Abstract
In regulated exocytosis, the core membrane fusion machinery proteins, the SNARE proteins, are assisted by a group of regulatory factors in order to couple membrane fusion to an increase of intracellular calcium ion (Ca2+) concentration. Complexin-I and synaptotagmin-I have been shown to be key elements for this tightly regulated process. Many studies suggest that complexin-I can arrest the fusion reaction and that synaptotagmin-I can release the complexin-I blockage in a calcium-dependent manner. Although the actual molecular mechanism by which they exert their function is still unknown, recent in vivo experiments postulate that domains of complexin-I produce different effects on neurotransmitter release. Herein, by using an in vitro flipped SNARE cell fusion assay, we have identified and characterized the minimal functional domains of complexin-I necessary to couple calcium and synaptotagmin-I to membrane fusion. Moreover, we provide evidence that other isoforms of complexin, complexin-II, -III, and -IV, can also be functionally coupled to synaptotagmin-I and calcium. These correspond closely to results from in vivo experiments, providing further validation of the physiological relevance of the flipped SNARE system.
Vesicle trafficking comprises a highly orchestrated series of protein-protein interactions that culminate in the membrane fusion event (1). During the late steps of membrane fusion reaction, the vesicle docks closely and firmly, within molecular contact distance of the target bilayer, as the cognate SNARE3 proteins zip up to form a four-helix bundle between the two membranes (termed a trans-SNARE complex or SNAREpin). When the SNARE complex fully zips up, the bilayers are merged, and the SNARE complex now emanates from the single, combined membrane.
In regulated exocytosis, there are additional proteins added to this “core” fusion machinery that appear to freeze the process at intermediate stages and allow rapid mobilization from these intermediates (e.g. upon calcium entry) to allow a prompt bolus of secretion (2, 3). Three proteins in particular are known to directly affect the vesicle release probabilities and kinetics at various synapses: Munc18-1 (4-9), synaptotagmin (SYT), and complexin (CPX). SYT is now well described as the primary calcium sensor for SNARE-dependent fusion (reviewed in Refs. 10 and 11). SYTs are vesicle-associated integral membrane proteins with two cytosol-facing calcium-binding C2 domains (12). Knock-out of mouse SYT1 abolishes the fast synchronous component of calcium-mediated release (13). Mutations of SYT reduce or eliminate the apparent cooperativity of calcium in exocytosis, alter the vesicle release probability, and shift the calcium sensitivity (14, 15). A series of genetic and electrophysiological experiments place the small soluble protein CPX at the same step in synaptic release as SYT (reviewed in Ref. 16). As with SYT, changing the expression level of CPX impairs evoked exocytosis (17, 18), whereas the knock-out of CPX abolishes synchronous release (19).
A sudden influx of calcium into the presynaptic terminal of a neuron leads to a synchronized, rapid release of neurotransmitter via the fusion of docked and primed vesicles (10, 11, 20). We had suggested, with the discovery of the SNARE proteins, that such a rapid synchronized release might result from a calciumsensitive clamp preventing the complete assembly of SNARE proteins (21). In this way, the vesicles can be brought to the brink of fusion and a relatively mild energetic or kinetic switch could drive full neurotransmitter secretion. We have identified CPX as a protein that can clamp the fusion apparatus and SYT as the calcium sensor that releases this clamp (22). Studies from three other groups working in liposomes, cortical neurons (by overexpression), and knock-out cells (from Drosophila) are consistent with this clamp hypothesis (23-25). A further prediction of the clamp hypothesis would be that removal of one or the other clamp components (SYT or CPX) should result in an increase in unfettered or spontaneous exocytosis. In fact, spontaneous release is increased in Drosophila knockouts of SYT and CPX (14, 25, 26) as well as mammalian knockouts of SYT-I and -II (where interneuronal synapses are considered) (27, 28).
How CPX clamps the SNARE complex and at what stage of assembly are open questions. CPX binds the cis-SNARE complex with a short helical sequence in the middle of the protein (29). This sequence is sufficient to block exocytosis when appended directly to a SNARE (24), and mutations in this sequence reduce or eliminate the clamping capacity of CPX (22, 24). Calcium-mediated exocytosis also relies on sequences in other parts of the protein (notably the membrane-proximal N terminus) (30), although how these sequences might act functionally is unknown. In this paper, we used an in vitro flipped SNARE cell fusion assay in order to identify and characterize functional domains on CPX-I required either to clamp the fusion reaction or to couple the fusion reaction to calcium and SYT-I.
Although the expression of SNAREs and other normally cytoplasmic proteins on the outside of the cell is a decidedly nonphysiologic maneuver, it has the advantage of providing a highly simplified environment to which potential regulatory proteins can be added in a controlled manner. The artificial nature of the system raises potential questions about the physiological relevance of results from this assay. Comparison of results from this system with those obtained from more reductionist or even physiological systems have to date been favorable, and here we allow this comparison to be extended to include functional analysis of domains of CPX. Here we present direct evidence that CPX-I-(26-83) represents the minimal functional domain of CPX-I that preserves both the clamping activity and the calcium/SYT-I sensitivity in vitro. In addition, we show that other isoforms of CPX-glycosylphosphatidylinositol (GPI) (CPX-II-, CPX-III-, and CPX-IV-GPI) can be functionally coupled to SYT-I. We compare these results with those obtained with in vivo systems.
