Abstract
The Rho GTPases are critical regulators of the actin cytoskeleton and are required for cell adhesion, migration, and polarity. Among the key Rho regulatory proteins in the context of cell migration are the p190 RhoGAPs (p190A and p190B), which function to modulate Rho signaling in response to integrin engagement. The p190 RhoGAPs undergo complex regulation, including phosphorylation by several identified kinases, interactions with phospholipids, and association with a variety of cellular proteins. Here, we have identified an additional regulatory mechanism unique to p190A RhoGAP that involves priming-dependent phosphorylation by glycogen synthase-3-β (GSK-3β), a kinase previously implicated in establishing cell polarity. We found that p190A-deficient fibroblasts exhibit a defect in directional cell migration reflecting a requirement for GSK-3β-mediated phosphorylation of amino acids in the C-terminal “tail” of p190A. This phosphorylation leads to inhibition of p190A RhoGAP activity in vitro and in vivo. These studies identify p190A as a novel GSK-3β substrate and reveal a mechanism by which GSK-3β contributes to cellular polarization in directionally migrating cells via effects on Rho GTPase activity.
Directional cell migration requires stringently coordinated regulation of the actin and microtubule cytoskeletal network. Among the key regulators of cytoskeleton dynamics in this context are the Rho GTPases, for which role in cell adhesion, migration, and polarity is well established. Recent studies have revealed that spatiotemporal regulation of Rho GTPase activity plays an important role in the formation of membrane protrusions and retractions in migrating adherent cells (1). The Rho GTPases are regulated by two major classes of proteins: the GTPase-activating proteins (GAPs),3 which promote Rho inactivation by stimulating the weak GTP hydrolyzing activity of Rho GTPases thereby converting them to an inactive GDP-bound form; and the guanine nucleotide exchange factors (GEFs), which promote Rho activation by stabilizing the nucleotide-free form of Rho GTPases thereby allowing them to rebind GTP, which is present at a high intracellular concentration (1). Several of the identified RhoGAPs and RhoGEFs have been implicated in cell migration and polarity (2).
Among the dozens of GAPs with predicted activity toward Rho GTPases that are encoded by vertebrate genomes are two highly homologous RhoGAPs, p190A and p190B, that are both widely expressed potent regulators of several of the Rho GTPases (3). Targeted disruption of the mouse genes encoding the p190 RhoGAPs has revealed that they are each essential for normal embryonic development, with knock-out mice exhibiting a variety of defects in tissue morphogenesis (4–7). Thus, p190A-deficient mice exhibit defects in axon guidance and fasciculation, neural tube closure, and eye development, and p190B-deficient mice exhibit reduced cell size and organism size, resulting from altered Rho-modulated insulin-like growth factor-1 signaling. Notably, although p190A and p190B are potent RhoGAPs, the mouse knock-outs are not associated with global changes in tissue levels of Rho-GTP, suggesting that they exert their biological effects through the context-dependent regulation of subcellular pools of Rho.
Accumulating evidence indicates that the p190 RhoGAPs mediate signals in response to the engagement of integrins by extracellular matrix components to promote Rho GTPase-dependent actin rearrangements (5, 8–12), which may account for some of the observed phenotypes in p190-deficient mice. Furthermore, p190A RhoGAP has been implicated in cell migration in several reports (9, 10, 13–17), suggesting that these GAPs may be critical for mediating integrin signals that drive cell motility in the context of tissue morphogenesis. However, p190A-deficient fibroblasts do not exhibit an obvious defect in their ability to adhere to and spread on fibronectin, indicating that p190A is not essential for adhesion to extracellular matrix (4).
The p190 RhoGAPs undergo complex regulation, including phosphorylation by several identified kinases, interactions with phospholipids, and association with several different cellular proteins (18). Here, we have established an additional regulatory mechanism unique to the p190A isoform that involves phosphorylation by glycogen synthase-3-β (GSK-3β), which has been implicated previously in cell polarity (19). We found that p190A-deficient fibroblasts exhibit a defect in directional cell migration and that GSK-3β-mediated phosphorylation of amino acids in the C terminus of p190A is required for proper polarized migration. These studies have identified p190A as a novel substrate for GSK-3β, revealing a mechanism by which GSK-3β contributes to cell polarity that involves the regulation of Rho GTPase activity in directionally migrating cells.
EXPERIMENTAL PROCEDURES
DNA Constructs—To generate the pGEX-KG-1252 and pGEX-KG-50AA constructs, DNA sequences corresponding to amino acids 1251–1500 and 1450–1500 of p190A were PCR-amplified and subcloned into the pGEX-KG. Site-directed mutagenesis (QuikChange mutagenesis kit, Stratagene) was used to generate S1472A, S1476A, T1480A, and S1483A mutations in the above plasmids. All plasmids were verified by DNA sequencing. The coding sequences of the above plasmids were subcloned into pEBG vectors using PCR to generate pEBG-1252 constructs and pEBG-50AA constructs. Full-length p190A cDNA was cloned into the pEGFP-C1 vector using standard PCR, and site-directed mutagenesis was used to generate the pEGFP-C1-p190A-1472A, pEGFP-C1-p190A-1476A, pEGFP-C1-p190A-1480A, and pEGFP-C1-p190A-1483A plasmids. p190(1191)ΔPBR was produced as a glutathione S-transferase fusion protein in Escherichia coli. The ΔPBR mutant of full-length p190A was created by overlap extension PCR followed by a three-part ligation. This plasmid was used as template for production of p190(1191)ΔPBR by PCR followed by subcloning into the EcoRI site of pGEX-4T-1 vector. This protein contains the sequence of p190A between amino acids 1191 and 1499. All plasmids were verified by DNA sequencing. The constructs are illustrated in supplemental Fig. 1.
Generation of Stable Cell Lines—p190A-/- fibroblasts derived from E14 homozygous mutant embryos were transfected with the pBabe puromycin-resistance vector and either the MFG expression vector (control) or an MFG-p190A-expressing vector at a 1:10 ratio and selected for puromycin resistance 2 days after transfection. After 2 weeks, single clones were isolated and expanded, and expression of p190A level was confirmed by immunoblotting. p190A-/- fibroblasts were transfected with pEGFP-C1, pEGFP-C1-p190A-WT, pEGFP-C1-p190A-1472A, pEGFP-C1-p190A-1476A, pEGFP-C1-p190A-1480A, or pEGFP-C1-p190A-1483A using Lipofectamine 2000 (Invitrogen). Two days after transfection the cells were replated in medium containing 3 μg/ml G418 (Sigma), and polyclonal cell lines were established. Exogenous p190A levels in the various stable cell lines were determined by immunoblotting, and cell lines expressing comparable levels of proteins were used for further analysis.
