Abstract
We present the data and the technology, a combination of which allows us to determine the identity of proprotein convertases (PCs) related to the processing of specific protein targets including viral and bacterial pathogens. Our results, which support and extend the data of other laboratories, are required for the design of effective inhibitors of PCs because, in general, an inhibitor design starts with a specific substrate. Seven proteinases of the human PC family cleave the multibasic motifs R-X-(R/K/X)-R↓ and, as a result, transform proproteins, including those from pathogens, into biologically active proteins and peptides. The precise cleavage preferences of PCs have not been known in sufficient detail; hence we were unable to determine the relative importance of the individual PCs in infectious diseases, thus making the design of specific inhibitors exceedingly difficult. To determine the cleavage preferences of PCs in more detail, we evaluated the relative efficiency of furin, PC2, PC4, PC5/6, PC7, and PACE4 in cleaving over 100 decapeptide sequences representing the R-X-(R/K/X)-R↓ motifs of human, bacterial, and viral proteins. Our computer analysis of the data and the follow-on cleavage analysis of the selected full-length proteins corroborated our initial results thus allowing us to determine the cleavage preferences of the PCs and to suggest which PCs are promising drug targets in infectious diseases. Our results also suggest that pathogens, including anthrax PA83 and the avian influenza A H5N1 (bird flu) hemagglutinin precursor, evolved to be as sensitive to PC proteolysis as the most sensitive normal human proteins.
The dual objective of our study was to compare the furin recognition pattern with the recognition patterns of the other proteinases of the proprotein convertase (PC)3 family and to identify the individual PCs, which are the most promising host cell drug targets in infectious diseases. Furin and related PCs are specialized serine endoproteinases that cleave the multibasic motifs R-X-(R/K/X)-R↓, and thus transform proproteins into biologically active proteins and peptides (1–3). Structurally and functionally, furin resembles its evolutionary precursor: the prohormone-processing kexin of yeast Saccharomyces cerevisiae. Furin, because of its ubiquity and biological importance, is currently the most studied enzyme of the PC family. Seven PCs (furin, PC1/3, PC2, PC4, PACE4, PC5/6, and PC7) have been identified in humans (4–6). Furin, incidentally, is expressed in all examined tissues, and cell lines and is mainly localized in the trans-Golgi network. Some proportion of the furin molecules cycles between the trans-Golgi and the cell surface (3). Because of the overlapping substrate preferences and cell/tissue expression, there is some redundancy in PC functionality, albeit certain distinct functions of individual PCs have also been demonstrated. For example, furin knock-out is lethal in mice (7).
In addition to normal cell functions, PCs, including furin, are implicated in many pathogenic states because they process to maturity membrane fusion proteins and pro-toxins of a variety of both bacteria and viruses, including anthrax and botulinum toxins, influenza A H5N1 (bird flu), flaviviruses, Marburg, and Ebola viruses (8–18). After processing by furin and the subsequent endocytic internalization in the complex with the respective cell surface receptor followed by acidification of the endosomal compartment, the processed, partially denatured, infectious proteins expose their membrane-penetrating peptide region and escape into the cytoplasm (8). The intact toxins and viral proteins, however, are incapable of accomplishing these processes, because they cannot penetrate the membrane and escape into the cytoplasm. Evidence suggests that the inhibition of cellular furin and other PCs prevents aggressive viral and bacterial diseases (19–23). This evidence leads to the suggestion that furin and related PCs are promising host cell drug targets in infectious diseases. The precise sensitivity of the individual pathogens to processing by the individual PCs, however, has not been determined and designing disease-specific antagonists is, therefore, difficult. Conversely, because furin and PCs are required for the processing and activation of multiple normal human proteins, one may reasonably expect that wide-range PC antagonists will interfere with normal cell functions and possibly elevate the level of side effects.
To shed more light on the selectivity and efficiency of the individual PCs in the physiological processing of normal human proteins compared with that of the pathogens, we evaluated the relative efficiency of furin, PC2, PC4, PC5/6, PC7, and PACE4 in cleaving over 100 decapeptide sequences. The peptides we used included the R-X-(R/K/X)-R↓ cleavage motifs of human proteins and proteins of bacterial and viral origin. The follow-on cleavage analysis of the selected full-length proteins by the individual PCs was then used to corroborate the peptide cleavage data we had generated. Our experimental results suggest that the anthrax protective antigen (PA83) and the influenza A H5N1 hemagglutinin (HA) evolved to increase their sensitivity to processing by the PCs. As a result, the sensitivity of these pathogens equals or exceeds that of normal human proteins allowing these pathogens to compete efficiently with physiological human targets for proteolytic processing by the PCs. We also identified the individual PCs that are most relevant to the processing of specific pathogens.
MATERIALS AND METHODS
Reagents—All reagents were purchased from Sigma unless indicated otherwise. An inhibitor of PCs (decanoyl-Arg-Val-Lys-Arg-chloromethylketone; dec-RVKR-cmk) and a hydroxamate inhibitor of matrix metalloproteinases (GM6001) were obtained from Bachem and Chemicon, respectively. Fmoc amino acids, N,N′-diisopropylcarbodiimide, and biotin resin, which we used in the peptide synthesis were purchased from Novabiochem-EMD Biosciences. The solvents used in peptide synthesis were from VWR International. Anthrax PA83 and Pseudomanas exotoxin A (PEx) were purchased from List Laboratories. The ectodomain of avian influenza A H5N1 hemagglutinin precursor (HA) was expressed in a baculoviral expression system and purified as described earlier (22).
Recombinant PCs—Recombinant PCs, including human furin, PC1/3, PC2, PC5/6, PC7, PACE4, and murine PC4 were prepared using the S2 Drosophila expression system (Invitrogen) and purified to homogeneity as described earlier (4, 24). One unit of activity is equal to the amount of the enzyme that is required to cleave 1 pmol/min of the pyroglutamic acid-Arg-Thr-Lys-Arg-methyl-coumaryl-7-amide (Pyr-RTKR-AMC) substrate at 37 °C. The Km value of furin, PC1/3, PC2, PC4, PC5/6, PC7, and PACE4 against Pyr-RTKR-AMC we determined was 6.5, 3.0, 6.6, 1.7, 2.0, 9.5, and 3.0 μm, respectively. We determined that the specific activity of furin, PC1/3, PC2, PC4, PC5/6, PC7, and PACE4 was 10.8, 2.8, 11.9, 1.4, 2.1, 3.0, and 3.7 units/μg, respectively.