EXPERIMENTAL PROCEDURES
Constructs and Stable Cell Lines—To generate GPI-anchored complexin-I deletion constructs, we performed PCR using as a template plasmid CPX-I-GPI (22) and the appropriate combination of the set of sense primers, which contains an XbaI site and encodes for an AU1 epitope (DTYRYI) (CPXI26AU1, CACCTCTAGAGACACATACCGATACATAAAGGACCCAGACGCCGCCAAGAAGGAGGAGGAG; CPXI34AU1, CACCTCTAGAGACACATACCGATACATAGAGGAGGAGCGGCAGGAGGCGCTGCGCCAG; CPXI48AU1, CACCTCTAGAGACACATACCGATACATACGCAAGGCCAAGTACGCCAAGATGGAGGCG) and the set of antisense primers, which contains an XhoI site (CPXI48-3′, TTTGGCTCGAGGCGCTCCTCCTCCGCCTGGCG; CPXI70-3′, TTTGGCTCGAGGTACTTGTCTCGGATGCCCTGGCG; CPXI86-3′, TTTGGCTCGAGCTGGGCCTCGGCCTCGCGCTC). The PCR product and the plasmid pCDNA3.1(+) CPX-I-GPI were digested with XbaI and XhoI. Both digestion products were ligated, yielding the indicated CPX-I-GPI deletion constructs. GPI-anchored constructs of human CPX-II and CPX-III or mouse CPX-IV fused to an AU1 epitope were generated using the same strategy as for CPX-I-GPI deletion constructs described above.
Site-specific mutations were introduced on CPX-I-GPI using the QuikChange site-directed mutagenesis kit-II (Stratagene). HeLa stable cell lines co-expressing flipped SNAREs with the fluorescent protein markers were generated using the Tet-Off gene expression system (31) (Clontech) as described (22). Two different types of SNARE-expressing HeLa cells were mainly used in this paper, cells lines stably expressing flipped VAMP2 and DsRed2-nuclear export signals (v-cells) or cell lines stably expressing flipped syntaxin 1, flipped SNAP-25, and cyan fluorescent protein-nls (t-cells).
Cell-Cell Fusion Assay—5.5 × 105 HeLa v-cells grown for at least 3 days in complete medium in the absence of doxycycline (to initiate flipped VAMP2 and DsRed expression) were seeded in a 6-well plate. The next day, the cells were used for transient transfection to express complexin, synaptotagmin, and YFP-nls marker (4 μg of CPX-I-GPI plasmid, 4 μg of flipped Syt-I plasmid, 4 μg of YFP-nls plasmid, and 15 μl of Lipofectamine 2000 (Invitrogen)), complexin and the YFP-nls marker (CPX-I-GPI plasmid, 4 μg of YFP-nls plasmid, and 10 μl of Lipofectamine 2000 (Invitrogen)), or YFP-nls marker alone for control experiments (4 μg of YFP-nls plasmid and 8 μl of Lipofectamine 2000). The same day as the transfection, 4.5 × 104 HeLa t-cells previously grown for at least 5 days in complete medium in the absence of doxycycline (to initiate expression of the flipped t-SNARE subunits and cyan fluorescent protein-nls) were seeded on sterile 12-mm glass coverslips contained in 24-well plates. The following day, transfected HeLa-v cells were detached from the dishes with sodium citrate cell-detaching solution (11 g/liter KCl, 4.4 g/liter sodium citrate). The detached cells were centrifuged at 200 × g and resuspended in HEPES-buffered Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and counted in a hemacytometer.
The assay was initiated by adding 5 × 104 v-cells to each coverslip already containing t-cells. After overnight incubation at 37 °C in 5% CO2, the coverslips were gently washed once with HEPES-buffered Dulbecco's modified Eagle's medium supplemented with 1% bovine serum albumin and 1 mm dithiothreitol (recovery solution).
The recovery from the CPX-I-GPI block was performed by adding prewarmed recovery solution containing 1 unit/ml phosphatidylinositol-specific phospholipase C (PI-PLC; Molecular Probes, Inc., Eugene, OR) and 20 μg/ml laminin, in the presence or absence of 1.8 mm EGTA (as indicated). The addition of PI-PLC is defined as t = 0. Cells were incubated for the indicated period of time at 37 °C in the water bath, and then the medium was aspirated, and the cells were fixed with 4% paraformaldehyde for 10 min. Cells were washed three times with phosphate-buffered saline supplemented with 0.1 g/liter CaCl2 and 0.1 g/liter MgCl2 (PBS++) and mounted with Prolong Antifade Gold mounting medium (Molecular Probes). Confocal images were collected as indicated under “Image Acquisition.”
