondary chemical shifts of the RhoGAP domain for both free and SycE-bound YopE (Fig. 2). 13C-α chemical shifts for this region were in close agreement with the crystal structure of this domain (23). Residues 79–101 connecting the Cb region to the RhoGAP domain lacked detectable secondary structure regardless of SycE binding (Fig. 2), indicating that these residues likely serve as an unstructured linker.
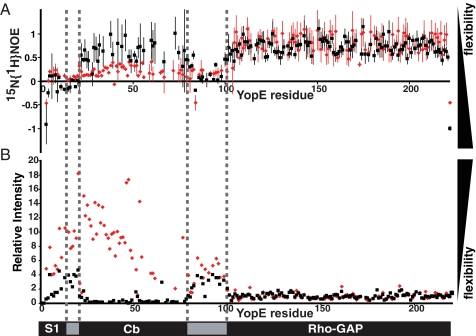
Disorder-to-Order Transition upon SycE Association—We next examined dynamic properties of free YopE to ask whether the Cb region, despite having random coil-like chemical shifts, was discretely structured. A heteronuclear 15N{1H} NOE experiment was carried out to examine the mobility of amide 1HN-15N bond vectors (28). In this experiment, values close to 1 correspond to 15N,1H moities that are well structured (i.e. values of 0.8–1.0 are indicative of a rotational correlation time that is greater than a few ns), and values <0.5 are indicative of an unstructured and flexible state (i.e. values <0 are indicative of a rotational correlation time of <1 ns). The 15N,1H moieties of the RhoGAP domain had 15N{1H} NOE values close to 1 in both free and SycE-bound YopE (Fig. 3A), indicating that the RhoGAP domain was well structured both in the presence and absence of SycE. In contrast, the N-terminal 98 residues of free YopE had markedly lower 15N{1H} NOE values (Fig. 3A), diagnostic of an unstructured and flexible state. The 15N{1H} NOEs of the Cb region were between 0.0 and 0.4, which indicated that this region had some local and limited ordering.
FIGURE 3.
Dynamics of free and SycE-bound YopE. A, 15N{1H} NOE intensity values are plotted for free (red) and SycE-associated (black) 15N,2H-labeled YopE. Error bars were calculated from the spectral noise of the reference experiments. B, 15N,1H TROSY cross-peak intensities are plotted for free (red) and SycE-bound (black) 15N-labeled YopE, with intensities normalized to the mean intensity of resonances from the RhoGAP domain. Samples were prepared by adding either phosphate buffer or unlabeled SycE to otherwise identical aliquots of free 15N-labeled YopE.
Upon addition of SycE to free YopE, 15N{1H} NOE values in the Cb region (i.e. residues 22–83) rose considerably (Fig. 3A), providing evidence that the Cb region had become structurally constrained by SycE. Similarly, the intensities of the cross-peaks in the 15N,1H TROSY spectrum throughout the Cb region were significantly elevated in free YopE as compared with SycE-bound YopE, consistent with a disordered and flexible state in this region when free but structurally constrained when SycE-bound (Fig. 3B). These results demonstrated that the Cb region was unstructured and flexible in free YopE but nevertheless competent to bind SycE and that SycE binding caused a pronounced disorder-to-order transition within this region.
Disorder of S1 with or without SycE—The cross-peaks of the 15N,1H TROSY spectra for the N-terminal 20 residues in free and SycE-bound forms of YopE superimposed well, indicating that SycE had no effect on the S1 region (Fig. 1). Based on the random coil-like chemical shifts (Fig. 2) and the negative 15N{1H} NOE values (Fig. 3), the S1 region appeared to be unstructured and flexible in both free and in SycE-bound YopE. It is noteworthy that in the flagellar export system, which is related to TTS systems, the N-terminal secretion signals of exported flagellar proteins are also disordered (29).
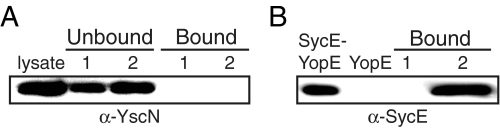
No Association between SycE-YopE and YscN—A disorder-to-order transition imposed by SycE supports a targeting mechanism of chaperone action. Because association of chaperones and effectors and chaperone-effector complexes with membrane-associated TTS ATPases has been detected in Salmonella and E. coli (15, 30), we sought to determine whether SycE-YopE, free YopE, or both associated with the Y. pseudotuberculosis TTS ATPase YscN (48 kDa). A detergent-solubilized Y. pseudotuberculosis lysate was prepared using a protocol similar to that used for detecting association in Salmonella and E. coli. The supernatant of this detergent-solubilized lysate was applied to streptavidin-agarose beads containing SycE-YopE complexes immobilized via site-specific biotinylation of YopE. Although YscN was evident in the supernatant of the detergent-solubilized lysate and in material that did not adhere to the beads, no YscN was detected bound to SycE-YopE (Fig. 4A). As a positive control, the supernatant of a detergent-solubilized lysate from Y. pseudotuberculosis (ΔyopE) was applied to biotinylated free YopE immobilized on streptavidin-agarose beads, and SycE was found to specifically associate with free YopE as expected (Fig. 4B). Again, no YscN was detected (data not shown), even though ∼13-fold more sample was applied to the YscN immunoblot as compared with the SycE immunoblot.