Cell Culture, Transfections, and Inhibitor Treatments— p190A+/+ and p190A-/- fibroblast cell lines were generated as described (4). 3T3 fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% calf serum. COS-7 cells were cultured in DMEM with 10% fetal bovine serum. Lipofectamine 2000 (Invitrogen) was used for transient transfection of COS-7 cells. Cells were maintained for 48 h in medium containing 10% fetal bovine serum after transfection. The following inhibitors were added to the cell culture as indicated in the text: SB203580 (Calbiochem), G06983 (Calbiochem), roscovitine (Calbiochem), U0126 (Calbiochem), and GSK-3β inhibitor XII (Calbiochem).
Protein Purification—pGEX-KG plasmids were transformed in E. coli BL21 cells. Log-phase BL21 cells expressing GST fusions were induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 5 h. Bacterial pellets were lysed by sonication in 50 mm Tris (pH 7.6), 150 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm EDTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. pEBG plasmids were transfected into COS-7 cells. Two days after transfection the cells were lysed in Triton lysis buffer (TLB) (50 mm Tris-HCl (pH 7.6), 1% Triton X-100, 150 mm NaCl, 2 mm Na3VO4, 10 mm NaF, 1 mm EDTA, 1 mm EGTA, and 1 mm DTT) with protease inhibitors. In each case, GST fusion proteins were purified on glutathione resin (Sigma) as suggested by the manufacturer. Full-length p190A was purified from baculovirus-infected Sf9 cells as described previously.
In Vitro Kinase Assays, Priming, and Phosphatase Treatment— For kinase priming studies, three 10-cm plates of confluent COS-7 cells were washed once with phosphate-buffered saline and resuspended in 1.5 ml of cold hypotonic lysis buffer (20 mm Tris, pH7.5, 5 mm KCl, 1.5 mm MgCl2, 1 mm DTT) by mechanical scraping. The cells were disrupted using a Dounce homogenizer on ice. Crude extracts were centrifuged twice (once at 10,000 × g for 10 min and then at 10,000 × g for 30 min) to remove the cell debris and nuclei. Aliquots were flash-frozen and stored at -80 °C before use (3–5 mg/ml as determined by Bradford assay). 360 μl of cell lysate and 40 μl of 10× energy-regenerating system (ERS) (20 mm Tris (pH 7.5), 10 mm ATP, 10 mm magnesium acetate, 300 mm creatine phosphate (Roche Applied Science), 0.5 mg/ml creatine phosphokinase (Roche Applied Science)) were added to the bacterially purified GST fusion proteins on glutathione beads, and the mixtures were incubated at 30 °C for 2–3 h before proceeding to the in vitro kinase assays. Where indicated, cell lysates were preincubated with either DMSO or 50 μm SB203580 for 2 h at 30 °C prior to incubation with 10× ERS as described above.
For kinase assays, freshly purified GST fusion proteins on glutathione beads (20 μl) were washed three times with wash buffer (20 mm Tris, pH 7.5, 5 mm KCl, 1.5 mm MgCl2, 1 mm DTT) and one time with kinase buffer and then incubated with 30 μl of kinase buffer (20 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm DTT, 0.1 mm EGTA), 10 μCi of [γ-32P]ATP, and recombinant kinase as indicated. The reaction was carried out at 30 °C for 30 min and stopped by washing twice with wash buffer and then boiled in 2× sample buffer (125 mm Tris, pH 6.8, 4% SDS, 30% glycerol, bromphenol blue). GSK-3β (Sigma) was used at 2 units/reaction; the GSK-3β kinase buffer. p38 MAPK (BIOSOURCE), p42 MAPK (New England Biolabs), and casein kinase I (CKI) (New England Biolabs) kinase assays were performed as described by the manufacturer.
For phosphatase studies, λ-phosphatase (New England Biolabs) was used. Purified proteins on glutathione beads, pre-treated with λ-phosphatase, λ-phosphatase with phosphatase inhibitors, or phosphatase inhibitors alone at 30 °C for 30 min, were then washed five times with wash buffer before proceeding with in vitro kinase assays.
Monolayer Wounding and Polarity Assays—3T3 fibroblasts were grown to confluency and then “wounded” using a single 200-μl pipette tip to create a narrow linear scratch in the cell monolayer. Microscopy images were captured immediately after and 15 h after wounding. For polarity assays, cells were seeded onto coverslips and grown to confluency before they were wounded as described above and then fixed at 0, 2, 4, and 6 h post-wounding. The cells were stained with anti-GM130 (1:200), to detect Golgi apparatus, and DAPI (0.2 μg/ml), and microscopy images were captured using a fluorescence microscope. The orientation of the Golgi was assessed as described previously (35); 200 leading edge cells were counted for each experiment. Data were expressed in graphical form reflecting the mean value and standard deviation from three independent experiments.
Transwell Boyden Chamber Assays—Migration assays were performed using 5-μm pore, 6.5-mm polycarbonate Transwell filters (Corning Costar). Cells were grown to confluency and serum-starved overnight. 5 × 104 cells in 100 μl of serum-free medium were then seeded on the upper surface of the filters, and 600 μl of medium containing 10% fetal bovine serum was introduced into the lower chambers. Migration was allowed to proceed for 15 h. Cells were then fixed in 4% formaldehyde and stained with 0.1% crystal violet solution for 30 min. The upper surfaces of the filters were cleaned well using cotton swabs. Each filter was then excised and destained in 10% acetic acid solution, and readings were obtained at 575 nm. The analysis was reproduced in triplicate for each sample, and mean values and standard deviations were determined and plotted as the percentage of cells that had migrated to the lower chamber as based on the absorbance reading of stained cells at 575 nm.
Western Blot and Immunoprecipitation—Cells were lysed in TLB with protease inhibitors. For immunoblotting, p190A RhoGAP antibody (BD Transduction Laboratories) was used at 1:1000; 12CA5 anti-hemagglutinin antibody, an ascites, was used at 1:1000; p120 RasGAP antibody B4F8 (Upstate) was used at 1:1000; GSK-3β antibody (BD Transduction laboratories) was used at 1:1000; and phospho-GSK-3β antibody (Cell Signaling Technology) was used at 1:1000.
Co-immunoprecipitation assays were performed by incubating lysates with primary antibody overnight at 4 °C. A mixture of protein G and protein A-Sepharose beads (Amersham Biosciences) was added for 2–3 h before the beads were washed three times with 1 ml of 50 mm Tris-HCl (pH 7.6), 0.1% Triton X-100, 150 mm NaCl, 2 mm Na3VO4, 10 mm NaF, 1 mm EDTA, 1 mm EGTA, and 1 mm DTT. Immunoprecipitates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
GAP Assays—In vivo assays of p190 RhoGAP activity were performed using COS-7 cells transiently transfected with pEGFP-p190A-WT or the indicated mutants as described previously (4). EGFP-positive cells were scored blindly on coverslips at 48 h post-transfection as demonstrating “strong” or “weak” GAP activity. Strong activity refers to cells demonstrating the extension of one or more cell processes that exceed 1 cell body in diameter. Other GFP-positive cells lacking such processes were scored as weak. For each sample, 200 cells were counted; the graphed data reflect the mean from three independent experiments including standard deviations. In vitro GAP assays using a p190A fragment containing the GAP catalytic domain and the C-terminal phosphorylation sites were performed as described previously using GSK-3β-primed or nonprimed p190A and purified recombinant Rho GTPase as described previously (36).