Protein Expression and Purification—Recombinant soluble CPX-I, CPX-I-(26-83), and CPX-III were purified following the protocol described in Ref. 22. The flow-through after benzamidine treatment was incubated with 50 μl of (100%) benzamidine-Sepharose for 20 min at room temperature to bind thrombin. Benzamidine-Sepharose beads were collected in a spin column, and the flow-through was saved. The flow-through was dialyzed at 4 °C against 1 liter of HEPES-buffered Dulbecco's modified Eagle's medium with two changes of media. The dialyzed pool was then concentrated using a Centricon 20 concentrator (Amicon, Millipore) at 4,000 r.p.m. for 10 min at 4 °C. The concentration of the soluble complexin-I was then determined using the protein determination assay (Bio-Rad), and the purity of the protein was analyzed by SDS-PAGE.
Image Acquisition—Images were acquired on a Leica TCS SP2 AOBS confocal microscope, equipped with Leica LCS software, and usually using an HCX PL APO ×40, 1.25 numerical aperture oil immersion objective. For higher magnification images, a HCX PL APO ×63, 1.4 numerical aperture oil immersion objective was used. The images were processed with Adobe Photoshop software.
Data Analysis—At each time point, the percentage of transfected cells (red cytoplasm expressing the YFP-nls construct in the nucleus) containing blue nuclei (appearing either as a separate blue nucleus, or as “white” nuclei expressing both YFP and cyan fluorescent protein) was determined. Each data point in the paper represents two independent coverslips from which 40-60 separate fields were randomly chosen and imaged. The total number of cells counted per coverslip was 2000-3000. Each experiment is representative of at least two independent experiments.
In previous studies using this assay, we recorded fusion as “contacting v- and t-cells that fused” (32, 33). This metric limits the data to cells that have the opportunity to fuse with a cognate SNARE-expressing cell. Unfortunately, this requires an additional color marker to define the plasma membrane of the t-cells (or to fill the cytosol of the t-SNARE such that the shape of the cell is obvious). In the experiments presented in this paper, no additional color was available in the t-cell, and as such the total efficiency should be considered an underestimate of the actual cell-cell fusion efficiency. In experiments following just the stable cells and utilizing a content marker to discern t-cell shape, the actual efficiency of contacting cells fusing was comparable with previous measures.
Binding Experiments—t-cells were transiently co-transfected with flipped Vamp2 and the indicated CPX-I-GPI mutant construct or mock-transfected, and they were incubated with or without 500 milliunits/ml PI-PLC for 30 min, washed, fixed, and stained with anti-AU1 antibody. Total fluorescent intensity from individual cells in images was determined.
RESULTS
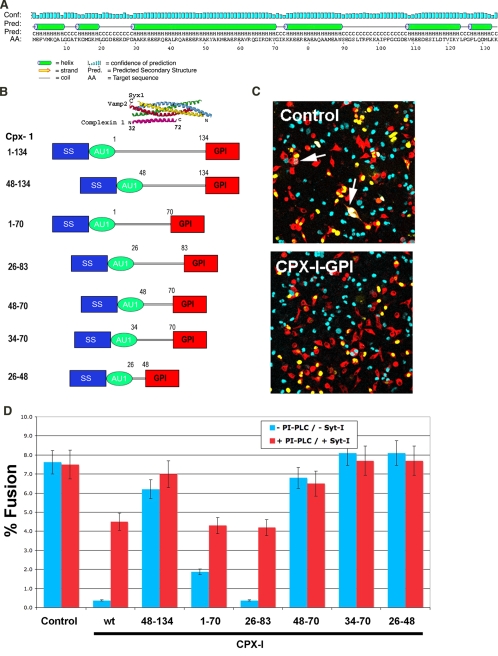
Functional Domains in Complexin-I That Control SNARE-mediated Fusion—CPX-I protein is a small (15-kDa) soluble protein composed of 134 residues with an overall acidic pH. Secondary structure predictions showed a high α-helix content, the central α-helix (aa 29-71) being the most prominent (Fig. 1A). The crystal structure data of CPX-I bound to the cis-SNARE complex (29, 34) reveals that within this central α-helix reside the amino acids that physically interact with the SNARE bundle (residues 48-70). To delineate functional requirements for domains of CPX-I and SYT-I in calcium control of SNARE-dependent fusion, we have designed a series of deletion constructs of CPX-I-GPI. The first sets of serial deletions span both the N terminus and C terminus of CPX-I, including the accessory α-helix (aa 29-48), which was partially solved in the crystal structure CPX-I·SNARE complex, and the central α-helix (aa 48-70) (Fig. 1B).
FIGURE 1.
