Abstract
The Forkhead box M1 (FoxM1) protein is a proliferation-specific transcription factor that plays a key role in controlling both the G1/S and G2/M transitions through the cell cycle and is essential for the development of various cancers. We show here that FoxM1 directly activates the transcription of the c-Jun N-terminal kinase (JNK1) gene in U2OS osteosarcoma cells. Expression of JNK1, which regulates the expression of genes important for the G1/S transition, rescues the G1/S but not the G2/M cell cycle block in FoxM1-deficient cells. Knockdown of either FoxM1 or JNK1 inhibits tumor cell migration, invasion, and anchorage-independent growth. However, expression of JNK1 in FoxM1-depleted cells does not rescue these defects, indicating that JNK1 is a necessary but insufficient downstream mediator of FoxM1 in these processes. Consistent with this interpretation, FoxM1 regulates the expression of the matrix metalloproteinases MMP-2 and MMP-9, which play a role in tumor cell invasion, through JNK1-independent and -dependent mechanisms in U2OS cells, respectively. Taken together, these findings identify JNK1 as a critical transcriptional target of FoxM1 that contributes to FoxM1-regulated cell cycle progression, tumor cell migration, invasiveness, and anchorage-independent growth.
The Forkhead box (Fox)4 family of transcription factors includes more than 50 mammalian members that share sequence homology in the winged helix DNA binding domain (1). The expression of one member of this family, FoxM1, is tightly associated with proliferation and is extinguished in differentiated or resting cells that have exited the cell cycle, suggesting that FoxM1 plays a key role in cell proliferation (2). Indeed, the requirement of FoxM1 for cell proliferation has been demonstrated in hepatocytes in vivo, and in endothelial cells and various mesenchymal cell types in vitro (3–6). FoxM1 is highly expressed in many tumor-derived cell lines, and promotes the development of various types of cancers, including hepatocellular carcinoma, prostate cancer, lung cancer, and colorectal cancer in established mouse models (7–11). In recent studies, FoxM1 has emerged as a critical regulator of both the G1/S and G2/M transitions through the cell cycle (12, 13). FoxM1–/– MEFs and human U2OS osteosarcoma cells depleted of FoxM1 by siRNA failed to proliferate in culture because of reduced DNA replication and a block in mitotic progression (12). Using transcription and chromatin immunoprecipitation assays, we and others (12–14) have shown FoxM1 is directly involved in the transcriptional activation of specific genes that regulate the entry into and progression through mitosis, including Cdc25B, cyclin B, polo-like kinase, aurora B kinase, and centromere protein (CENP)-A, CENP-B, and CENP-F. In addition, FoxM1 is essential for the transcription of Skp2 and Cks1, which encode subunits of the Skp-Cullin 1-F-box (SCF) ubiquitin ligase complex that targets the CDK inhibitor proteins p21Cip1 and p27Kip1 for degradation during the G1/S transition (12, 15). Therefore, FoxM1-deficient cells accumulate nuclear levels of p21Cip1 and p27Kip1, contributing to decreased entry into the S phase (16).
The JNK (also known as stress-activated protein kinases, SAPK) gene family is induced by extracellular stimuli (e.g. cytokines, mitogens, morphogens) and stresses (e.g. DNA damage, osmotic stress, oxidative stress, UV irradiation, γ-radiation, heat shock)(17, 18). Of the three JNK isoforms, JNK1 and JNK2 are expressed in most cell types, whereas JNK3 is specific to nerve cells. JNK-mediated signaling pathways control multiple aspects of cellular function, including cell proliferation, survival, apoptosis, differentiation, and the immune response (19, 20). Upon activation via phosphorylation by MKK4 or MKK7, JNKs phosphorylate their downstream targets, including the transcription factors c-Jun, ATF-2, Elk-1, Smad4, nuclear factor of activated T cells 4 (NFAT4), NF-ATc1, and p53, thus triggering the activation of gene transcription (21, 22). JNKs bind to the c-Jun N-terminal transactivation domain and phosphorylate it on serine 63 and serine 73, leading to c-Jun activation and heterodimerization with members of the Fos (c-Fos, FosB, Fra1, and Fra2) or Jun (c-Jun, JunB, and JunD) families or to ATF-2 to form the AP-1 transcription factors (23). Activated AP-1 is known to directly transactivate genes encoding cyclin D1, cyclin E, and cyclin A, which activate their corresponding Cdks and stimulate cell cycle progression through the G1/S transition (24–27). Deficiency in JNK can result in p53 accumulation, leading to premature senescence (28).