Mass Spectrometry Analysis—pEBG-50AA-WT was transfected into COS-7 cells. Two days after transfection, the cells were lysed in TLB and incubated with glutathione-agarose beads at 4 °C for 2 h, washed three times with 50 mm Tris-HCl (pH 7.6), 0.1% Triton X-100, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, and boiled in sample buffer. The proteins were resolved on a 10% SDS-polyacrylamide gel, and after Coomassie Blue staining the protein band was excised and sent for mass spectrometry analysis to identify phosphorylated peptides at the Taplin Mass Spectrometry Facility, Harvard Medical School.
RESULTS
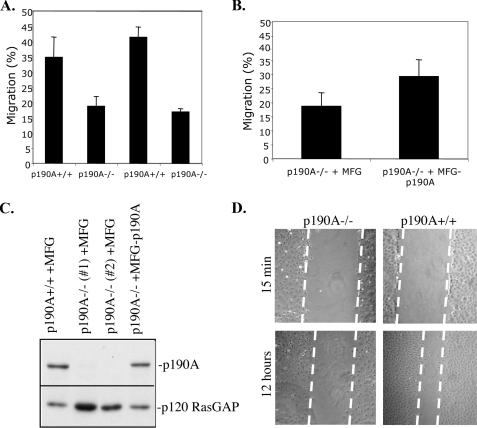
p190A RhoGAP Is Required for Polarized Cell Migration— We have previously generated mice deficient for the p190A RhoGAP by targeted gene disruption; homozygous mutant animals die shortly after birth, exhibiting several developmental defects in tissue morphogenesis (4, 5). However, the cellular role of p190A in tissue morphogenesis is still somewhat unclear. To begin to identify cell-autonomous requirements for p190A, we cultured embryo-derived fibroblasts from p190A-deficient embryos and their wild-type littermates. The p190A-deficient cells did not exhibit obvious differences relative to wild-type cells in proliferative potential or any gross morphologic phenotypes in culture (data not shown). Considering the important role for Rho GTPases in cell migration, as well as previous findings implicating p190A in cell migration (9, 10, 13–17), we utilized a standard Boyden chamber assay to examine the migration properties of p190A-deficient cells. In that assay, we observed a striking migration defect in multiple, independently derived lines of p190A-deficient fibroblasts (Fig. 1A). To verify that this defect reflected a specific loss of p190A function, we stably reintroduced into p190A-deficient cells an expression vector encoding full-length p190A, and we determined that these “rescued” cells exhibited migration properties similar to those seen in wild-type cells (Fig. 1, B and C). Then, using in vitro“wound healing” as an independent assay of cell migration, we similarly observed a clear defect in the migration potential of p190A-deficient cells (Fig. 1D). These results indicate that p190A is required for some aspect of cell migration in fibroblasts.
FIGURE 1.
p190A-deficient cells exhibit defects in cell migration. A, two pairs of p190A+/+ and p190A-/- embryo-derived fibroblasts were grown to confluency in DMEM + 10% calf serum, starved overnight in serum-free medium, and then assayed for migration using Transwells as described under “Experimental Procedures.” The graphs depict the mean values from three independent experiments with error bars describing S.D. Migration percentage refers to the percentage of cells that had migrated from the upper to the lower chamber of a Transwell unit after 15 h. B, single clones of p190A-/- cell lines stably transfected with either MFG or MFG-p190A were assayed for migration using Transwells. C, immunoblots of the lysates from cells used in B demonstrating the levels of p190A. p120 RasGAP was immunoblotted as a loading control. Two different p190-/- clones are indicated (#1 and #2, respectively). D, p190A+/+ and p190A-/- cells were grown to confluency in DMEM + 10% calf serum, and a monolayer “scratch wound” was induced as described under “Experimental Procedures.” Photomicrographs were taken 15 min and 12 h post-wounding. Note that the p190A-/- cells exhibit a clear defect in wound closure as highlighted by the superimposed white dashed lines along the wound edge.
A previous report revealed that p120 RasGAP, the major binding partner for p190A, is required for polarized cell migration in cultured fibroblasts (20). Moreover, expression of a putative dominant-negative mutant form of p190A is reported to interfere with cell polarity in a wounding assay (9). Therefore, we examined the ability of p190A-deficient cells to orient properly toward the migratory plane in a wounding assay. First, we noted that in wild-type cells, F-actin-containing protrusions are oriented toward the wound at the leading edge at both 2 and 6 h post-wounding, whereas in p190A-deficient cells, the overall F-actin staining appears more intense, yet relatively disorganized, and the membrane protrusions are generally disoriented relative to the migration plane (Fig. 2A). These results suggest a role for p190A in regulating and orienting the actin cytoskeleton in migrating cells. Then, using a Golgi apparatus marker as a means of specifically monitoring cell polarity in cells along a wound edge, we determined that p190A-deficient cells exhibit a striking defect in their ability to orient toward the plane of cell migration (Fig. 2, B and C). Taken together, these observations indicate a requirement for p190A RhoGAP in polarized cell migration.
FIGURE 2.
p190A-deficient fibroblasts are defective for polarized cell migration. A, F-actin staining (phalloidin) of p190A+/+ and p190A-/- cells 2 h and 6 h post-wounding. The white arrows indicate membrane protrusions oriented toward the migration plane in the wild-type cells. The dashed white line indicates the wound plane. B, a scratch-wounded monolayer illustrating Golgi orientation in p190A+/+ and p190A-/- cells at 6 h post-wounding. Blue, nuclear DAPI staining; red, anti-GM130. The dashed white line indicates the wound edge. The superimposed “bent” white lines correspond to the 120° arc at each cell location that faces the plane of the wound. C, graph depicting the quantification of Golgi orientation in p190A+/+ and p190A-/- cells at 0, 2, 4, and 6 h post-wounding. The dashed horizontal line indicates the 33% value that would be expected with random orientation. 200 cells were counted for each genotype, and graphs depict the mean value obtained from three independent experiments, with error bars representing S.D.
p190A Is a Phosphorylation Substrate for GSK-3β—Among the critical regulators of cell polarity is glycogen synthase kinase-3-β; we had previously detected a direct interaction between p190A and GSK-3β in a yeast two-hybrid screen (21). Moreover, a high stringency Scansite (22) analysis of p190A RhoGAP revealed the presence of three high-scoring candidate sites for GSK-3β phosphorylation (Ser-1472, Ser-1476, and Thr-1480) within a nine-amino acid peptide stretch in the C-terminal “tail” of p190A, suggesting that p190A may be a substrate of GSK-3β (Fig. 3A). A fourth nearby site (Ser-1483) scored highly as a potential p38 MAP kinase substrate site; MAP kinase sites have previously been implicated in priming subsequent GSK-3β-mediated phosphorylation. These sites are all conserved in mouse, human, and rat p190A sequences, but they are not present in the functionally distinct but highly homologous p190B RhoGAP protein. Notably, this peptide stretch is remarkably similar to the GSK-3β-phosphorylated region of β-catenin with regard to high proline content and the spacing of candidate phosphorylation sites (23).