In Vitro Cleavage of PA83, HA, and PEx—The cleavage reactions (22 μl each) were performed using the following buffers. The buffer for furin was 100 mm HEPES, pH 7.5, containing 1 mm CaCl2 and 1 mm β-mercaptoethanol. The buffer for PC1/3, PC4, PC5/6, PC7, and PACE4 was 20 mm Bis-Tris, pH 6.5, supplemented with 1 mM CaCl2. The PC2 buffer was 20 mm Bis-Tris, pH 5.6, supplemented with 1 mM CaCl2 and 0.1% (w/v) Brij30. The indicated amounts of furin, PC1/3, PC2, PC4, PC5/6, PC7, and PACE4 were incubated for 60–180 min at 37 °C with anthrax PA83, PEx, and HA (1 μm each). The cleavage reactions were stopped using 5× SDS sample buffer. The digests were analyzed by SDS-PAGE followed by Coomassie Blue staining. The images were scanned using an AlphaImager (Alpha Innotech), and the band density was digitized using the MultiGauge image analysis and processing software (FujiFilm).
Cleavage of Synthetic Peptides Followed by Mass Spectrometry—The peptides 102VRRRRRYALS (1332.6 Da) and 157VRRRRRYSLS (1348.6 Da) that span the furin cleavage sites in the human and mouse prodomain of MT6-MMP, respectively, were synthesized by GenScript. The peptides (1.5 μg each) were incubated for 2 h at 37 °C in the cleavage buffer supplemented with furin (two and five units of activity) and PC2 (five units of activity). The masses of the intact peptides and the cleavage products were determined by MALDI-TOF MS (matrix-assisted laser-desorption ionization-time-of-flight mass spectrometry) using an Autoflex II mass spectrometer (Brucker Daltonics) at the Center for Proteolytic Pathways at the Burnham Institute. The predicted mass of the 102VRRRRR and 157VRRRRR cleavage products is 898.1 Da. Where indicated, dec-RVKR-cmk (1 μm) was added to the reactions to inhibit the PCs.
Synthesis of Peptide-Oligonucleotide Conjugates—The synthesis, the conjugation, and the follow-on procedures were described in detail earlier (25). Briefly, high throughput peptide synthesis was performed in wells of a 96-well flat bottom polypropylene microtiter plate (Evergreen Scientific) in a custom built centrifugal peptide synthesizer (26, 27). The resin (Nova Tag, Novabiochem-EMD Bioscience) modified with Fmoc-Gly-biotin-PEG was used for the peptide synthesis. Peptides were synthesized using Fmoc chemistry and benzotriazole-1-yl-oxy-tris-(dimethylamino)-phosphoniumhexafluorophosphate as the coupling reagent and 4-methylpiperidine as the deprotection reagent (28, 29). The N terminus of each peptide molecule contained a (His)5 tag and hydroxyaminoacetic acid, while the C-terminal end of the peptides was linked to biotin. The peptides were cleaved from the resin by a trifluoroacetic acid/thioanisol/water/phenol/1,2-ethanedithiol mixture (82.5:5:5:5:2.5 v/v) (30) and precipitated with ether. The ether wash was then repeated three times. The peptides were dried using a SpeedVac centrifuge. The purity of the peptides was confirmed by reverse-phase HPLC on a μBondapak C18 column (10 μ, 125 Å, 3.9 × 150 mm) using a gradient of solvent (A) 0.05% trifluoroacetic acid/water and solvent (B) 0.05% trifluoroacetic acid/70% acetonitrile (from 5 to 60% of B in 15 min) on an Agilent 1100 HPLC instrument and also by MALDI-TOF MS performed at HT-Labs (San Diego, CA) and at the Center for Proteolytic Pathways at the Burnham Institute.
The oligonucleotide tags were synthesized using a high-throughput oligonucleotide synthesizer at Illumina, San Diego, CA (27, 31). Oligonucleotides were modified by coupling a phosphoramidite containing a formylindole group (Link Technologies, Bellshill, Scotland) as the last building block in DNA synthesis. Hydroxylamine-modified peptide substrates and aldehyde-modified oligonucleotides were incubated in 0.2 m sodium citrate buffer, pH 5.0, to form an oxime linkage. The reactions were then quenched using 8 m urea. The peptide-oligonucleotide conjugates were pooled and purified from the residual free reactants by polyacrylamide gel electrophoresis. The concentration of the purified peptide-oligonucleotide conjugates was determined using a spectrophotometer and adjusted to result in a master pool with a 5 nm concentration of the individual conjugates.
Proteinase Assay of the Peptide-Oligonucleotide Conjugates— The aliquots of the peptide-oligonucleotide conjugate master pool were diluted 10-fold using an appropriate buffer. The diluted aliquots (20 μl each) were mixed with an equal volume of the PC sample in the same buffer. The buffer composition of the cleavage reactions containing furin, PC2, PC4, PC5/6, PC7, and PACE4 is shown above under “In Vitro Cleavage of PA83, HA, and PEx.” The reactions containing an aliquot of the master pool, and the buffer only were used as a negative control. The reactions containing an aliquot of the master pool, the proteinase (0.2–2 units), and, in addition, trypsin (40 nm) were used as a positive control. Reactions were incubated at 37 °C for 45 min in the wells of a 96-well plate. Reactions were then heated for 10 min at 95 °C to inactivate the proteolytic activity. For the complete pull-down of the peptides a 10-μl aliquot (0.6% slurry (w/v); 100 pmol streptavidin)) of streptavidin-coated magnetic beads (Seradyn) was added to each reaction. A positive control received a 10-μl buffer aliquot. After a 15-min incubation at ambient temperature, magnetic beads were sedimented by placing the plate into a magnetic particle concentrator Dynal MPC-96S (Invitrogen) for 2 min. Following a 10-fold dilution with the hybridization buffer (100 mm K2HPO4, 1 m sodium chloride, 20% formamide, pH 7.6), the reaction supernatants (50 μl each) were employed for the peptide-oligonucleotide conjugate hybridization using Sentrix Array Matrices (SAMs).
Hybridization and Read-out—SAMs comprising 96-fiber optic bundles, each with 1624 different bead types, were used for hybridization and data measurement. Every bead type of the 1624 contained a unique oligonucleotide sequence that was complementary to the respective oligonucleotide of the peptide-oligonucleotide conjugate. SAMs were removed from the packaging and washed for 1 min with 95% formamide and with the 100 mm potassium phosphate, 1 m sodium chloride, and 20% formamide, hybridization buffer, pH 7.6. The PC cleavage samples were each diluted 1:10 in the hybridization buffer. Aliquots (45 μl each) of the diluted samples were transferred to the wells in a 384-well black-bottom plate and in the controlled-humidity hybridization chamber where they were allowed to hybridize at 48 °C overnight to the individual bundles of SAMs. Each sample was analyzed in triplicate.