Functional domains on CPX-I. A, protein secondary structure prediction of CPX-I sequence (G. P. S. Raghava, personal communication). B, domain structure of CPX-I-GPI deletion constructs compared with the solved crystal structure of CPX-I bound to SNARE pin (29) that comprises aa 32-72 of CPX1.C, representative image of a cell fusion assay. Control, not transfected with CPX-I-GPI; CPX-I-GPI, transfected with CPX-I-GPI. D, effect of different CPX-I-GPI deletion constructs on cell fusion (blue bars) and efficiency of cell fusion recovery after the addition of PI-PLC in the presence of SYT-I and calcium (red bars). Experiments are the mean ± S.E. of three independent experiments.
Each shortened deletion variant was fused to a GPI anchor motif in order to concentrate CPX-I on the cell surface (22). Expression of these constructs on the cell surface was confirmed by immunofluorescence (data not shown), and their effect on cell fusion activity was analyzed using the flipped SNARE assay (22). In this assay, HeLa v-cells expressing flipped Vamp2 and DsRed protein fused to nuclear export signals were transfected with YFP-nls (co-transfection marker) and with or without a CPX-I-GPI wild type or deletion construct. After 24 h of expression, cells were seeded on top of HeLa t-cells attached to coverslips expressing flipped syntaxin-1, flipped SNAP-25, and the cyan fluorescent protein fused to nuclear localization signals. Cells were incubated overnight, and the fusion efficiency was quantified as a percentage of transfected v-cells that fused. Deletion of the first 47 residues of CPX-I (CPX-I-(48-134)-GPI) resulted in a nearly complete loss of the clamping effect of CPX-I (Fig. 1, C and D). In stark contrast, deletion of the C-terminal half of CPX-I (CPX-I-(1-70)-GPI) produced only a modest loss of function. Similar to previous observations with full-length CPX-I, the clamping effect of CPX-I-(1-70)-GPI could be fully reversed by cleaving CPX-I off of the GPI anchor, in the presence of calcium and SYT-I with PI-PLC. Likewise, the construct CPX-I-(26-83)-GPI (the domain previously used for the crystallization studies (29)) could both clamp cell fusion and be reversed with PI-PLC, Ca2+/SYT-I. Further deletion at the amino terminus (CPX-I-(48-70)-GPI and CPX-I-(34-70)-GPI) resulted in inactive CPX-I fragments (i.e. no clamping). Thus, the region 26-83 of CPX-I constitutes the minimal clamping domain for coupling calcium control with SNARE-mediated membrane fusion. This domain consists of a SNARE binding region or central α-helix (aa 48-70) and of an accessory α-helix (aa 26-48). Both regions were mutually necessary to impart calcium sensitivity to the SNARE-mediated cell fusion, since constructs of other helix alone (CPX-I-(48-70)-GPI and CPX-I-(26-83)-GPI) did not affect the extent of cell fusion.
Functional Domains in Complexins and Synaptotagmin-I That Interact in Calcium Control—In addition to CPX-I and CPX-II, which are mainly enriched in neurons, two other CPX isoforms were identified: CPX-III and CPX-IV, which are mainly expressed in retina, hippocampus, olfactory bulb, cortex, inferior colliculus, thalamus, and striatum (19, 35). The CPX-III and -IV subfamily share a CAAX C-terminal farnesylation motif and a 10-12-aa insertion at the N-terminal region. Despite these differences, CPX-III and CPX-IV can functionally rescue the calcium-evoked release in neurons derived from CPX-I/II double knock-out mice, thus emphasizing a high level of functional redundancy.
To better define functional domains of SYT-I and CPX-I that interact in calcium control, we studied the specificity of SYT-I for controlling calcium-stimulated fusion, using different combinations of the various CPXs and neuronal v- and t-SNAREs. First, we generated and characterized the expression of GPI-anchored versions of all known mammalian CPXs (CPX-I through -IV) (Fig. 2A). Then we compared the effect of CPX-I through -IV on cell-cell fusion using the flipped SNARE cell fusion assay. In agreement with previous rescue experiments (35), our results showed that all four CPXs inhibited the cell fusion reaction mediated by neuronal SNAREs to approximately the same extent. Moreover, the inhibition exerted by all CPX isoforms was partially reverted after cleavage of the GPI anchor. In all cases, SYT-I enhanced the cell fusion recovery in a Ca2+-dependent manner to levels comparable with CPX-I. These results not only provide direct evidence that the neuronal SNARE complexes can be the target for CPX-I through -IV but also provide evidence that SYT-I can confer calcium sensitivity to the fusion reaction halted by each isoform.
FIGURE 2.
GPI-anchored constructs of CPX-I, -II, -III, and -IV inhibit cell fusion. A, domain structure of GPI-anchored constructs of CPX-I, -II, -III, and -IV compared with the solved crystal structure of CPX-I bound to SNARE pin. B, effect of different CPX-GPI (CPX-I through -IV; Cpx1-Cpx4) constructs on cell fusion (blue bars) and efficiency of cell fusion recovery after the addition of PI-PLC in the absence of SYT-I (yellow bars) and in the presence of SYT-I and calcium (red bars). Experiments are the mean ± S.E. of three independent experiments. C, dose-dependent inhibition of the cell fusion reaction using different soluble CPXs, sCPX-I, sCPX-I-(26-83), and sCPX-III. Results are the mean ± S.E. of two independent experiments.