FIGURE 3.
GSK-3β phosphorylates the C-terminal domain of p190A in vitro. A, p190A protein sequences from human, mouse, and rat were analyzed using Scansite software (high stringency), and GSK-3 kinase consensus sites were identified corresponding to amino acids Ser-1472, Ser-1476, and Thr-1480. Amino acid Ser-1483 was also identified as a potential p38 MAPK site. The percentile scores correspond to the probability that these putative sites are related to documented consensus sites by chance. B, full-length p190A baculovirus-expressed protein purified from Sf9 insect cells can be phosphorylated by GSK-3β in vitro. The GSK-3β kinase assay was performed in vitro as described under “Experimental Procedures,” and reaction products were resolved by SDS-PAGE and visualized by autoradiography. C, pEBG, pEBG-50AA-WT, pEBG-50AA-1472A, pEBG-50AA-1476A, pEBG-50AA-1480A, and pEBG-50AA-1483A were transfected into COS-7 cells, and the corresponding GST fusion proteins were purified on glutathione beads. In vitro kinase assays, with or without GSK-3β, were performed as indicated. The left panel corresponds to an immunoblot demonstrating levels of the various fusion protein proteins captured from the cell lysates. The right panel corresponds to the kinase reaction products following SDS-PAGE and autoradiography. D, phosphorylation of a domain containing the C-terminal 50 amino acids of p190A in vivo. The GST-50AA fusion proteins derived from p190A were purified either from E. coli (pGEX-KG-encoded) or from COS-7 cells (pEBG-encoded). The proteins on the glutathione beads were mock-treated, treated with λ-phosphatase, or treated with λ-phosphatase in the presence of phosphatase inhibitors (PPI). Then the beads were washed and boiled in sample buffer, and the proteins were resolved by SDS-PAGE and stained with Coomassie Blue. The faster migrating species are presumed to reflect degradation products derived from the GST fusion proteins. E, mass spectrometry analysis identified sites of phosphorylation within the C-terminal 50 amino acids of p190A. pEBG-50 AA-WT was transfected into COS-7 cells and purified as described under “Experimental Procedures.” The proteins were gel-purified, and proteolytically cleaved peptides were analyzed by mass spectrometry. The phosphorylation sites detected in vivo are underscored in three different peptide species that were detected, and these correspond to amino acids 1472, 1476, and 1483 in p190A.
Therefore, we pursued studies to test the possibility that the role of p190A in polarized cell migration involves regulation by GSK-3β. We first determined that full-length recombinant p190A can indeed be directly phosphorylated in vitro by purified GSK-3β (Fig. 3B). Then, to determine whether the C-terminal region of p190A is the target of GSK-3β phosphorylation, we expressed a GST fusion protein encoding the C-terminal 50 amino acids of p190A (which contain the putative phosphorylation sites) in transfected COS-7 cells and then captured the fusion protein on glutathione beads and performed in vitro kinase assays using purified GSK-3β and [γ-32P]ATP (Fig. 3C). In this assay, we observed potent phosphorylation of this region of p190A by GSK-3β. Next, we performed this assay using fusion proteins containing this same region of p190A but with site-directed mutants in which each of the four putative phosphorylation sites in this domain was substituted with alanine. We found that substitution of sites Ser-1476, Thr-1480, or Ser-1483 completely eliminated phosphorylation by GSK-3β, whereas substitution of Ser-1472 reduced phosphorylation by about 50%, thereby directly implicating these sites in GSK-3β-mediated phosphorylation of p190A (Fig. 3C).
Consistent with in vivo phosphorylation of this C-terminal region of p190A, we noted that the gel mobility of a GST fusion encoding the 50-amino acid p190A fragment captured from COS-7 cells was significantly reduced relative to the mobility of an identical fusion protein produced in bacteria (which would not be expected to be phosphorylated) (Fig. 3D). Moreover, treatment of the isolated COS-7-expressed protein with λ-phosphatase increased the mobility of this fragment (which could be prevented by including a general phosphatase inhibitor) such that it co-migrated with the bacterially produced protein, suggesting that the COS-7-expressed protein is phosphorylated (Fig. 3D).
To confirm that these sites within the p190A C-terminal region are indeed phosphorylated in vivo, we utilized matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. The GST fusion protein containing the 50-amino acid tail of p190A was expressed in transfected COS-7 cells, and the fusion protein captured from lysates on glutathione beads was subjected to proteolytic peptide fragmentation and mass spectrometry analysis to identify phosphorylated peptides (Fig. 3E). In this analysis, three phosphorylated peptides were identified: the first corresponds to a p190A species that was doubly phosphorylated on Ser-1472 and Ser-1476; the second corresponds to a species phosphorylated either on Ser-1472 or Ser-1476 (it is not possible to resolve these two species by mass spectrometry); and the third corresponds to a peptide species triply phosphorylated on Ser-1472, Ser-1476, and Ser-1483. The phosphorylation of Thr-1480 was not detected in this experiment due to the technical limitations of this methodology. These findings indicate that these putative phosphorylation sites of p190A do in fact undergo phosphorylation in vivo.
Phosphorylation of p190A Rho-GAP by GSK-3β Requires Priming by MAP Kinases—Phosphorylation of protein substrates by GSK-3β often requires “priming” of a neighboring residue by a distinct kinase, leading to subsequent phosphorylation by GSK-3β (24). Consistent with such a priming mechanism for p190A, we observed that the C-terminal tail fragment of p190A expressed in bacteria could not be detectably phosphorylated by purified GSK-3β in an in vitro kinase assay; however, preincubation of that fragment (immobilized on glutathione beads) with a cytoplasmic extract from COS-7 cells yielded a protein that could readily be subsequently phosphorylated by GSK-3β (Fig. 4A). To further confirm a role for a priming phosphorylation on p190A, we determined that, whereas the GST-p190A tail fragment captured from transfected COS-7 cell lysates can be phosphorylated by GSK-3β without further priming, phosphatase treatment of this fragment following capture from COS-7 cell lysates completely prevents subsequent GSK-3β-mediated phosphorylation in vitro (Fig. 4A). Together, these results indicate that a priming phosphorylation of the C-terminal region of p190A is required prior to phosphorylation by GSK-3β.