After the hybridization, SAMs were extensively washed, initially using the hybridization buffer and then PBS, 0.1% Tween-20. After washing, SAMs were incubated for 2 h at ambient temperature in 45 μl of PBS-0.1% Tween-20, 0.1% casein containing 0.1 μg/ml murine (His)5 antibody (Qiagen). After washing four times with PBS-0.1% Tween-20, the SAMs were incubated for 1 h at ambient temperature in 45 μl of PBS, 0.1% Tween-20, 0.1% casein containing 1 μg/ml Alexa 555-conjugated goat anti-mouse IgG and then washed twice with PBS, 0.1% Tween-20 and once with PBS. To measure the fluorescence, SAMs were scanned using an Illumina Bead Array Reader that measured the fluorescence intensity of each bead on each bundle. The Illumina software matched the fluorescent signal of the individual beads with the sequence of the hybridized peptide-oligonucleotide conjugate (32). The peptide cleavage was calculated using the following equation: 100 × [(assay signal intensity - negative control signal intensity)/(assay signal intensity - positive control signal intensity)]. To eliminate errors, the peptide-oligonucleotide conjugates with a positive-to-negative ratio below 2 were not considered.
The Analysis of Cleavage Data—The sequence logos were obtained by calculating the average cleavage efficiency for each enzyme over the entire set of substrates and then selecting those substrates that had cleavage efficiency above the average value for the enzyme. The actual logos were created by a web-based Weblogo program (33, 34).
An additional analysis of the cleavage efficiency was performed by a specialized computer program we had developed. The program determines the contribution of each amino acid residue at each of the positions near to the scissile bond to the efficiency of the peptide proteolysis by a proteinase. First, the program calculates the average value of the cleavage efficiency for each proteolytic enzyme over the whole set of substrates. The program then calculates the average cleavage efficiency associated with each amino acid residue type at each of the substrate subsites (in our case, at each of the P6-P4′ positions of the decapeptides we used). An average cleavage efficiency for an amino acid residue at a given substrate subsite position is determined by averaging the cleavage efficiency of all substrates that have this specific amino acid at this given substrate subsite. This calculation is performed for all amino acid residue types and their positions in the peptide substrate. The averages obtained in this way yield the effective cleavage efficiency value for each amino acid residue type, and this value is not affected by the occurrence of other amino acids at other positions. The effective cleavage efficiency values of the individual amino acids are next compared with the global average of the cleavage efficiency for a proteolytic enzyme. The differences between these two values indicate the specific contribution, which could be either negative or positive, of an amino acid at a specific subsite position of the substrate to the cleavage efficiency of the peptide substrate by a proteinase. This approach yields highly reliable results if a large number of substrates is taken into consideration. Here, there are ∼100 natural substrates and, therefore, the level of representation of amino acid types at the certain peptide substrate subsites is limited. The results of our calculations are presented in Fig. 3. The plots represent the differences between the effective cleavage efficiency values calculated for each amino acid type at a given position of the peptide substrate, and the average cleavage efficiency global value calculated for the entire set of substrates for a given enzyme. The 0 number in the plots denotes that the effective cleavage efficiency value of an amino acid residue is equal to the global value of the entire set of substrates for a given enzyme.
FIGURE 3.
Analysis of the cleavage preferences of the individual PCs in a radar-plot format. The plots represent the differences between the effective cleavage efficiency values calculated for each amino acid type at a given position of the peptide substrate and the average cleavage efficiency global value calculated for the entire set of substrates for a given PC. The 0 number in the plots denotes that the effective cleavage efficiency value of an amino acid residue is equal to the global value of the entire set of substrates for a given PC.
RESULTS
In general, an efficient approach to develop selective, safe and potent inhibitors of proteinases is to start an inhibitor design with a known substrate. Many previous studies have established that furin and related PCs exhibit similar cleavage preferences and that the presence of a minimal (K/R)-R motif at the P1-P2 positions is required to cleave the protein and peptide substrates by the individual PCs. A detailed comparative analysis of the cleavage preferences of the PCs, however, has never been performed, and this hole in our knowledge makes the design of effective inhibitors of the individual PCs exceedingly difficult. To facilitate the determination of the cleavage preferences of the PCs we synthesized those decapeptides, whose sequence spans the R-X-(R/K/X)-R↓ cleavage motif of ∼100 known cleavage targets of furin. These targets represent human proteins and human pathogens of bacterial and viral origin. Earlier studies have proved the efficiency and the accuracy of the custom-built centrifugal synthesizer, and the synthetic scheme we used and the high quality of the synthesized peptides (22, 35–37). These synthetic peptides were used in the cleavage reactions to identify the cleavage preferences of furin, PC2, PC4, PC5/6, PC7, and PACE4. To advance our cleavage study, we have used a miniaturized and multiplexed solution assay methodology for the measurement of proteinase activity.
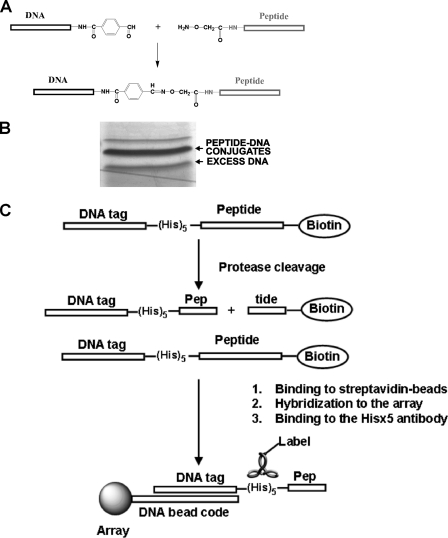
Multiplexed Cleavage Assay— The multiplexed profiling assay methodology we used has been validated by using over 1000 sequences representing substrates for several proteinase classes, including trypsin, chymotrypsin, thrombin, factor Xa, caspases, matrix metalloproteinases, enterokinase, and many other enzymes (25). This multiplexed methodology greatly accelerates a determination of the relative specificity and cleavage efficiency of the proteinases, including the PCs we employed in our study. Fig. 1 shows the principal steps of the methodology we used. The cleavage assay employs a biotin labeled peptide sequence C-terminally linked to a (His)5 tag and then additionally conjugated with an oligonucleotide tag. The peptide-oligonucleotide conjugates were then separated from the residual free reactants by polyacrylamide gel electrophoresis. The purified peptide-oligonucleotide conjugates were used as substrates of the individual PCs in the cleavage reactions. Following the proteolytic cleavage both the cleaved biotin-labeled C-terminal portion of the peptide-oligonucleotide conjugate and the residual of the intact conjugate were removed by incubation with streptavidin-coated magnetic beads. To screen the peptides and to distinguish the peptides, which are either resistant or are relatively insensitive to the PCs, we specifically chose exhaustive proteolysis conditions for our cleavage reactions. The cleaved parts of the conjugates that remained in solution were captured by hybridization of their oligonucleotide sequence to Sentrix Bead Arrays and detected using a labeled antibody against a (His)5 tag of the conjugate. The recorded cleavage data are summarized in supplemental Table S1. These additional data verified that the synthetic method and the follow-on peptide cleavage screening are applicable to the analyses of many endoproteinase types instead of for PCs alone. We are now confident that the peptide synthesis and peptide cleavage assay methods we have designed will be used by other laboratories interested in employing a time-saving, efficient method of rapidly and precisely determining the cleavage preferences of proteolytic enzymes.