In this study, we show that FoxM1 directly activates transcription of JNK1 in U2OS osteosarcoma cells, and both FoxM1 and JNK1 are necessary for cell cycle progression, tumor cell migration, invasion, and anchorage-independent growth. Expression of JNK1 efficiently rescues the G1/S cell cycle block in FoxM1-depleted cells, indicating that JNK1 functions as the pivotal mediator of FoxM1 functions in the G1/S transition. Furthermore, FoxM1 regulates the expression of MMP-2 and MMP-9, which is associated with tumor cell migration and invasion, through JNK1-independent and -dependent pathways, respectively. These findings identify JNK1 as a crucial transcriptional target of FoxM1 that contributes to FoxM1-regulated G1/S transition, tumor cell migration, invasion, and anchorage-independent growth.
MATERIALS AND METHODS
Cell Culture and siRNA Transfection—Human U2OS osteosarcoma cells and FoxM1–/– MEFs were cultured as described (12). FoxM1 siRNA (GGACCACUUUCCCUACUUU) and control siRNA (GGACCUGUAUGCGUACAUU) were synthesized by Dharmacon Research, and duplexes (100 nm) were transfected into U2OS cells using Lipofectamine™ 2000 (Invitrogen) as described (8, 12). U2OS cells were harvested at 48 or 72 h after transfection for flow cytometry and RNA and protein analyses.
PCR Amplification of the Human JNK1 Promoter—We amplified the –1309/+41 region of the human JNK1 promoter from U2OS genomic DNA by PCR using the primers 5′-GTACCCGGGTTGTCCTCGGTTTTACAGC-3′ and 5′-GGACTCGAGTTGTCACGCTTGCTTCTGCTC-3′. The PCR product was digested with SmaI, and blunt-end ligated into SmaI-digested pGL3-Basic firefly luciferase reporter plasmid (Promega). Correct orientation and sequences were verified by DNA sequencing.
Dual Luciferase Assays of JNK1 Promoter-Luciferase Constructs—U2OS cells were plated at 2 × 105 cells per well in a 6-well plate and transfected via Lipofectamine™ 2000 with 200 ng of either CMV-FoxM1B expression construct or CMV empty vector, 1.5 μg of –1308/+41 JNK1 promoter-firefly luciferase reporter, and 10 ng of CMV-Renilla luciferase as an internal control. Cells were harvested 24 h after transfection, and protein extracts were subjected to Dual luciferase assays (Promega), with firefly luciferase activity normalized to Renilla luciferase activity as described (29). Promoter activity was expressed as fold induction of transcription by the FoxM1b expression vector ± S.D., where the promoter activity resulting from transfection with CMV empty vector was set at one. Experiments were performed in triplicate, and statistical analysis was performed with Microsoft Excel tools.
Real-Time RT-PCR—Total RNA from early passage WT, FoxM1+/–, and FoxM1–/– MEFs, immortalized MEFs, or siRNA-transfected U2OS cells was prepared and subjected to RT-PCR reaction as described (12). The following sense (S) and antisense (AS) primer sequences and annealing temperature (Ta) were used to amplify and measure the amount of human mRNA by real-time RT-PCR: JNK1-S: 5′-TCT TCC CTG ATG TCC TTT TCC-3′ and JNK1-AS: 5′-TCT TCT ATT GTG TGT TCC CTT TC-3′ (Ta, 61.3 °C); JNK2-S: 5′-AAG CCC CAC CAC CTC AAA-3′ and JNK2-S: 5′-TTC CAT CAA CTC CCA AGC A-3′ (Ta, 58.7 °C); ATF2-S: 5′-TTT CCT CCA GGG GTG CTT TG-3′ and ATF2-AS: 5′-GCA GTC CTT TCT CAA GTT TCC ATC-3′ (Ta, 55.8 °C); MMP-2-S: 5′-CCA ACT ACA ACT TCT TCC CTC GC-3′ and MMP-2-AS: 5′-AGC AAA GGC ATC ATC CAC TGT C-3′ (Ta, 57 °C); MMP-9-S: 5′-TGC CAC TTC CCC TTC ATC TTC-3′ and MMP-9-AS: 5′-TTT CCC ATC AGC ATT GCC G-3′(Ta, 58.7 °C). For measuring mouse mRNAs, the following primers were used: JNK1-S: 5′-AGG AGG TAA TGG ATT TGG AGG AAC-3′ and JNK1-AS: 5′-GAA GAC GAT GGA TGC TGA GAG C-3′(Ta, 58.2 °C). These real time RT-PCR RNA levels were normalized to human or mouse cyclophilin mRNA levels as described (12).