FIGURE 4.
Priming-dependent phosphorylation of p190A by GSK-3β. A, priming is required for p190A phosphorylation by GSK-3β. Bacterially purified p190A-50AA (pGEX-KG-50AA-WT) on beads was treated either with or without priming by COS-7 lysate. The beads were washed extensively and then subjected to in vitro phosphorylation by GSK-3β. p190A-50AA (pEBG-50AA-WT) expressed in transfected COS-7 cells was purified on glutathione beads, pretreated with either λ-phosphatase, λ-phosphatase plus phosphatase inhibitors (PPI), or phosphatase inhibitors alone. The beads were washed extensively and then subjected to in vitro phosphorylation by GSK-3β. Reaction products were resolved by SDS-PAGE and autoradiography. B, p38 or p42 MAPK, but not CKI, can prime the C-terminal domain of p190A for subsequent phosphorylation by GSK-3β in vitro. Bacterially purified p190A-50AA (pGEX-KG-50AA-WT) was first phosphorylated in vitro by purified recombinant p38MAPK, p42 MAPK, or CKI in the presence of cold ATP. Proteins were then subjected to GSK-3β kinase assays using [γ-32P]ATP, and products were resolved by SDS-PAGE and visualized by autoradiography. Note the absolute requirement for priming by MAPKs.
To identify the kinase responsible for priming GSK-3β-mediated phosphorylation of p190A, we tested several purified recombinant kinases that have been implicated previously in the priming of GSK-3β-mediated phosphorylation of other substrates. For this analysis, we used the bead-immobilized purified GST-p190A tail fragment produced in bacteria in kinase assays in which the protein was initially incubated with a candidate priming kinase and “cold” ATP, and then the kinase was washed away, and GSK-3β and [γ-32P]ATP were introduced for a secondary kinase reaction. Using this assay, we found that p38 and p42 MAP kinases could effectively prime the C-terminal fragment of p190A for subsequent phosphorylation by GSK-3β, whereas CKI, which primes phosphorylation of β-catenin for GSK-3β (24), could not prime p190A for phosphorylation, thereby confirming a priming-dependent mechanism and suggesting that MAP kinases may be the relevant priming kinases (Fig. 4B).
To establish the priming sites within the p190A tail, we examined the ability of the purified MAP kinases to phosphorylate the various substitution mutants within the C-terminal domain using in vitro kinase assays with [γ-32P]ATP. Although none of the single-site mutants eliminates phosphorylation by either p38 or p42 MAP kinase, a significant reduction in phosphorylation is seen when Ser-1476, Thr-1480, or Ser-1483 is substituted (Fig. 5A). This finding confirms that these kinases can indeed directly phosphorylate this region of p190A and suggests that priming may involve more than one of those three sites or that substitution of those residues leads to a conformational effect on closely neighboring residues that impacts their ability to be efficiently phosphorylated. The fact that the COS-7-expressed p190A tail can be phosphorylated on these sites in vitro despite the fact that these sites are phosphorylated on the protein at the time of isolation from cells suggests that the sites are only partially phosphorylated in vivo and can therefore undergo further phosphorylation. Further analysis of the priming sites, using a primary cold ATP priming assay followed by a secondary “hot” ATP kinase assay with GSK-3β, revealed that the Ser-1476 and Thr-1480 sites seem to be the most critical for MAP kinase-dependent priming of GSK-3β-mediated phosphorylation (Fig. 5B). Thus, substituting either of those amino acids virtually eliminates priming-dependent phosphorylation by GSK-3β.
FIGURE 5.
Mapping the p190A priming sites. A, mapping the sites of p38 and p42 MAPK phosphorylation on p190A. pEBG (vector) or pEBG-50AA-WT and the indicated phosphorylation site mutant proteins were captured from transfected COS-7 cells, and the proteins on beads were incubated with either p38MAPK or p42MAPK and [γ-32P]ATP. The leftmost lane corresponds to the pEBG-WT protein with buffer only. Reaction products were analyzed by SDS-PAGE and autoradiography (upper panels) and Coomassie Blue staining (lower panels). B, mapping the priming sites on p190A in vivo. pEBG-50AA-WT and the various phosphorylation site mutant proteins were purified from transfected COS-7 cells as indicated. The proteins on beads were then either mock-treated or prephosphorylated in vitro by either p38 or p42 MAPK in the presence of cold ATP. They were then incubated with GSK-3β in the presence of [γ-32P]ATP. Reaction products were analyzed by SDS-PAGE and autoradiography. C, inhibiting p38MAPK in the priming lysate disrupts priming activity. pEBG-50AA-WT proteins were purified from COS-7 cells as described under “Experimental Procedures.” For priming, following cell lysis the lysates were preincubated with either DMSO (vehicle control) or the p38 MAPK inhibitor SB203580. The lysates were then used to prime the pEBG-50AA-WT proteins on beads. The beads were then processed for GSK-3β kinase assays using [γ-32P]ATP. Reaction products were analyzed by SDS-PAGE and autoradiography (upper panel) and Coomassie Blue staining (lower panel).
To confirm that priming-dependent phosphorylation of p190A occurs in vivo, we preincubated purified bacterially produced GST-p190A with COS-7-derived cell lysates that were either pretreated or not with the pharmacologic p38 MAPK inhibitor SB203580. After priming, in vitro kinase assays were performed with GSK-3β. As expected, GST-p190A that was not preincubated with COS-7 lysate was not detectably phosphorylated with GSK-3β (Fig. 5C). GST-p190A that was preincubated with untreated COS-7 lysate (DMSO only) could be efficiently phosphorylated with GSK-3β, whereas preincubation of COS-7 lysate with SB203580 substantially reduced the ability of p190A to be phosphorylated by GSK-3β, confirming that a SB203580-sensitive kinase is responsible for priming p190A for phosphorylation by GSK-3β in COS-7 cells (Fig. 5C).