FIGURE 1.
Multiplexed oligonucleotide-peptide cleavage assay. A, synthesis of peptide-oligonucleotide conjugates. Peptides were synthesized using Fmoc chemistry. The N terminus of each peptide molecule contained a (His)5 tag and hydroxyaminoacetic acid, while the C-terminal end of the peptides was linked to biotin. Synthetic oligonucleotides were modified by coupling a phosphoramidite containing a formylindole group. Hydroxylamine-modified peptides and aldehyde-modified oligonucleotides were co-incubated to form an oxime linkage. B, purification of the peptide-oligonucleotide conjugates using polyacrylamide gel electrophoresis. C, peptide cleavage assay and hybridization. The purified peptide-oligonucleotide conjugates were incubated with the individual PCs. After an exhaustive proteolysis, the biotin-labeled cleavage products and the residual intact peptides were pulled-down from the cleavage reactions using streptavidin-coated magnetic beads. The reaction supernatants were hybridized using SAMs containing unique oligonucleotide sequences that were complementary to the respective oligonucleotide of the peptide-oligonucleotide conjugates. The hybridized samples were incubated with a murine (His)5 antibody followed by incubation with Alexa 555-conjugated goat anti-mouse IgG. The resulting fluorescence signal was measured using an Illumina Bead Array Reader. Each sample was analyzed in triplicate.
The Cleavage Data Analysis—It is clear from a review of the data presented in supplemental Table S1 that the peptides, which represented the R-X-(R/K/X)-R↓ cleavage motif of the known human targets of furin, vary widely in their sensitivity to proteolysis by an individual PC. It should be pointed out that a comparison of the efficiency of hydrolysis of the peptides by a specific enzyme is valid but a comparison between the PCs is not meaningful because of our assay conditions different amounts of the individual enzymes were used in the cleavage reactions.
We established a relationship between the cleavage efficiency of the PCs and the frequency of the occurrence of the individual amino acids at specific positions relative to the scissile bond. For this purpose, we calculated the average cleavage efficiency for each PC and then presented the cleavage peptides as groups with the cleavage above and below the average cleavage efficiency. The P6-P4′ peptide sequence of the groups is presented in the form of sequence logos using the Weblogo program (Fig. 2) (33). The height of a character is proportional to the frequency of the amino acid residue at the individual position of the cleavage peptide. Obviously, because the peptide substrates were derived from known protein targets of furin, the R-(R/K)-(R/K)-R pattern is predominant at the P4-P1 positions of the peptides. Note that PC2 strongly prefers the presence of Lys instead of Arg at P2 and that the cleavage preferences of PC2 are significantly distinct from those of all other PCs, including furin. The promiscuity of the amino acid representation is significantly higher at the P6, P5, P3, and P1′-P4′ positions of the cleavage peptides. The efficient substrates most frequently exhibit Ser, Ala, and Glu or Asp at the P1′ position. The presence of Met at P5, Gly at P2′, Val at P3′, and Pro at P4′ is the common feature that decreases the efficiency of PC proteolysis of the natural substrate sequences. It is also clear that the cleavage preferences of PC2 are significantly distinct from those of all other PCs, including furin.
FIGURE 2.
Frequency plot of the cleavage sequence of the individual PCs in a Weblogo format. The size of the symbol indicates the frequency of the individual residue occurrence at individual substrate positions relative to the P1-P1′ scissile bond.
We continued our analysis by determining the effect of each amino acid residue at each of the P6-P4′ positions of the peptide substrates on the efficiency of the peptide cleavage by the individual PCs. For this purpose, we first calculated the average efficiency of each PC using the entire set of substrates. We then calculated an effective cleavage efficiency for each amino acid at each of the P6′-P4 positions. This value is the average cleavage efficiency of the peptide in which this amino acid type is present. The identical efficiency of the substrate hydrolysis by the individual PCs, which we determined in our multiplexed cleavage assays, was assigned to each of the P6-P4′ residues of the peptide substrate. This approximation established an effective cleavage efficiency value for each amino acid residue at each position. We then compared the values of the individual amino acid types with the average efficiency of the proteolysis of all peptides by each of the individual PCs. The difference between these two values indicates if the presence of a specific amino acid type at the specific substrate position either decreases or increases the efficiency of the cleavage of the peptide substrate by the individual PC. We know from experience that this approach produces highly reliable data, especially when many hundreds or thousands of substrates are analyzed. The limited number of peptide substrates we analyzed for this project results in a limited number of occurrences of the individual amino acid types at the individual substrate subsites. Regardless, our analysis provides a valuable insight into how the presence of the individual amino acid types affects the hydrolytic efficiency of the peptidic substrates by the individual PCs.
Fig. 3 shows the results of our calculations in the form of radar-type plots. These plots clearly show the effect of the amino acid type at the individual P6-P4′ positions of the peptides on the hydrolytic efficiency of the individual PCs. A zero value was assigned to represent an average cleavage efficiency for each enzyme. A deviation from this value reflects the effect of the amino acid type at this particular subsite: a negative value indicates suppression of the proteolysis when compared with the average, while a positive value indicates that the presence of this specific amino acid residue at this subsite favors the proteolysis of the substrate by the individual PC. A number in parentheses next to an amino acid symbol shows the number of amino acid occurrences at this specific substrate subsite in all of the substrates we analyzed. Based on our analysis, the cleavage preferences of PCs can be divided into three groups. The first group includes only furin, the second, closely related group includes PC4, PC5/6, PC7, and PACE4, and the third, distantly related group, is comprised of PC2.