To further characterize the minimal-clamping domain of CPX-I and different isoforms of CPX, we compared the effect of soluble forms of CPX (i.e. without a GPI anchor; sCPX-I) on the cell fusion assay. Soluble CPX-I also inhibited cell-cell fusion when applied at sufficient concentrations and did so in a dose-dependent manner (22). Although CPX-III is naturally prenylated, it also inhibited as a soluble protein, as did the minimal clamping domain of CPX-I. Both sCPX-I-(26-83) and sCPX-III inhibited the cell fusion reaction in a dose-dependent manner (Fig. 2C). The efficiency of inhibition did not differ significantly from sCPX-I.
Site-directed Mutagenesis of Conserved Residues in CPX-I, -II, -III, and -IV—Previous biochemical and structural studies have identified the central α-helical region as responsible for the physical interaction with the SNARE complexes (29, 34, 36, 37). Crystal structure data show how a truncated form of CPX (comprising residues 32-72) intercalates in an antiparallel fashion in the groove between VAMP2 and syntaxin-1 (Fig. 3, A and B). The interaction of CPX-I with the SNARE complex involves hydrogen bonds (residues Tyr52 and Tyr70), salt bridges (Arg48, Arg59, Arg63, and Lys69), and hydrophobic interactions (Met62 and Ile66). It is yet unknown whether these residues are essential to the clamped state, where trans-SNARE complexes embedded in biological membranes are the target rather than truncated, isolated cis-SNARE bundles. Here, we tested the importance of conserved residues within the central helical region of CPX-I through CPX-IV that may play a role in either the clamping effect of CPX-I or in the SYT-I/Ca2+ release.
FIGURE 3.
Effect of site-specific CPX-I-GPI mutants on SNARE-mediated cell fusion. Schematic diagram of the three-dimensional structure of the CPX·SNARE complex (29). The color code used to label each protein is as follows: CPX (magenta), Vamp2 (red), syntaxin1 (green), and SNAP25 N-terminal helix and SNAP25 C-terminal helix (blue and yellow, respectively). A, the residues in CPX-I that face the SNAREpin have been labeled in orange, and those that are conserved among different CPXs were highlighted in white. B, the residues in CPX-I that face out from the SNAREpin have been labeled in light blue, and those that are conserved among different CPXs are highlighted in white. C, sequence alignment of different CPXs, showing the identical residues in yellow, the conserved residues in light blue, and the similar residues in green. The intended mutations that will be analyzed are also shown on the alignment. The SNAREpin binding region is displayed in a red box. D, effect of different CPX-I-GPI mutant constructs on cell fusion (blue bars) and efficiency of cell fusion recovery after the addition of PI-PLC in the presence of SYT-I and calcium (red bars). Experiments are the mean ± S.E. of three independent experiments.
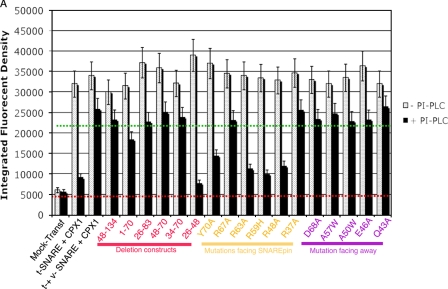
Residues conserved within the central helical region across the four CPXs were identified in an alignment plot of CPX sequences from different species (Fig. 3C). The conserved amino acids within the central region of CPX-I where three-dimensional structure data are available were classified either as residues facing the SNAREpin (and therefore potentially playing a role in stabilizing SNARE-complexin interactions) or residues facing away from the SNARE complex (and potentially involved in complexin-lipid or complexin-synaptotagmin interactions) (Fig. 3, A and B, respectively). First, we established whether the CPX-I mutants or deletion constructs preserved their ability to bind to cis-SNARE complexes present on the cell surface. For this purpose, HeLa t-cells were co-transfected with the desired CPX-I deletion/mutant construct and with a plasmid encoding for flipped VAMP2 in order to form cis-SNARE complexes on the cell surface. Afterward, CPX-I-GPI was cleaved off from its GPI anchor motif using PI-PLC, and the remaining CPX-I bound on the cell surface was determined by immunofluorescence using an anti-AU1 antibody (22).
As predicted by the structural data, the central α-helical region (aa 48-70) was indispensable for the interaction with SNARE complexes (30, 36, 37). Site-specific mutations of the amino acids on CPX-I that are thought to contact the SNARE complex (R59H, R48A, R63A, and Y70A) showed a strong reduction in the binding capacity (Fig. 4); however, mutation of other residues facing the SNAREpin but not making contact with (R37A or R67A) or residues facing away from the SNAREpin (E36A, Q43A, E46A, A50W, A57W, or D68A) did not alter the binding capacity to SNARE complexes (Fig. 4). Therefore, membrane-incorporated cis-SNAREs bind CPX in a similar manner as soluble cis-SNARE bundles.