Promoter Chromatin Immunoprecipitation (ChIP) Assay—ChIP assay was performed using in situ cross-linked U2OS cells as described (12). The primers used to amplify the following human gene promoter fragments were annotated with the binding position upstream of the transcription start site, annealing temperature (Ta) and whether in the sense (S) or antisense (AS) orientation: JNK1–1770S: 5′-ACC ATC AAC TTT GTG CTC AGC C-3′ and JNK1–1670 AS: 5′-CAT TTA TCC TGG AAA CTG GGT CAG-3′, (Ta, 58 °C); ATF2–4340S: 5′-TGG ATG CCT TCA TTT CTT GAG C-3′ and ATF2–4151AS: 5′-CTG TAT GTA AGT TCC CAC CCA CG-3′, (Ta, 61.2 °C).
Western Blot Analysis and Antibodies—U2OS cells were harvested 72 h following FoxM1 siRNA transfection, whereas FoxM1–/– MEFs were harvested at passage 4. Cell lysates were subjected to immunoblot analysis using rabbit polyclonal anti-FoxM1 antibodies (1:5000), or mouse anti-β-actin (AC-15; 1:5000; Sigma), anti-cyclin A (H-432; 1:1000; Santa Cruz Biotechnology), rabbit anti-JNK (1:1000), rabbit anti-c-Jun (1:1000), rabbit anti-phospho-c-Jun (Ser-63; 1:1000), rabbit anti-ATF-2 (20F1; 1:1000), rabbit anti-phospho-ATF-2 (Thr-71; 1:1000; Cell signaling) as described (12).
EMSA—Nuclear protein extracts were prepared from U2OS cells and used to bind a double-stranded oligonucleotide probe spanning –1275/–1248 of the human JNK1 promoter (5′-TTTCCTGTTTAGTTTTGTTTATTTTTAA-3′) that was labeled with [γ-32P]ATP and T4 kinase as described (29). Poly(dI-dC) was added as a nonspecific DNA competitor (1 μg/ml). Where indicated, unlabeled oligonucleotides (100-fold excess) were added to the nuclear extracts for 20 min on ice before addition of radiolabeled probes. DNA-protein complexes were resolved by non-denaturing polyacrylamide gel (5%) ran in 0.5× TBE buffer, followed by autoradiography.
Flow Cytometry—U2OS cells were transfected with 100 nm siRNA, with or without pHA-JNK1 (30) and then fixed and stained for flow cytometry 72 h thereafter (12). Where indicated, cells were treated with 10 μm JNK-specific inhibitor SP600125 (Calbiochem) for 48 h prior to flow cytometry.
Cell Proliferation Assay—U2OS cells were either left untreated, or transfected with siFoxM1 or siJNK1(31), or treated with SP600125 (10 μm). Viable cells were harvested at 1, 2, or 4 days, and cell numbers were measured using the Cell Titer-Glow luminescent cell viability assay kit (Promega). For BrdU incorporation, cells were serum-starved with 0.1% fetal bovine serum (FBS) for 48 h and restimulated with 10% FBS for 16 h with a 1-h pulse of BrdU. Cells were fixed and immunostained with the BrdU Labeling and Detection Kit II (Roche Applied Science). Cell nuclei were counterstained with DAPI and both BrdU-positive and -negative cells were counted in 10 random fields (200×). Experiments were performed in triplicates; shown are average numbers of cells ± S.D.
Soft Agar Assay—U2OS cells were transfected siRNA (12) or treated with 10 μm SP600125 in triplicate, and cells were harvested and plated on soft agar 24 h after treatment as described (7, 9). The culture medium containing 10% fetal calf serum or 10 μm SP600125 was replaced every 3 days. Colonies containing more than 50 cells were scored after 2 weeks.
Cell Invasion and Migration Assay—Cell invasion assay was performed by using the Matrigel-coated transwell invasion chamber (BD Bioscience) using the vendor's protocol. Briefly, cells were pretreated with siRNAs or JNK inhibitor for 48 h and were trypsinized and resuspended into Dulbecco's modified Eagle's medium (DMEM) containing 0.1% bovine serum albumin. Cells were then added to the upper chambers at 2.5 × 104 cell per well. DMEM containing 5% FBS was added to the lower chambers as chemoattractant. After 18 h, the invaded cells on the membrane's undersurface were stained with Giemsa's solution and counted in 5 fields per well. All experiments were conducted in triplicate and repeated twice.