Ser-1476 and Thr-1480 Phosphorylation Sites in p190A RhoGAP Are Required for Polarized Cell Migration—To determine whether the identified sites of GSK-3β-mediated phosphorylation of p190A are required for p190A function in polarized cell migration, we reintroduced full-length p190A proteins containing the various phosphorylation site mutants into p190A-deficient fibroblasts and assayed the migration. The mutations were subcloned into a vector expressing a p190A fusion protein that also includes green fluorescence protein (GFP) to facilitate protein detection in live cells by immunofluorescence. Stably transfected polyclonal cell lines expressing either wild-type p190A or each of the four substitution mutants were established, and they each expressed comparable levels of p190A protein (Fig. 6A). First, they were each tested in the wound healing assay. As expected, transfected wild-type p190A protein completely rescues the ability of p190A-deficient cells to rapidly close a scratch wound (Fig. 6B). Similarly, p190A mutants S1472A and S1483A effectively rescue the wound healing defect in p190A-deficient cells. However, mutants S1476A and T1480A appear to be completely defective in restoring migration to cells lacking p190A, suggesting that phosphorylation of these two sites is critical for the function of p190A in cell migration (Fig. 6B).
FIGURE 6.
GSK-3β phosphorylation-defective p190A mutants fail to rescue the directional migration defects in p190A-deficient cells. A, stable cell lines derived from p190A-/- fibroblasts, expressing either pEGFP-C1 vector alone or with pEGFP-C1-p190A-WT (full-length) or the various phosphorylation site mutants, were established. Immunoblots demonstrate expression of p190A. p120 RasGAP was also detected as a loading control. B, wound closure assays of the various stable cell lines as indicated. Photomicrographs were taken immediately after and 15 h after wounding. The dashed lines correspond to the boundaries of cell monolayers along the wound edge. Note that the wound closure defect can be rescued by wild-type p190A and the 1472A and 1483A mutants but not by the 1476A and 1480A mutants. C, Golgi reorientation 6 h post-wounding with the indicated p190A-expressing cell lines as indicated. Cells were seeded on coverslips and grown to confluence. A single wound was induced, and the cells were fixed at 6 h post-wounding. They were then double-stained with anti-GM130 and DAPI, and immunofluorescence microscopy was performed. For each genotype, 200 cells were counted for two individual lines. Graphs depict the mean from three independent experiments, and S.D. are indicated by the error bars. The dashed horizontal line corresponds to the 33% value, which would reflect random polarization. Note that the 1476A and 1480A mutants are defective for Golgi reorientation. D, F-actin staining (phalloidin) at 6 h post-wounding of the indicated p190A-expressing cell lines. White arrows highlight the oriented protrusions along the leading edge.
Next, we examined these same cell lines for their ability to polarize toward the migration plane, using the Golgi reorientation assay. As seen in the wound closure assay, the wild-type, S1472A, and S1483 mutant proteins restore normal Golgi orientation to cells along the wound edge, whereas the S1476A and T1480A are significantly impaired in this function (Fig. 6C). Notably, cells expressing those two mutants are not completely defective for Golgi reorientation, suggesting that additional elements of p190A may also contribute to its role in polarization. By staining the various cell lines for F-actin, we also observed that the accumulation of F-actin at the leading edge of cells along the wound is specifically impaired in cells expressing the S1476A and T1480A p190A mutants (Fig. 6D). Taken altogether, our findings suggest that Ser-1476 and Thr-1480 correspond to two critical phosphorylation sites within the C-terminal region of p190A required for proper oriented cell migration and that phosphorylation of these sites influences the ability of p190A to regulate actin dynamics and Golgi reorientation at the leading edge of migrating cells.
GSK-3β-mediated Phosphorylation of p190A Regulates Its RhoGAP Activity in Vivo—To begin to address the mechanism by which GSK-3β-mediated phosphorylation of p190A could influence its function in polarized cell migration, we first considered a potential role in the regulation of protein stability, a property that can be affected by GSK-3β-mediated phosphorylation (25). However, we found that phosphorylation-defective p190A mutants are not detectably affected in their half-life in vivo and, similarly, that treatment of cells with cell-permeable GSK-3 inhibitors does not affect p190A protein levels (data not shown).
Next, we considered a potential effect of GSK-3β-mediated phosphorylation on p190A GAP activity. For this analysis, we prepared a recombinant C-terminal portion of p190A containing amino acids 1191–1499, which includes the C-terminal GSK-3β phosphorylation sites as well as the entire GAP catalytic domain. By comparing the in vitro GAP activity of this protein with or without prior phosphorylation by GSK-3β, we determined that priming-dependent GSK-3β phosphorylation significantly inhibits the RhoGAP activity of p190A (Fig. 7, A and B).
FIGURE 7.
GSK-3β phosphorylation inhibits p190A GAP activity. A, in vitro assay of p190A GAP activity toward the Rho GTPase, demonstrating that MAPK priming-dependent phosphorylation by GSK-3β reduced GAP activity as reflected in an increase in the percentage of [32P]GTP-loaded GTPase retained on filters. Error bars reflect S.D. from four independent assays. B, in vitro phosphorylation reactions using [32P]ATP, demonstrating the kinetics of priming and subsequent phosphorylation of p190A by GSK-3β following SDS-PAGE and autoradiography (lower panel). The upper panel demonstrates equivalent levels of p190A as shown by Coomassie Blue staining. Similar conditions (leading to maximal phosphorylation; 60 min priming and 60 min GSK-3β phosphorylation) were used to prepare the material for use in the assays presented in A. C, two distinct types of morphologies are seen in COS-7 cells transiently transfected with pEGFP-p190A-WT or the various indicated mutants. The left panel exemplifies a cell demonstrating strong RhoGAP activity, as evidenced by the rounded morphology and thin cellular processes. The fluorescent signal is derived from the GFP portion of the p190A fusion protein. D, the indicated p190A constructs were transiently transfected into COS-7 cells. Immunoblots demonstrate the protein levels of p190A in transfected cells. GSK-3β was immunoblotted as a loading control. E, quantification of the percentage of transfected (GFP-positive) cells exhibiting the morphology characteristic of strong RhoGAP activity for each of the various p190A constructs. Error bars indicate S.D. (n = 200 for each construct). F, COS-7 cells were transiently transfected with either GFP or pEGFP-p190A-WT. At 40 h post-transfection, the cells were treated with DMSO, GSK-3β inhibitor XII, SB203580, U0126, or roscovitine for 4 h as indicated. The cells were then fixed and counted. The graph illustrates the average percentage of transfected cells exhibiting the strong RhoGAP phenotype (“GAP cells”) from three independent experiments, with the error bars representing S.D.