FIGURE 4.
Binding of different CPXs deletion/mutant constructs to SNARE complexes. HeLa t-cells were co-transfected with the desired CPX-I deletion/mutant construct and with a plasmid encoding for flipped VAMP2 in order to form cis-SNARE complexes on the cell surface. Afterward, CPX-I-GPI was cleaved off from its GPI anchor motif using PI-PLC for 30 min at 37 °C, and the remaining CPX-I bound on the cell surface was determined by immunofluorescence using anti-AU1 antibody (22). The dashed red line corresponds to the basal fluorescent intensity background quantified when neither SNAREs nor CPXs are present (negative control). The dashed green line corresponds to the mean fluorescent intensity of constructs that bind SNARE complexes.
Next, we tested the capacity of each of these point mutants to clamp cell-cell fusion. Conserved residues facing the SNARE-pin were Arg37, Arg48, Arg59, Arg63, Arg67, and Tyr70 (Fig. 3A). Of the SNAREpin-facing mutants, four (Arg48, Arg59, Arg63, and Tyr70) are involved in the interaction with the cis-SNARE complex, whereas Arg67 and Arg37 are not. Data from our functional flipped SNARE fusion assay correlated well with the structural data and showed a profound decrease in the clamping effect of the SNARE interaction mutants R48A, R63A, and Y70A. On the other hand, clamping effects of CPX-I mutants R67A and R37A were functionally indistinguishable from that of the CPX-I wild type (Fig. 3D). Neither R67A nor R37A mutants showed an altered Ca2+/SYT-I fusion recovery after the addition of PI-PLC. These results support the fact that the functional approach is sensitive and specific enough to detect minor changes in the regulatory machinery.
When we introduced the outward facing mutations E36A, Q43A, E46A, A50W, A57W, or D68A (Fig. 3B), neither the clamping effect of CPX-I nor the calcium sensitivity conferred by SYT-I was significantly affected (Fig. 3C).
The Minimal Clamping Domain of CPX-I Preserves the SYT-I/Calcium Sensitivity—Recent rescue experiments using neurons derived from CPX-I/-II double knock-out mice have suggested that the first 26 residues of CPX-I have a crucial facilitating function in vesicle release that may be associated with a novel interaction (30). To further test in vitro the requirement of the N-terminal region of CPX-I for SYT-I/calcium sensitivity, we compared the cell fusion recovery of the minimum clamping domain of CPX-I (residues 26-83) with the wild type form of CPX-I-GPI. To this end, we performed a classical cell fusion experiment (22). After overnight incubation, we incubated the cells with EGTA for 5 min and then incubated for an additional 5 min with PI-PLC. Finally, cells were treated with or without calcium, and the cell fusion recovery was analyzed over time. Results showed that the calcium/SYT-I-dependent recovery of the SNARE-mediated fusion of CPX-I-(26-83)-GPI was almost indistinguishable from that of CPX-I (wild type)-GPI (Fig. 5). These results established that CPX-I-(26-83) can fully recapitulate the calcium-regulated SNARE-mediated membrane fusion.
FIGURE 5.
Calcium-dependent release of the complexin block. v- and t-cells were incubated overnight before the addition of 1 unit/ml PI-PLC and 1.8 mm EGTA (t = 0, where indicated) to release the complexin GPI anchor and reduce free calcium in the media to 10 μm. At t = 5 min, free calcium was raised to 200 μm (▪, CPX-I (wild type)-GPI; ○, CPX-I-(26-83)-GPI) or kept constant low at 10 μm (□, CPX-I-(26-83)-GPI). Cells were fixed, and data were quantified as in Fig. 1.
DISCUSSION
In regulated exocytosis, the core membrane fusion machinery, SNARE proteins, is assisted by a group of regulatory factors in order to couple membrane fusion to particular stimuli, such as an increase of intracellular calcium ion (Ca2+) concentration. The functions of these factors in regulated exocytosis are largely unknown due to a lack of mechanistic studies in minimal functional fusion systems. Here, by using the cell fusion system in which “flipped” v- and t-SNAREs needed in exocytosis are co-expressed on the cell surface with different regulatory factors, we have identified and characterized the minimal functional domain of CPX-I that confers calcium sensitivity to the fusion reaction. The cell fusion system provides several key advantages. First, the fusion involves a biological membrane but without the complexity of the cytosol, so the functions of individual regulatory proteins can be assessed in a biologically relevant environment. Second, the regulators can be co-expressed with SNAREs such that the topology is maintained. Third, individual fusion events can be imaged to detect subpopulations of distinct fusion outcomes, and fusion can be followed in real time to allow the observation of relatively stable transient fusion intermediates, such as hemifusion (33). Fourth, we have previously shown that the calcium-dependent membrane fusion reaction can be fully recapitulated using this in vitro fusion system (22).