FIGURE 4.
Lack of association between SycE-YopE and YscN. A, YscN was present in a Y. pseudotuberculosis detergent-solubilized lysate, as detected by immunoblotting with anti-YscN polyclonal antibodies. The lysate was applied to unmodified streptavidin-agarose beads (1) or streptavidin-agarose beads with biotinylated SycE-YopE attached (2). YscN was present in unbound fractions but not in bound fractions for beads 1 and 2. B, SycE, as detected by immunoblotting with anti-SycE polyclonal antibodies, from a detergent-solubilized lysate of Y. pseudotuberculosis (ΔyopE) did not bind unmodified streptavidin-agarose beads (1) but did bind streptavidin-agarose beads with biotinylated free YopE attached (2). As controls, SycE was detected in biotinylated SycE-YopE complexes but not in biotinylated free YopE.
DISCUSSION
TTS effectors have been observed to associate with their specific chaperones through a conserved binding mode. The most distinctive feature of this mode is the highly extended conformation of the effector Cb region, as seen for fragments of Salmonella SptP (4) and SipA (13) and Yersinia YopE (6), YscM2 (9), and YopN (12) bound to their specific chaperones. However, the functional importance of this binding mode has remained unclear. To inform models of chaperone action, we investigated the consequences of SycE binding on the dynamics and structure of intact YopE through NMR studies.
Three major conclusions derived from these studies. First, the effect of SycE binding was strictly localized and isolated to the Cb region of YopE, as deduced from comparison of 15N,1H TROSY spectra (Fig. 1). Binding by SycE had no effect on the YopE S1 region, the RhoGAP domain, or the linker region connecting Cb to the RhoGAP domain. Second, the Cb region in the absence of SycE appeared to be flexible and disordered, as evidenced by chemical shift analysis, 15N{1H} NOEs, and 15N,1H TROSY signal intensities (Fig. 3). It must be noted that the Cb region of free YopE had 15N{1H} NOE values between 0 and 0.4 (Fig. 3), which suggests some amount of local and limited ordering in this region. Such features may contribute to reducing the entropic cost of binding SycE. These data also revealed that nearly half of free YopE was unstructured and flexible (Fig. 5), thus identifying YopE as a member of the natively unfolded protein family (31). In host cells, the disordered state of the N-terminal region of YopE may be required for association with host cell membranes (32), as has been suggested for several effectors (33). Third, as also revealed by chemical shifts, 15N{1H} NOEs, and 15N,1H TROSY NMR signal intensities, a pronounced disorder-to-order transition in the YopE Cb region was brought about by SycE binding.
FIGURE 5.
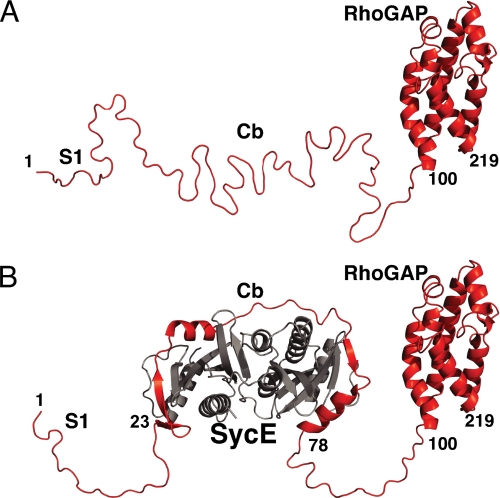
Free and SycE-bound YopE. A, a ribbon representation of free YopE, with the crystal structure of the RhoGAP domain (23) depicted and other portions in modeled random coil conformation. B, a ribbon representation of the SycE-YopE complex, with the conformation of SycE (gray) and the YopE Cb region corresponding to the crystal structure of SycE-YopE(Cb) (6) and the conformation of the YopE RhoGAP domain corresponding to its crystal structure (23); other portions are depicted in modeled random coil conformation.