There are many preliminary conclusions that can be drawn from the data we have gathered and presented. Some of these conclusions could also be drawn, albeit in a less clear way, from the logos-type analysis (Fig. 2). A sample of these conclusions follows. At P6, the presence of Gly, Asp, Asn, Glu, Gln, and Leu produces the most significant effect on the efficiency of the proteolysis by the PCs. The presence of the hydrophobic amino acid residues (Val, Ile, and Leu) and either Gln or Asn at P5 is favorable for all of the PCs. The P4 position is predominantly occupied by Arg in the natural cleavage substrates. It appears that the occurrence in Gln and Ile, albeit infrequent in the natural substrate subset we analyzed, stimulates the activity of PC2, PC4, PC5/6, and PACE4. The presence of the charged Glu, Lys, and His, the hydrophobic Phe and Tyr and the small-size Gly does not favor PC proteolysis of the peptide substrates.
The presence of Lys, Arg, Gln, and Ser dominates the P3 position of the substrates. Our data suggest that Arg, Val, and Leu are favorable for multiple PCs except PC2 that is more efficient if Glu occupies the P3 position of the substrate. In agreement with the logos data analysis (Fig. 2), the presence of Pro at P2 negatively affects the efficiency of the substrate cleavage by the PCs. In contrast to other PCs, which favor Arg, the presence of Lys at P2 is favorable for the high sensitivity of the peptide substrate to PC2 proteolysis. A few substrates with Glu and Ser are poorly cleaved by all of the PCs. The P2 Val increases the activity of PC4, PC5/6, and PACE4 but not furin, PC2 and PC7.
Only positively charged Arg and Lys are allowed at the P1 position. Lys, however, is associated with the low hydrolysis efficiency of all of the PCs. Ala, Cys, Ser, Asp, and Glu are the most frequent residues at P1′ but apparently Thr and Arg increase the sensitivity of the substrate to PC2 proteolysis while Glu selectivity increases the activity of the PCs other than PC2. The presence of either Val or Pro and Cys at P1′ decreases the efficiency of the PC proteolysis of the peptides. The P2′, P3′, and P4′ positions appear less sensitive to amino acid substitutions than other substrate subsites to the occupying residue type. The P2′ Ile is likely to stimulate the proteolysis of the peptides by the PCs. The significant stimulatory effect of Asp, and especially Pro at P3′ on the activity of PCs (except PC2) is also evident. The P4′ Met enhances the peptide cleavage by PC4, PC5/6, PC7, and PACE4 but not by furin and PC2.
Cleavage of the MT6-MMP Peptides—According to our cleavage data, the P2′ Ala is favorable for furin proteolysis, while the P2′ Ser is unfavorable. Both Ala and Ser at P2′ are acceptable for PC2, which, however, strongly prefers the presence of Lys (but not Arg) at the P2 position. To corroborate these preliminary conclusions derived from our peptide cleavage data, we elected to use the peptide derived from the known furin cleavage sites of human and murine MT6-MMP (102VRRRRR↓YALS and 157VRRRRR↓YSLS, respectively). The presence of Ala at P2′ discriminates the human proteinase from the murine enzyme that exhibits Ser at this position. We expected that PC2 would be less efficient in cleaving both the human and murine MT6-MMP peptides when compared with furin and that furin would be more efficient in specifically cleaving the human MT6-MMP peptide.
Both peptides were subjected to digestion with furin and PC2. The digest samples were analyzed by MALDI-TOF MS to determine the molecular mass of the cleavage products (Fig. 4). Consistent with our expectations, the residual levels of the intact peptides and the levels of the degradation product VRRRRR (898 Da) we determined in the cleavage reactions demonstrated that the murine peptide with the P2′ Ser was significantly more resistant to furin proteolysis and that PC2 was significantly less potent than furin in cleaving both peptides.
FIGURE 4.
Mass spectrometry analysis of the cleavage peptides. The human MT6-MMP peptide VRRRRR↓YALS and the mouse MT6-MMP peptide VRRRRR↓YSLS were incubated for 2 h at 37 °C with furin and PC2 (5 activity units each). The additional furin reactions, in which 2 units of furin were used, are indicated in the panels. The molecular mass of the peptides was determined by MALDI-TOF MS. There was no difference between the calculated and the estimated molecular mass of the peptides. Where indicated, dec-RVKR-cmk was added to the cleavage reactions. Insets, in the results of furin reactions, which contained dec-RVKR-cmk are presented as insets. The molecular mass of the intact human and mouse peptide is 1332 and 1348 Da; respectively (the numbers are underlined in the figure panels). The molecular mass of the N-terminal cleavage product VRRRRR is 898 Da.
Protein Cleavage Studies—To further support our cleavage data, HA, PA83, and PEx were each co-incubated for 1–3 h with increasing amounts (1–10 activity units) of furin, PC1/3, PC2, PC4, PC5/6, PC7, and PACE4. The digest samples were separated by SDS-PAGE. The residual undigested precursor and the respective mature proteins, which were generated because of the precursor proteolysis, were then identified using Coomassie Blue staining of the gels (HA, PA83, and PEx). The images were scanned, the band density was digitized, and graphed. An example of the proteolysis of HA by the individual PCs is shown in Fig. 5. Multiple additional cleavage reactions, which we analyzed, are presented in supplemental Fig. S1.
FIGURE 5.
Cleavage of HA and PA83 by PCs. A, individual PCs cleave the HA precursor. The HA precursor (1 μm) was incubated for 1 h at 37 °C with the individual PCs (1 activity unit each). Top panel, digest reactions were analyzed by SDS-PAGE. Bottom panel, the gels were scanned, the band density was digitized and the conversion of the precursor into the mature HA was expressed in percent. B, furin efficiently cleaves PA83. PA83 and the HA precursor were each incubated for 1 h at 37 °C with the indicated amounts of furin. The asterisks indicate the amounts of furin that were required for accomplishing a 50% conversion of the precursor into the mature species. DEC, dec-RVKR-cmk.