Comparison of results from the flipped SNARE system with those obtained from physiological systems has to date been favorable, and here we allow this comparison to be extended to include functional analysis of domains of CPX with broad agreement with in vivo results (30). Our studies using CPX-I truncation constructs revealed that the N-terminal region of CPX-I is crucial for both the clamping effect on cell fusion and for the SYT-I/calcium-dependent fusion recovery. More precisely, the minimal functional domain of CPX-I we have identified that remains able to reconstitute the SYT-I/calcium-dependent fusion comprised residues 26-83 (Fig. 1D). Recent studies proposed that calcium-regulated exocytosis relies also on sequences in other parts of the protein (notably the membrane-proximal N terminus) (30). These studies, in agreement with our results, attributed an inhibitory effect to an accessory helix of CPX-I because of the failure of a CPX-I-(27-134) construct to rescue the CPX-I/-II double knock-out phenotype. However, the authors speculate that the first 26 residues of CPX-I have a positive role on the fast calcium-regulated exocytosis in the absence of SYT-I. They also hypothesize that the positive role of the N-terminal region of CPX-I is mediated by interaction with a missing factor. Nonetheless, the authors cannot rule out the compensation of other SYT and or CPX isoforms in vivo. In stark contrast with this hypothesis, our results provide direct evidences that the minimal functional domain of CPX-I (residues 26-83) can recapitulate the SNARE-mediated calcium/SYT-I-dependent fusion in vitro without the need of any other accessory proteins.
Combining our functional data from the cell fusion system together with the information from the crystal structure of the CPX-I·SNARE complex (29) strongly supports the model that binding of CPX-I to SNARE complex through residues 48-70 is a requisite for CPX-I to function but is not sufficient. The SNARE-facing residues Arg48, Arg59, Arg63, and Tyr70 are all conserved among different CPXs from different species and are essential to both binding and clamping, suggesting that the binding mechanism of other CPX isoforms to the SNAREpin may be conserved.
Rescue experiments in CPX-I/-II double knock-out neurons using CPX-III and -IV constructs suggest a functional redundancy of different CPX isoforms (35). In agreement with previous results, data presented here establish that CPX-I through -IV can each block a cell fusion reaction involving neuronal SNAREs with similar efficiencies. In addition, each CPX-I through -IV fusion blockage can be reverted by the same calcium sensor protein, SYT-I.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1-GM071458 (to J. E. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SNARE, soluble N-ethylmaleimide-sensitive attachment protein receptor; CPX, complexin; SYT, synaptotagmin; GPI, glycosylphosphatidylinositol; YFP, yellow fluorescent protein; PI-PLC, phosphatidylinositol-specific phospholipase C; aa, amino acids; nls, nuclear localization signal.
References
- 1.Mellman, I., and Warren, G. (2000) Cell 100 99-112 [DOI] [PubMed] [Google Scholar]
- 2.Brunger, A. T. (2005) Q. Rev. Biophys. 38 1-47 [DOI] [PubMed] [Google Scholar]
- 3.Jackson, M. B., and Chapman, E. R. (2006) Annu. Rev. Biophys. Biomol. Struct. 35 135-160 [DOI] [PubMed] [Google Scholar]
- 4.Rizo, J., and Sudhof, T. C. (2002) Nat. Rev. Neurosci. 3 641-653 [DOI] [PubMed] [Google Scholar]
- 5.Gallwitz, D., and Jahn, R. (2003) Trends Biochem. Sci. 28 113-116 [DOI] [PubMed] [Google Scholar]
- 6.Peng, R. W. (2005) Sci. World J. 5 471-477 [Google Scholar]
- 7.Toonen, R. F., and Verhage, M. (2003) Trends Cell Biol. 13 177-186 [DOI] [PubMed] [Google Scholar]
- 8.Dulubova, I., Sugita, S., Hill, S., Hosaka, M., Fernandez, I., Sudhof, T. C., and Rizo, J. (1999) EMBO J. 18 4372-4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhage, M., Maia, A. S., Plomp, J. J., Brussaard, A. B., Heeroma, J. H., Vermeer, H., Toonen, R. F., Hammer, R. E., van den Berg, T. K., Missler, M., Geuze, H. J., and Sudhof, T. C. (2000) Science 287 864-869 [DOI] [PubMed] [Google Scholar]