In Vitro Wound Closure Assay—U2OS cells were either treated with JNK inhibitor (SP600125) or transfected with indicated siRNAs. Cells were plated 48 h after treatment onto 24-well plates (3 × 105 cells per well) and allowed to grow into confluent monolayers. Scrape wounds were generated using a micro tip, and cell media were replaced. 5 h after wounding, cells were fixed and stained with Giemsa's solution. Phase-contrast images of the cells were taken at 0 and 5 h after wounding, and the average wound closure rate was measured. Experiments were done in triplicate.
Statistical Analysis—Statistical significance was calculated by the Student's t test using the Prism statistics software. Statistically significant changes (p values <0.05) are indicated with asterisks.
RESULTS
Coordinate Expression of FoxM1 and JNK1—Although FoxM1 is known to regulate a genetic program essential for mitotic progression, how it controls the G1/S transition is not completely understood (12). Thus, we analyzed the expression of FoxM1 in serum-starved, quiescent mouse fibroblasts stimulated to reenter the cell cycle by serum addition. In MEFs immortalized by the expression of GSE56, a dominant negative suppressor of p53 (32, 33), FoxM1 mRNA levels increased by 8-fold within 8 h upon serum stimulation (Fig. 1A). The expression of JNK1 followed similar kinetics, suggesting that FoxM1 might regulate JNK1 expression. To test this possibility, we examined the effects of FoxM1 depletion by RNAi. Transfection of U2OS osteosarcoma cells with siFoxM1, but not control siRNA, effectively depleted FoxM1 mRNA and protein (Fig. 1, B and C). Knockdown of FoxM1 resulted in the reduction of JNK1 mRNA by 60%, whereas JNK2 mRNA was not affected (Fig. 1B). The level of ATF-2 mRNA, which encodes a transcription factor implicated in the regulation of cyclin D1 and cyclin A (25, 34, 35), was also significantly reduced in FoxM1-depeleted cells.
FIGURE 1.
FoxM1 and JNK1 expression in response to serum stimulation. A, MEFs immortalized by dominant negative p53 were serum-starved for 72 h and restimulated by the addition of FBS to 10%. FoxM1 and JNK1 mRNA levels were detected by qRT-PCR. B, diminished levels of endogenous JNK1, ATF-2, and FoxM1 mRNAs were found in U2OS cells transfected with FoxM1 siRNA as determined by qRT-PCR. No significant change in JNK2 mRNA was detected. C, U2OS cells transfected with siFoxM1 exhibited diminished JNK1, ATF-2, cyclin A2 protein levels, and reduced phosphorylation of c-Jun and ATF2 as determined by immunoblotting. D, RNA preparations and total cell extracts from early passage of FoxM1+/+, FoxM1+/–, and FoxM1–/– MEFs were used for qRT-PCR (upper) and immunoblot (lower) analysis. FoxM1–/– MEFs expressed reduced levels of JNK1 mRNA and protein compared with WT MEFs. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Knockdown of FoxM1 also significantly decreased the levels of JNK1 and ATF-2 proteins (Fig. 1C). JNK1 is known to phosphorylate and activate c-Jun and ATF-2, which promote entry into S-phase (37). Consistent with the diminished JNK1 expression, FoxM1-depleted cells showed reduced phosphorylation of c-Jun and ATF-2 (Fig. 1C). The expression of cyclin A2, a known transcription target of c-Jun (27), was also diminished by 50% (Fig. 1C). Furthermore, JNK1 expression was significantly reduced in FoxM1–/– MEFs compared with FoxM1+/+ or FoxM1+/– MEFs (Fig. 1D). Thus, consistent with the notion that FoxM1 regulates JNK1, FoxM1 deficiency leads to reduced JNK1 mRNA and protein, curtailed phosphorylation of the JNK1 substrates c-Jun and ATF2, and diminished expression of the c-Jun target gene cyclin A2.