To verify that this in vitro effect on GAP activity could be observed in vivo, we used a cell-based assay to examine the role of the various phosphorylation site p190A mutants in RhoGAP function. We utilized an established assay for p190 RhoGAP activity in which transiently transfected cells that overexpress p190A become rounded and extend long thin processes, as seen in cells exposed to the botulinum C3 anti-Rho toxin (26) (Fig. 7C). By determining the proportion of transfected COS-7 cells that undergo this morphologic transformation, we were able to effectively quantify the in vivo RhoGAP activity. We observed that the wild-type, S1472A, and S1483A mutants of p190A exhibit similar levels of RhoGAP function in vivo, with each protein promoting a clear morphologic transformation of 30–35% of the transfected cells, associated with the extension of thin processes (Fig. 7, D and E). Interestingly, the S1476A and T1480A mutants exhibited significantly increased RhoGAP activity in vivo, causing the morphologic transformation of about 55% of the transfected cells and reinforcing the importance of these two sites as revealed in the wound healing assays. Consistent with these findings, we observed that treatment of wild-type p190A-transfected cells with a pharmacologic cell-permeable inhibitor of GSK-3β led to an increase in the percentage of morphologically transformed cells from 32 to 61% (Fig. 7F). Taken altogether, these results suggest that GSK-3β-mediated phosphorylation of the p190A C-terminal domain inhibits its RhoGAP catalytic activity in vitro and leads to diminished RhoGAP function in vivo.
DISCUSSION
We have described findings that establish p190A RhoGAP as a novel substrate for GSK-3β. Based on in vitro kinase assays, site-directed mutagenesis studies, and mass spectrometry analysis of protein in vivo, we conclude that three or four closely spaced amino acids within the p190A C-terminal region undergo phosphorylation by GSK-3β. Further analysis indicated that these phosphorylations are strictly dependent on priming by a second kinase, a property of many documented GSK-3β substrates. Moreover, the priming kinase is likely to be a member of the MAP kinase family. Our studies with site-directed p190A mutants also confirmed an essential biological requirement for two of these sites, namely, S1476A and T1480A. Thus, although the other sites can also undergo phosphorylation in vivo, it is possible that they serve alternative functions. It is also possible that the ability of MAP kinases to phosphorylate this domain of p190A serves a distinct function in addition to the priming of subsequent phosphorylations by GSK-3β. MAP kinases have been implicated previously in directional cell migration, raising the possibility that priming of p190A by a MAP kinase for subsequent phosphorylation by GSK-3β provides a mechanism by which p190A serves as a “coincidence sensor” for regulating Rho GTPase activity in an appropriate signaling context. Significantly, these identified GSK-3β phosphorylation sites in p190A are not present in the otherwise closely related p190B protein, possibly explaining the lack of redundant function for these two proteins in fibroblasts, where they are both expressed.
Although many GSK-3β substrates have been identified previously, the substrates that play a role in the ability of GSK-3β to regulate cell polarity have been somewhat elusive. The best documented GSK-3β substrate in this context is the adenomatous polyposis coli tumor suppressor (27). Thus, in response to activation of the CDC42 GTPase, which also plays an important role in cell polarity, PKCζ phosphorylates GSK-3β leading to its inactivation. Adenomatous polyposis coli phosphorylation by GSK-3β promotes its interaction with the plus ends of microtubules, and consequently, GSK-3β inactivation by PKCζ leads to spatially restricted interaction of adenomatous polyposis coli with microtubules, thereby affecting centrosome polarization (27). In the context of directional cell migration, the accumulation of GSK-3β at the leading edge of migrating cells appears to account for the regional restriction of adenomatous polyposis coli-microtubule association. Our findings implicate p190A RhoGAP as a second GSK-3β substrate that contributes to cell polarity. Notably, the GSK-3β phosphorylation sites within p190A are not at all conserved within the closely related p190B protein, and p190B-deficient fibroblasts exhibit a distinct migration phenotype that appears to reflect a role in cell adhesion but does not involve polarity.4 Interestingly, p190A is also a PKC phosphorylation substrate (4), raising the possibility that the role of p190A in cell polarity is also under the regulation of the atypical PKC isoform PKCζ, an established regulator of cell polarity via its role in the protein complex with PAR3 and PAR6 (28). Indeed, a recent report identified p190A as an effector of PAR6 in the regulation of dendritic spine morphogenesis (29).
The most thoroughly studied consequence of GSK3β-mediated protein phosphorylations is an effect on protein stability, and the regulation of β-catenin signaling by GSK-3β is a good example of the ability of GSK-3β to affect protein degradation via the proteasome pathway. Notably, we did not observe any change either in the in vivo half-life of p190A mutants that are unable to undergo phosphorylation by GSK-3β or in wild-type p190A in cells treated with pharmacologic inhibitors of GSK-3β, indicating that phosphorylation by GSK-3β does not detectably affect p190A protein levels.
The mechanism by which p190A phosphorylation by GSK-3β contributes to polarized cell movement appears to involve inhibitory consequences of such phosphorylation on the RhoGAP activity of p190A in directionally migrating cells. The ability of GSK-3β phosphorylation of p190A to inhibit RhoGAP catalytic activity in vitro implicates a conformational effect of phosphorylation on the catalytic domain, although further structural studies would be required to confirm such a mechanism. Consistent with a role for GSK-3β phosphorylation in regulating p190A RhoGAP activity, we observed that the phosphorylation-defective S1476A and T1480A p190A mutants exhibited a substantially increased ability to promote morphologic changes in cells that resemble those seen when Rho GTPase activity is inhibited. Notably, the fact that cells expressing p190A mutants exhibiting excessive RhoGAP activity and cells deficient for p190A RhoGAP activity are similarly defective for polarized migration suggests that fine modulation of Rho GTPase activity is critical for this process. This is consistent with previous findings demonstrating that both p190A knockdown and p190A overexpression lead to a disruption in cell migration (9, 14).
Recent findings have implicated p190A in caveolin-1-dependent directional cell migration (15, 30). Caveolin-1-deficient fibroblasts were found to be defective for polarized migration, and interestingly, this defect can be rescued by knockdown of p190A (15). The caveolin-1-deficient cells exhibit increased Src kinase activity as well as consequently increased Src-dependent p190A tyrosine phosphorylation, which has been shown previously to promote p190A RhoGAP activity (10, 31, 32). A model was proposed wherein caveolin-1 promotes p190A-mediated Rho inhibition via Src kinase activation. Taken together with our findings, these observations suggest that p190A is not absolutely essential for directional cell migration but, rather, is probably required for the fine modulation of Rho activity that facilitates this complex process in response to upstream signals. Because caveolins specifically accumulate within the cavelolae/lipid raft subdomain of plasma membranes and have been implicated as scaffolds that promote signaling events within membrane subregions, it is possible that caveolin-dependent p190A regulation of directional cell migration gives rise to regional restriction of RhoGAP activity within leading edge membrane protrusions. Notably, a recent report demonstrated a role for filamin in recruiting p190A to lipid rafts of endothelial cells to promote Rho inactivation during cell spreading (33).
Although the studies described here have addressed a role for p190A in fibroblasts, it is interesting to consider the major phenotypes identified in p190A knock-out mice, which largely involve developmental defects in neuronal axon outgrowth and guidance (5). Taken together with accumulating evidence indicating that GSK-3 plays a critical role in neuronal polarity and axon growth (34) and the recent report demonstrating a role for p190A in dendritic spine morphogenesis downstream of PAR6 (29), our findings raise the possibility that GSK-3β-mediated phosphorylation of p190A also plays an essential role in the polarization of neurons.