- 10.Sudhof, T. C. (2004) Annu. Rev. Neurosci. 27 509-547 [DOI] [PubMed] [Google Scholar]
- 11.Koh, T. W., and Bellen, H. J. (2003) Trends Neurosci. 26 413-422 [DOI] [PubMed] [Google Scholar]
- 12.Perin, M. S., Fried, V. A., Mignery, G. A., Jahn, R., and Sudhof, T. C. (1990) Nature 345 260-263 [DOI] [PubMed] [Google Scholar]
- 13.Geppert, M., Goda, Y., Hammer, R. E., Li, C., Rosahl, T. W., Stevens, C. F., and Sudhof, T. C. (1994) Cell 79 717-727 [DOI] [PubMed] [Google Scholar]
- 14.Littleton, J. T., Stern, M., Perin, M., and Bellen, H. J. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 10888-10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Chacon, R., Konigstorfer, A., Gerber, S. H., Garcia, J., Matos, M. F., Stevens, C. F., Brose, N., Rizo, J., Rosenmund, C., and Sudhof, T. C. (2001) Nature 410 41-49 [DOI] [PubMed] [Google Scholar]
- 16.Melia, T. J., Jr. (2007) FEBS Lett. 581 2131-2139 [DOI] [PubMed] [Google Scholar]
- 17.Tokumaru, H., Umayahara, K., Pellegrini, L. L., Ishizuka, T., Saisu, H., Betz, H., Augustine, G. J., and Abe, T. (2001) Cell 104 421-432 [DOI] [PubMed] [Google Scholar]
- 18.Ono, S., Baux, G., Sekiguchi, M., Fossier, P., Morel, N. F., Nihonmatsu, I., Hirata, K., Awaji, T., Takahashi, S., and Takahashi, M. (1998) Eur. J. Neurosci. 10 2143-2152 [DOI] [PubMed] [Google Scholar]
- 19.Reim, K., Mansour, M., Varogueaux, F., McMahon, H. T., Sudhof, T. C., Brose, N., and Rosenmund, C. (2001) Cell 104 71-81 [DOI] [PubMed] [Google Scholar]
- 20.Wojcik, S. M., and Brose, N. (2007) Neuron 55 11-24 [DOI] [PubMed] [Google Scholar]
- 21.Sollner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., and Rothman, J. E. (1993) Nature 362 318-324 [DOI] [PubMed] [Google Scholar]
- 22.Giraudo, C. G., Eng, W. S., Melia, T. J., and Rothman, J. E. (2006) Science 313 676-680 [DOI] [PubMed] [Google Scholar]
- 23.Schaub, J. R., Lu, X., Doneske, B., Shin, Y. K., and McNew, J. A. (2006) Nat. Struct. Mol. Biol. 13 748-750 [DOI] [PubMed] [Google Scholar]
- 24.Tang, J., Maximov, A., Shin, O. H., Dai, H., Rizo, J., and Sudhof, T. C. (2006) Cell 126 1175-1187 [DOI] [PubMed] [Google Scholar]
- 25.Huntwork, S., and Littleton, J. T. (2007) Nat. Neurosci. 10 1235-1237 [DOI] [PubMed] [Google Scholar]
- 26.Broadie, K., Bellen, H. J., DiAntonio, A., Littleton, J. T., and Schwarz, T. L. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 10727-10731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang, Z. P., Melicoff, E., Padgett, D., Liu, Y., Teich, A. F., Dickey, B. F., Lin, W., Adachi, R., and Sudhof, T. C. (2006) J. Neurosci. 26 13493-13504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maximov, A., and Sudhof, T. C. (2005) Neuron 48 547-554 [DOI] [PubMed] [Google Scholar]
- 29.Chen, X., Tomchick, D. R., Kovrigin, E., Arac, D., Machius, M., Sudhof, T. C., and Rizo, J. (2002) Neuron 33 397-409 [DOI] [PubMed] [Google Scholar]
- 30.Xue, M., Reim, K., Chen, X., Chao, H. T., Deng, H., Rizo, J., Brose, N., and Rosenmund, C. (2007) Nat. Struct. Mol. Biol. 14 949-958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron, U., Freundlieb, S., Gossen, M., and Bujard, H. (1995) Nucleic Acids Res. 23 3605-3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, C., Ahmed, M., Melia, T. J., Sollner, T. H., Mayer, T., and Rothman, J. E. (2003) Science 300 1745-1749 [DOI] [PubMed] [Google Scholar]
- 33.Giraudo, C. G., Hu, C., You, D., Slovic, A. M., Mosharov, E. V., Sulzer, D., Melia, T. J., and Rothman, J. E. (2005) J. Cell Biol. 170 249-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bracher, A., Kadlec, J., Betz, H., and Weissenhorn, W. (2002) J. Biol. Chem. 277 26517-26523 [DOI] [PubMed] [Google Scholar]
- 35.Reim, K., Wegmeyer, H., Brandstatter, J. H., Xue, M., Rosenmund, C., Dresbach, T., Hofmann, K., and Brose, N. (2005) J. Cell Biol. 169 669-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pabst, S., Margittai, M., Vainius, D., Langen, R., Jahn, R., and Fasshauer, D. (2002) J. Biol. Chem. 277 7838-7848 [DOI] [PubMed] [Google Scholar]
- 37.Pabst, S., Hazzard, J. W., Antonin, W., Sudhof, T. C., Jahn, R., Rizo, J., and Fasshauer, D. (2000) J. Biol. Chem. 275 19808-19818 [DOI] [PubMed] [Google Scholar]