To generate additional in-depth data, we then determined how much of an individual PC is required for a 50% conversion of the most efficient protein targets, PA83 and HA, into the respective processed mature proteins. Fig. 5 demonstrates the proteolysis of HA and PA83 using increasing concentrations of furin. These data show that in our experimental conditions ≈0.125 activity units of furin are sufficient for a 50% cleavage of HA while an ∼2-fold smaller amount is required for a 50% cleavage of PA83, thus confirming that PA83 is highly sensitive to furin proteolysis.
The analysis of the selected additional cleavage reactions of PA83 by the individual PCs is shown in supplemental Fig. S2. Our results are summarized in Fig. 6, and they clearly show that PA83 was efficiently cleaved by multiple PCs, distinct from PC2. As calculated on the basis of the specific activity against Pyr-RTKR-AMC, PC2 was 10–20-fold less efficient in PA83 processing when compared with the other individual PC types. According to our analysis, PC5/6 is the most efficient proteinase in the processing of PA83. Furin, PC1/3, PC4, and PACE4 exhibited a similar efficiency of PA83 processing, while PC7 was ∼2-fold less efficient.
FIGURE 6.
The amounts of the individual PCs required for accomplishing a 50% conversion of PA83 into PA63. PA83 was incubated for 1 h at 37 °C with increasing amounts of the individual PCs. The digest reactions were analyzed by SDS-PAGE, the gels were scanned, and the band density was digitized to determine the amount of each PC that is required for accomplishing a 50% conversion of PA83 into PA63. Inset, PC2 was omitted to demonstrate the differences among furin, PC1/3, PC4, PC5/6, PC7, and PACE4.
To compare directly the efficiency of the cleavage of the peptides derived from HA, PA83, and PEx with that of the corresponding proteins, we re-calculated the peptide cleavage efficiencies relative to that of the NSRKKR↓STSA PA83 peptide (100%) and the protein cleavage efficiencies relative to that of the PA83 protein (100%). The data are summarized in Fig. 7 and demonstrate that there is a good general correlation between the peptide and the protein cleavage data. The PEx peptide RHRQPR↓GWEQ and the PEx protein were the two most resistant to processing by the PCs. Among the proteins we have analyzed, PA83 was the most efficient target for all of the PCs.
FIGURE 7.
Direct comparison of the peptide and protein cleavage efficiency by the individual PCs. A, efficiency of the cleavage of the peptides (sequences of the peptides are shown on the right) derived from PA83, HA, and PEx. The cleavage efficiency of the NSRKKR↓STSA PA83 peptide by each PC was taken as 100%. B, efficiency of the cleavage of the PA83, HA, and PEx proteins. The cleavage efficiency of PA83 by each PC was taken as 100%.
DISCUSSION
Human PCs are multidomain proteinases, the catalytic domains of which are similar in structure to bacterial subtilisin. PCs function in the Golgi apparatus, in the secretory vesicles and also on cell surfaces. These unique specificity proteinases cleave the multibasic R-X-(R/K/X)-R↓ motif in many functionally important cellular proteins, including soluble and membrane-tethered metalloproteinases, integrins, signaling receptors, growth factors, hormones, and neuropeptides, into their respective mature forms (3, 38–43). In addition to processing cellular precursor proteins, PCs are also exploited by dozens of pathogens. Pathogenic viruses and bacterial toxins usurp host PCs to become fully functional and to allow entry into host cells and to cause disease onset.
Because the design of inhibitors, especially of proteinase antagonists, frequently starts with a known substrate, the precise knowledge of the cleavage preferences of human PCs is mandatory. The cleavage preferences of PCs, however, have not been known at a level of detail sufficient for designing specific inhibitors of the individual enzymes. This lack of knowledge made designing specific inhibitors exceedingly difficult. The relative contribution of the individual PCs in the cleavage of the individual cleavage proteins was also not precisely known. Therefore, it has been enormously difficult to predict if a specific drug that targets the individual PC or a wide range inhibitor that blocks the activity of multiple PC types was required for protecting the host cell from the specific pathogens.
To shed more light on the cleavage preferences and the substrate specificity of the PC family members, we synthesized over 100 decapeptide sequences, which represented the multibasic cleavage motifs and which were derived from the known cleavage target of furin. These peptides were cleaved by the individual purified PCs, and the cleavage data were analyzed and presented using a specialized computer program we had developed. We confirmed our peptide analysis data by using the cleavage data of the proteins from which the peptides were derived. There was a good correlation between the results of our assays and the predictions we had made.
We find that multiple pathogens evolved to develop a high level of sensitivity to PC hydrolysis. This level is comparable with that of the most sensitive normal PC targets of human origin. We believe that our suggestion explains why multiple pathogens, including anthrax, diphtheria, and influenza A successfully compete for processing by the host cell PCs with normal human proteins.
Multiple preliminary conclusions concerning the relative significance of the individual PCs in the processing of the individual proteins of human and pathogen origin can be drawn from the supplemental Table S1 data. For example, it is highly likely that PC5/6, PC7, and PACE4 significantly contribute to the processing of BACE1. It is highly unlikely that PC2 plays any significant role in the processing and activation of Notch, and the α3 and α7 integrin subunits. It appears entirely possible that PC2 does not play a major role in the processing of Ebola virus glycoprotein but it does appear that among the PC family members PC2 is the specific proteinase that most efficiently cleaves the fusion (F) protein precursor of parainfluenza and, therefore, PC2 is a principal drug target in a parainfluenza infection. Because PC2 is a dense core secretory granule enzyme (44), we believe that the cell permeable-specific PC2 inhibitor is required to inhibit parainfluenza while the PCs distinct from PC2 may be spared, and potential side effects of the drug may be reduced. It becomes also clear that a wide-range cell-impermeable inhibitor of PCs is required for treating anthrax because PA83 is cleaved equally efficiently by all of the PCs we tested, and because the processing of PA83 involves the cell surface-expressed PCs but not the PCs of the intracellular milieu (8). It is highly likely that PC2 plays an unexpectedly significant role in the processing of HIV-1 gp160. Because both HIV-1 gp160 and Pseudomonas PEx are predominantly processed within the endosomal compartment (12, 18), cell-permeant specific inhibitors are required to block the infectivity of these pathogens.
Overall, our data correlate well with the results of several laboratories, which extensively studied the individual PCs and their role in the processing of PA83, HA, HIV-1 gp160, MMPs and many other functionally important proteins (9, 10, 13, 15, 18, 19, 24, 45–60).