These conclusions are at odds with models of chaperone action that require consequences outside the Cb region, for example in priming unfolding of C-terminal host-interaction domains or ensuring proper presentation of the S1 region. These conclusions are also inconsistent with models in which chaperone binding maintains the Cb region in an unfolded state (4). Rather than maintaining an unfolded state in the effector, SycE was found to promote structuring of the YopE Cb region. Taken together with structural conservation of chaperone-effector complexes, our results are most consistent with a targeting model of action in which the Cb region in association with the chaperone constitutes a three-dimensional targeting signal (6). The strongest experimental support for the targeting model comes from a Yersinia strain deleted of most of its effectors (ΔHOPEM) (34). In this strain, neither SycE nor the Cb region of YopE is required for translocation, whereas in wild-type strains both are required. Dispensability of SycE and the Cb region in the ΔHOPEM strain suggests that chaperones have a role in enhancing interaction between effectors and a TTS component that is required for translocation and that is also limiting in a wild-type background (i.e. in the presence of other chaperone-effector complexes). Additional evidence for competition between chaperone-effector complexes exists (35).
Whether this potentially limiting component is the TTS ATPase is not clear. In Salmonella, the SicP-SptP chaperone-effector complex interacts with the TTS ATPase InvC, but the chaperone SicP is sufficient for this interaction (15). Likewise, in E. coli the chaperone CesT is sufficient for interaction with the TTS ATPase EscN and so too is the effector Tir (30). Based on these data, it seems improbable that the TTS ATPase is the receptor for a putative translocation signal carried by the chaperone-effector complex, unless such complexes bind more strongly to the TTS ATPase than isolated chaperones and effectors. The relative affinities of chaperone-effector complexes as compared with individual components are not known. In Yersinia, interaction between SycE-YopE and the TTS ATPase YscN was not detected. Although multiple explanations exist for this result, it brings up the possibility that the putative receptor for a targeting signal in the SycE-YopE complex is a bacterial component other than YscN.
Supplementary Material
Acknowledgments
We thank G. Royappa and W. Kwiatkowski for help with NMR experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 GM007240 (to L. R.) and R01 AI061452 (to P. G.). This work was also supported by grants from the Universitywide AIDS Research Program (to L. R.) and the National Science Foundation Bridges to the Doctorate Program (to A. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: TTS, type III secretion; Cb, chaperone-binding; S1, signal 1; TROSY, transverse relaxation optimized spectroscopy; NOE, nuclear Overhauser effect.
L. Rodgers and S. Birtalan, manuscript in preparation.
References
- 1.Cornelis, G. R. (2006) Nat. Rev. Microbiol. 4 811-825 [DOI] [PubMed] [Google Scholar]
- 2.Ghosh, P. (2004) Microbiol. Mol. Biol. Rev. 68 771-795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birtalan, S., and Ghosh, P. (2001) Nat. Struct. Biol. 8 974-978 [DOI] [PubMed] [Google Scholar]
- 4.Stebbins, C. E., and Galan, J. E. (2001) Nature 414 77-81 [DOI] [PubMed] [Google Scholar]
- 5.Luo, Y., Bertero, M. G., Frey, E. A., Pfuetzner, R. A., Wenk, M. R., Creagh, L., Marcus, S. L., Lim, D., Sicheri, F., Kay, C., Haynes, C., Finlay, B. B., and Strynadka, N. C. (2001) Nat. Struct. Biol. 8 1031-1036 [DOI] [PubMed] [Google Scholar]
- 6.Birtalan, S. C., Phillips, R. M., and Ghosh, P. (2002) Mol. Cell 9 971-980 [DOI] [PubMed] [Google Scholar]
- 7.Singer, A. U., Desveaux, D., Betts, L., Chang, J. H., Nimchuk, Z., Grant, S. R., Dangl, J. L., and Sondek, J. (2004) Structure (Lond.) 12 1669-1681 [DOI] [PubMed] [Google Scholar]