Supplementary Material
Acknowledgments
We are grateful to members of the Settleman laboratory for many helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA62142 (to J. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GSK-3β, glycogen synthase kinase-3-β; DMEM, Dulbecco's modified Eagle's medium; DMSO, dimethyl sulfoxide; DTT, dithiothreitol; DAPI, 4′,6-diamidino-2-phenylindole; MAP, mitogen-activated protein; MAPK, MAP kinase; CKI, casein kinase I; PKC, protein kinase C; WT, wild type; GST, glutathione S-transferase; GFP, green fluorescent protein; EGFP, enhanced green fluorescent protein.
J. Settleman, unpublished observation.
References
- 1.Jaffe, A. B., and Hall, A. (2005) Annu. Rev. Cell Dev. Biol. 21 247-269 [DOI] [PubMed] [Google Scholar]
- 2.Fukata, M., Nakagawa, M., and Kaibuchi, K. (2003) Curr. Opin. Cell Biol. 15 590-597 [DOI] [PubMed] [Google Scholar]
- 3.Bernards, A. (2003) Biochim. Biophys. Acta 1603 47-82 [DOI] [PubMed] [Google Scholar]
- 4.Brouns, M. R., Matheson, S. F., Hu, K. Q., Delalle, I., Caviness, V. S., Silver, J., Bronson, R. T., and Settleman, J. (2000) Development (Camb.) 127 4891-4903 [DOI] [PubMed] [Google Scholar]
- 5.Brouns, M. R., Matheson, S. F., and Settleman, J. (2001) Nat. Cell Biol. 3 361-367 [DOI] [PubMed] [Google Scholar]
- 6.Sordella, R., Classon, M., Hu, K. Q., Matheson, S. F., Brouns, M. R., Fine, B., Zhang, L., Takami, H., Yamada, Y., and Settleman, J. (2002) Dev. Cell 2 553-565 [DOI] [PubMed] [Google Scholar]
- 7.Sordella, R., Jiang, W., Chen, G. C., Curto, M., and Settleman, J. (2003) Cell 113 147-158 [DOI] [PubMed] [Google Scholar]
- 8.Bradley, W. D., Hernandez, S. E., Settleman, J., and Koleske, A. J. (2006) Mol. Biol. Cell 17 4827-4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur, W. T., and Burridge, K. (2001) Mol. Biol. Cell 12 2711-2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthur, W. T., Petch, L. A., and Burridge, K. (2000) Curr. Biol. 10 719-722 [DOI] [PubMed] [Google Scholar]
- 11.Liang, X., Draghi, N. A., and Resh, M. D. (2004) J. Neurosci. 24 7140-7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burbelo, P. D., Miyamoto, S., Utani, A., Brill, S., Yamada, K. M., Hall, A., and Yamada, Y. (1995) J. Biol. Chem. 270 30919-30926 [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. S., Kim, J. G., Moon, M. Y., Jeon, C. Y., Won, H. Y., Kim, H. J., Jeon, Y. J., Seo, J. Y., Kim, J. I., Kim, J., Lee, J. Y., Kim, P. H., and Park, J. B. (2006) Blood 108 1821-1829 [DOI] [PubMed] [Google Scholar]
- 14.Kusama, T., Mukai, M., Endo, H., Ishikawa, O., Tatsuta, M., Nakamura, H., and Inoue, M. (2006) Cancer Sci. 97 848-853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grande-Garcia, A., Echarri, A., de Rooij, J., Alderson, N. B., Waterman-Storer, C. M., Valdivielso, J. M., and del Pozo, M. A. (2007) J. Cell Biol. 177 683-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peacock, J. G., Miller, A. L., Bradley, W. D., Rodriguez, O. C., Webb, D. J., and Koleske, A. J. (2007) Mol. Biol. Cell 18 3860-3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sastry, S. K., Rajfur, Z., Liu, B. P., Cote, J. F., Tremblay, M. L., and Burridge, K. (2006) J. Biol. Chem. 281 11627-11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernards, A., and Settleman, J. (2004) Trends Cell Biol. 14 377-385 [DOI] [PubMed] [Google Scholar]
- 19.Kim, L., and Kimmel, A. R. (2006) Curr. Drug Targets 7 1411-1419 [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni, S. V., Gish, G., van der Geer, P., Henkemeyer, M., and Pawson, T. (2000) J. Cell Biol. 149 457-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, R., Hu, K. Q., Lu, Y., Nolan, K. M., Thissen, J., and Settleman, J. (1996) Mol. Cell. Biol. 16 2689-2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635-3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinfeld, B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S., and Polakis, P. (1996) Science 272 1023-1026 [DOI] [PubMed] [Google Scholar]
- 24.Harwood, A. J. (2002) Dev. Cell 2 384-385 [DOI] [PubMed] [Google Scholar]
- 25.Yost, C., Torres, M., Miller, J. R., Huang, E., Kimelman, D., and Moon, R. T. (1996) Genes Dev. 10 1443-1454 [DOI] [PubMed] [Google Scholar]
- 26.Tatsis, N., Lannigan, D. A., and Macara, I. G. (1998) J. Biol. Chem. 273 34631-34638 [DOI] [PubMed] [Google Scholar]
- 27.Etienne-Manneville, S., and Hall, A. (2003) Nature 421 753-756 [DOI] [PubMed] [Google Scholar]
- 28.Etienne-Manneville, S., and Hall, A. (2003) Curr. Opin. Cell Biol. 15 67-72 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, H., and Macara, I. G. (2008) Dev. Cell 14 216-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickstrom, S. A., Alitalo, K., and Keski-Oja, J. (2003) J. Biol. Chem. 278 37895-37901 [DOI] [PubMed] [Google Scholar]
- 31.Haskell, M. D., Nickles, A. L., Agati, J. M., Su, L., Dukes, B. D., and Parsons, S. J. (2001) J. Cell Sci. 114 1699-1708 [DOI] [PubMed] [Google Scholar]
- 32.Dumenil, G., Sansonetti, P., and Tran Van Nhieu, G. (2000) J. Cell Sci. 113 71-80 [DOI] [PubMed] [Google Scholar]
- 33.Mammoto, A., Huang, S., and Ingber, D. E. (2007) J. Cell Sci. 120 456-467 [DOI] [PubMed] [Google Scholar]
- 34.Li, R. (2005) Curr. Biol. 15 R198-R200 [DOI] [PubMed] [Google Scholar]
- 35.Nobes, C. D., and Hall, A. (1999) J. Cell Biol. 144 1235-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ligeti, E., and Settleman, J. (2006) Methods Enzymol. 406 104-117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.