Regardless of the limitations of the peptide cleavage assays, which employ the unfolded short peptides, the data and the experimental and software tools we have generated can provide the foundation for developing a precise understanding of the specificity and the biological functions of cellular PCs. Knowledge of the mode of action of the individual PCs and their relation to the proteolytic processing of normal proteins and viral and bacterial pathogens will reveal potential novel applications in medicine, including unconventional approaches to treating multiple pathogens.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI061139 (to A. Y. S.), RR020843 (to J. W. S. and A. Y. S.), and AI055789 (to R. C. L. and A. Y. S.). This work was also supported by CIHR Grants (to R. Day). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
Footnotes
The abbreviations used are: PC, proprotein convertase; dec-RVKR-cmk, decanoyl-Arg-Val-Lys-Arg-chloromethylketone; HA, hemagglutinin; MALDI-TOF MS, matrix-assisted laser-desorption ionization-time-of-flight mass spectrometry; PA83, anthrax protective antigen precursor with the 83 kDa molecular mass; PACE4, paired basic amino acid cleaving enzyme 4; PEx, Pseudomonas exotoxin A; Pyr-RTKR-AMC; pyroglutamic acid-Arg-Thr-Lys-Arg-methyl-coumaryl-7-amide; SAM, Sentrix array matrix.
References
- 1.Seidah, N. G., Day, R., and Chretien, M. (1993) Biochem. Soc. Trans. 21685 -691 [DOI] [PubMed] [Google Scholar]
- 2.Steiner, D. F. (1998) Curr. Opin. Chem. Biol. 231 -39 [DOI] [PubMed] [Google Scholar]
- 3.Thomas, G. (2002) Nat. Rev. Mol. Cell. Biol. 3753 -766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fugere, M., Limperis, P. C., Beaulieu-Audy, V., Gagnon, F., Lavigne, P., Klarskov, K., Leduc, R., and Day, R. (2002) J. Biol. Chem. 2777648 -7656 [DOI] [PubMed] [Google Scholar]
- 5.Taylor, N. A., Van De Ven, W. J., and Creemers, J. W. (2003) FASEB J. 171215 -1227 [DOI] [PubMed] [Google Scholar]
- 6.Nakayama, K. (1997) Biochem. J. 327625 -635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scamuffa, N., Calvo, F., Chretien, M., Seidah, N. G., and Khatib, A. M. (2006) FASEB J. 201954 -1963 [DOI] [PubMed] [Google Scholar]
- 8.Collier, R. J., and Young, J. A. (2003) Annu. Rev. Cell Dev. Biol. 19 45-70 [DOI] [PubMed] [Google Scholar]
- 9.Feldmann, H., Volchkov, V. E., Volchkova, V. A., and Klenk, H. D. (1999) Arch. Virol. 15 159-169 [DOI] [PubMed] [Google Scholar]
- 10.Garten, W., Hallenberger, S., Ortmann, D., Schafer, W., Vey, M., Angliker, H., Shaw, E., and Klenk, H. D. (1994) Biochimie (Paris) 76217 -225 [DOI] [PubMed] [Google Scholar]
- 11.Gordon, V. M., and Leppla, S. H. (1994) Infect. Immun. 62333 -340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moulard, M., and Decroly, E. (2000) Biochim. Biophys. Acta 1469121 -132 [DOI] [PubMed] [Google Scholar]
- 13.Rockwell, N. C., Krysan, D. J., Komiyama, T., and Fuller, R. S. (2002) Chem. Rev. 1024525 -4548 [DOI] [PubMed] [Google Scholar]
- 14.Rott, R., Klenk, H. D., Nagai, Y., and Tashiro, M. (1995) Am. J. Respir. Crit. Care Med. 152 16-19 [DOI] [PubMed] [Google Scholar]
- 15.Stadler, K., Allison, S. L., Schalich, J., and Heinz, F. X. (1997) J. Virol. 718475 -8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decha, P., Rungrotmongkol, T., Intharathep, P., Malaisree, M., Aruksakunwong, O., Laohpongspaisan, C., Parasuk, V., Sompornpisut, P., Pianwanit, S., Kokpol, S., and Hannongbua, S. (2008) Biophys. J. 95128 -134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zambon, M. C. (2001) Rev. Med. Virol. 11227 -241 [DOI] [PubMed] [Google Scholar]
- 18.Chiron, M. F., Fryling, C. M., and FitzGerald, D. (1997) J. Biol. Chem. 27231707 -31711 [DOI] [PubMed] [Google Scholar]
- 19.Basak, A., Zhong, M., Munzer, J. S., Chretien, M., and Seidah, N. G. (2001) Biochem. J. 353537 -545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, J., Lee, K. H., Steinhauer, D. A., Stevens, D. J., Skehel, J. J., and Wiley, D. C. (1998) Cell 95 409-417 [DOI] [PubMed] [Google Scholar]
- 21.Sarac, M. S., Cameron, A., and Lindberg, I. (2002) Infect. Immun. 707136 -7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiryaev, S. A., Remacle, A. G., Ratnikov, B. I., Nelson, N. A., Savinov, A. Y., Wei, G., Bottini, M., Rega, M. F., Parent, A., Desjardins, R., Fugere, M., Day, R., Sabet, M., Pellecchia, M., Liddington, R. C., Smith, J. W., Mustelin, T., Guiney, D. G., Lebl, M., and Strongin, A. Y. (2007) J. Biol. Chem. 28220847 -20853 [DOI] [PubMed] [Google Scholar]
- 23.Jiao, G. S., Cregar, L., Wang, J., Millis, S. Z., Tang, C., O'Malley, S., Johnson, A. T., Sareth, S., Larson, J., and Thomas, G. (2006) Proc. Natl. Acad. Sci. U. S. A. 10319707 -19712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denault, J. B., Lazure, C., Day, R., and Leduc, R. (2000) Protein Expr. Purif. 19 113-124 [DOI] [PubMed] [Google Scholar]
- 25.Kozlov, I. A., Melnyk, P. C., Hachmann, J. P., Srinivasan, A., Shults, M., Zhao, C., Musmacker, J., Nelson, N., Barker, D. L., and Lebl, M. (2008) Comb. Chem. High Throughput. Screen. 1124 -35 [DOI] [PubMed] [Google Scholar]
- 26.Lebl, M. (1999) Bioorg. Med. Chem. Lett. 91305 -1310 [DOI] [PubMed] [Google Scholar]