- 8.van Eerde, A., Hamiaux, C., Perez, J., Parsot, C., and Dijkstra, B. W. (2004) EMBO Rep. 5 477-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan, J., Tropea, J. E., and Waugh, D. S. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1591-1599 [DOI] [PubMed] [Google Scholar]
- 10.Buttner, C. R., Cornelis, G. R., Heinz, D. W., and Niemann, H. H. (2005) Protein Sci. 14 1993-2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locher, M., Lehnert, B., Krauss, K., Heesemann, J., Groll, M., and Wilharm, G. (2005) J. Biol. Chem. 280 31149-31155 [DOI] [PubMed] [Google Scholar]
- 12.Schubot, F. D., Jackson, M. W., Penrose, K. J., Cherry, S., Tropea, J. E., Plano, G. V., and Waugh, D. S. (2005) J. Mol. Biol. 346 1147-1161 [DOI] [PubMed] [Google Scholar]
- 13.Lilic, M., Vujanac, M., and Stebbins, C. E. (2006) Mol. Cell 21 653-664 [DOI] [PubMed] [Google Scholar]
- 14.Blocker, A., Jouihri, N., Larquet, E., Gounon, P., Ebel, F., Parsot, C., Sansonetti, P., and Allaoui, A. (2001) Mol. Microbiol. 39 652-663 [DOI] [PubMed] [Google Scholar]
- 15.Akeda, Y., and Galan, J. E. (2005) Nature 437 911-915 [DOI] [PubMed] [Google Scholar]
- 16.Sory, M. P., Boland, A., Lambermont, I., and Cornelis, G. R. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11998-12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd, S. A., Norman, M., Rosqvist, R., and Wolf-Watz, H. (2001) Mol. Microbiol. 39 520-531 [DOI] [PubMed] [Google Scholar]
- 18.Ramamurthi, K. S., and Schneewind, O. (2005) J. Bacteriol. 187 707-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Pawel-Rammingen, U., Telepnev, M. V., Schmidt, G., Aktories, K., Wolf-Watz, H., and Rosqvist, R. (2000) Mol. Microbiol. 36 737-748 [DOI] [PubMed] [Google Scholar]
- 20.Keller, R. (2004) The Computer Aided Resonance Assignment Tutorial, CANTINA Verlag, Goldau, Switzerland
- 21.Guntert, P., Salzmann, M., Braun, D., and Wuthrich, K. (2000) J. Biomol. NMR 18 129-137 [DOI] [PubMed] [Google Scholar]
- 22.Blaylock, B., Riordan, K. E., Missiakas, D. M., and Schneewind, O. (2006) J. Bacteriol. 188 3525-3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evdokimov, A. G., Tropea, J. E., Routzahn, K. M., and Waugh, D. S. (2002) Protein Sci. 11 401-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bax, A., and Grzesiek, S. (1993) Acc. Chem. Res. 26 131-138 [Google Scholar]
- 25.Salzmann, M., Pervushin, K., Wider, G., Senn, H., and Wuthrich, K. (1999) J. Biomol. NMR 14 85-88 [DOI] [PubMed] [Google Scholar]
- 26.Salzmann, M., Wider, G., Pervushin, K., Senn, H., and Wuthrich, K. (1999) J. Am. Chem. Soc. 121 844-848 [Google Scholar]
- 27.Xia, Y., Sze, K., and Zhu, G. (2000) J. Biomol. NMR 18 261-268 [DOI] [PubMed] [Google Scholar]
- 28.Kay, L. E., Torchia, D. A., and Bax, A. (1989) Biochemistry 28 8972-8979 [DOI] [PubMed] [Google Scholar]
- 29.Daughdrill, G. W., Chadsey, M. S., Karlinsey, J. E., Hughes, K. T., and Dahlquist, F. W. (1997) Nat. Struct. Biol. 4 285-291 [DOI] [PubMed] [Google Scholar]
- 30.Gauthier, A., and Finlay, B. B. (2003) J. Bacteriol. 185 6747-6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fink, A. L. (2005) Curr. Opin. Struct. Biol. 15 35-41 [DOI] [PubMed] [Google Scholar]
- 32.Krall, R., Zhang, Y., and Barbieri, J. T. (2004) J. Biol. Chem. 279 2747-2753 [DOI] [PubMed] [Google Scholar]
- 33.Letzelter, M., Sorg, I., Mota, L. J., Meyer, S., Stalder, J., Feldman, M., Kuhn, M., Callebaut, I., and Cornelis, G. R. (2006) EMBO J. 25 3223-3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd, A. P., Grosdent, N., Totemeyer, S., Geuijen, C., Bleves, S., Iriarte, M., Lambermont, I., Octave, J. N., and Cornelis, G. R. (2000) Eur. J. Cell Biol. 79 659-671 [DOI] [PubMed] [Google Scholar]
- 35.Wulff-Strobel, C. R., Williams, A. W., and Straley, S. C. (2002) Mol. Microbiol. 43 411-423 [DOI] [PubMed] [Google Scholar]
- 36.Schwarzinger, S., Kroon, G. J., Foss, T. R., Chung, J., Wright, P. E., and Dyson, H. J. (2001) J. Am. Chem. Soc. 123 2970-2978 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.