- 27.Lebl, M. (2003) J. Assoc. Lab. Autom. 830 -35 [Google Scholar]
- 28.Hachmann, J., and Lebl, M. (2006) J. Comb. Chem. 8149. [DOI] [PubMed] [Google Scholar]
- 29.Hachmann, J., and Lebl, M. (2006) Biopolymers 84340 -347 [DOI] [PubMed] [Google Scholar]
- 30.King, D. S., Fields, C. G., and Fields, G. B. (1990) Int. J. Pept. Protein Res. 36 255-266 [DOI] [PubMed] [Google Scholar]
- 31.Lebl, M., Burger, C., Ellman, B., Heiner, D., Ibrahim, G., Jones, A., Nibbe, M., Thompson, J., Mudra, P., Pokorný, V., Poncar, P., and Ženíšek, K. (2001) Collect. Czech. Chem. Commun. 661299 -1314 [Google Scholar]
- 32.Gunderson, K. L., Kruglyak, S., Graige, M. S., Garcia, F., Kermani, B. G., Zhao, C., Che, D., Dickinson, T., Wickham, E., Bierle, J., Doucet, D., Milewski, M., Yang, R., Siegmund, C., Haas, J., Zhou, L., Oliphant, A., Fan, J. B., Barnard, S., and Chee, M. S. (2004) Genome Res. 14870 -877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004) Genome Res. 141188 -1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider, T. D., and Stephens, R. M. (1990) Nucleic Acids Res. 186097 -6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlov, I. A., Melnyk, P. C., Zhao, C., Hachmann, J. P., Shevchenko, V., Srinivasan, A., Barker, D. L., and Lebl, M. (2006) Comb. Chem. High Throughput Screen. 9481 -487 [DOI] [PubMed] [Google Scholar]
- 36.Shiryaev, S. A., Kozlov, I. A., Ratnikov, B. I., Smith, J. W., Lebl, M., and Strongin, A. Y. (2007) Biochem. J. 401743 -752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiryaev, S. A., Ratnikov, B. I., Aleshin, A. E., Kozlov, I. A., Nelson, N. A., Lebl, M., Smith, J. W., Liddington, R. C., and Strongin, A. Y. (2007) J. Virol. 814501 -4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mains, R. E. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D. & Woessner, J. F., eds), 2nd Ed., pp. 1871-1874, Elsevier, London
- 39.Seidah, N. G., and Chretien, M. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D. & Woessner, J. F., eds) 2nd Ed., pp.1861 -1864, Elsevier, London
- 40.Seidah, N. G., and Chretien, M. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D. & Woessner, J. F., eds), 2nd Ed., pp.1865 -1868, Elsevier, London
- 41.Seidah, N. G., and Chretien, M. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D. & Woessner, J. F., eds), 2nd Ed., pp.1868 -1870, Elsevier, London
- 42.Seidah, N. G., and Chretien, M. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D. & Woessner, J. F., eds), 2nd Ed., pp.1874 -1877, Elsevier, London
- 43.Seidah, N. G., and Chretien, M. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D. & Woessner, J. F., eds), 2nd Ed., pp.1877 -1880, Elsevier, London
- 44.Seidah, N. G., Mayer, G., Zaid, A., Rousselet, E., Nassoury, N., Poirier, S., Essalmani, R., and Prat, A. (2008) Int. J. Biochem. Cell Biol. 401111 -1125 [DOI] [PubMed] [Google Scholar]
- 45.Basak, A., Toure, B. B., Lazure, C., Mbikay, M., Chretien, M., and Seidah, N. G. (1999) Biochem. J. 343 29-37 [PMC free article] [PubMed] [Google Scholar]
- 46.Bassi, D. E., Fu, J., Lopez de Cicco, R., and Klein-Szanto, A. J. (2005) Mol. Carcinog. 44 151-161 [DOI] [PubMed] [Google Scholar]
- 47.Beinfeld, M. C. (1998) Endocrine 81 -5 [DOI] [PubMed] [Google Scholar]
- 48.Creemers, J. W., Jackson, R. S., and Hutton, J. C. (1998) Semin. Cell Dev. Biol. 9 3-10 [DOI] [PubMed] [Google Scholar]
- 49.Decroly, E., Benjannet, S., Savaria, D., and Seidah, N. G. (1997) FEBS Lett. 405 68-72 [DOI] [PubMed] [Google Scholar]
- 50.Dey, A., Norrbom, C., Zhu, X., Stein, J., Zhang, C., Ueda, K., and Steiner, D. F. (2004) Endocrinology 1451961 -1971 [DOI] [PubMed] [Google Scholar]
- 51.Gensberg, K., Jan, S., and Matthews, G. M. (1998) Semin. Cell Dev. Biol. 9 11-17 [DOI] [PubMed] [Google Scholar]
- 52.Golubkov, V. S., Chekanov, A. V., Shiryaev, S. A., Aleshin, A. E., Ratnikov, B. I., Gawlik, K., Radichev, I., Motamedchaboki, K., Smith, J. W., and Strongin, A. Y. (2007) J. Biol. Chem. 28236283 -36291 [DOI] [PubMed] [Google Scholar]
- 53.Lissitzky, J. C., Luis, J., Munzer, J. S., Benjannet, S., Parat, F., Chretien, M., Marvaldi, J., and Seidah, N. G. (2000) Biochem. J. 346133 -138 [PMC free article] [PubMed] [Google Scholar]
- 54.Munzer, J. S., Basak, A., Zhong, M., Mamarbachi, A., Hamelin, J., Savaria, D., Lazure, C., Hendy, G. N., Benjannet, S., Chretien, M., and Seidah, N. G. (1997) J. Biol. Chem. 27219672 -19681 [DOI] [PubMed] [Google Scholar]
- 55.Paquet, L., Zhou, A., Chang, E. Y., and Mains, R. E. (1996) Mol. Cell. Endocrinol. 120161 -168 [DOI] [PubMed] [Google Scholar]
- 56.Patel, Y. C., and Galanopoulou, A. (1995) Ciba Found. Symp. 19026 -40 [DOI] [PubMed] [Google Scholar]
- 57.Pei, D., and Weiss, S. J. (1995) Nature 375244 -247 [DOI] [PubMed] [Google Scholar]
- 58.Peinado, J. R., Li, H., Johanning, K., and Lindberg, I. (2003) J. Neurochem. 87 868-878 [DOI] [PubMed] [Google Scholar]
- 59.Stawowy, P., and Fleck, E. (2005) J. Mol. Med. 83865 -875 [DOI] [PubMed] [Google Scholar]
- 60.Wang, P., Tortorella, M., England, K., Malfait, A. M., Thomas, G., Arner, E. C., and Pei, D. (2004) J. Biol. Chem. 27915434 -15440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.