FoxM1 Transcriptionally Activates the JNK1 Promoter—The FoxM1 mRNAs are differentially spliced into three isoforms: FoxM1b, which is the predominant isoform and the isoform detected in our studies; FoxM1c, which contains an additional exon in the DNA binding domain; and the transcriptionally inactive FoxM1a (38, 39). We found three consecutive consensus FoxM1 binding sites (38, 40) between –1275 and –1251 bp and a fourth site between –330 and –316 bp of the human JNK1 promoter, suggesting the possibility that JNK1 is a direct transcriptional target of FoxM1. We used EMSA to address whether these consensus FoxM1 binding sites indeed bind nuclear factors. Nuclear extracts of U2OS cells bound labeled oligonucleotides of the sequence from –1275/–1248 bp of the JNK1 promoter to form a protein-DNA complex, which was efficiently competed by cold oligonucleotide (Fig. 2A). To test the transcriptional activity of FoxM1 upon the JNK1 promoter, we cotransfected U2OS cells with a FoxM1b expression construct (pCMV-FoxM1b) and a reporter plasmid containing 1.3 kb of the human JNK1 promoter driving luciferase expression. Overexpression of FoxM1b resulted in an 11-fold increase in JNK1 promoter activation, whereas depletion of FoxM1 using siRNA abrogated transcriptional activation of the JNK1 promoter (Fig. 2B). To determine whether FoxM1 binds directly to the human JNK1 promoter, we performed ChIP assays using PCR primers to DNA sequences (–1770/–1670 bp) in the vicinity of the potential FoxM1 binding sites (–1275/–1251 bp). Indeed, ChIP assays showed that FoxM1 protein is directly bound to the human JNK1 promoter region (Fig. 2C). Moreover, knockdown of FoxM1 by siRNA obliterated the binding of FoxM1 and RNA polymerase II to the JNK1 promoter (Fig. 2C). Because the CBP coactivator can be recruited by a number of other transcription factors, depletion of FoxM1 did not greatly diminish the recruitment of CBP to the promoter. We also found five FoxM1 consensus binding sites between –4419 and –3806 bp of the human ATF-2 promoter region. As shown by ChIP assay, FoxM1 protein is also associated with the human ATF-2 promoter, an association that is abrogated by siRNA depletion of FoxM1 (Fig. 2C). Taken together, these results demonstrate that FoxM1 binds directly to the human JNK1 promoter and transcriptionally activates JNK1 expression. FoxM1 also binds the ATF-2 promoter and is required for ATF-2 expression (Figs. 1B and 2C), suggesting that FoxM1 may transcriptionally regulate ATF-2 as well.
FIGURE 2.
FoxM1 transcriptionally activates JNK1. A, EMSA was performed using U2OS cell nuclear lysate to bind 32P-labeled double-stranded oligonucleotide probe made to human JNK1 promoter (–1275/–1248 bp) (lane 2). For DNA competitions, 100-fold molar excess of cold double-stranded oligonucleotide probe was added to the binding reaction (lane 3). Radioactivity-labeled probe only was used as negative control (lane 1). B, U2OS cells were transfected with the CMV-FoxM1b expression vector, a reporter construct with the –1308 bp human JNK1 promoter linked to firefly luciferase, and CMV-Renilla luciferase as the internal control. Cells were cotransfected with siFoxM1 where indicated. Cell extracts were prepared 24 h after transfection; dual luciferase enzyme activity was measured and expressed as fold induction of transcription. C, binding of FoxM1 to JNK1 and ATF2 promoters as determined by quantitative promoter ChIP assays. Untreated or FoxM1-depleted U2OS cells were cross-linked and sonicated, and chromatin fragments were immunoprecipitated with antibodies specific to FoxM1, CBP, or RNA polymerase II. The promoter DNA associated with the IP chromatin was quantified by qRT-PCR with primers for the human JNK1 (–1770/–1670) or ATF-2 (–4340/–4151) promoter, and normalized to the amount of DNA bound to FoxM1 in untreated cells. Error bars represent ± S.D. from experiments done in triplicate. *, p < 0.05, and ***, p < 0.001.
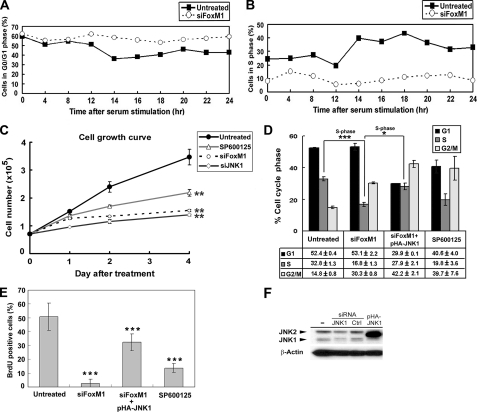
FoxM1-induced JNK1 Is Essential for the G1/S Transition—We have previously shown that FoxM1 depletion inhibits cell cycle progression at both the G1/S and G2/M boundaries (12). To determine whether FoxM1 is required for the re-entry of serum-deprived cells into the cell cycle after re-stimulation with serum, untreated or siFoxM1-transfected U2OS cells were maintained in 0.1% FBS for 48 h to drive cells into a G0/G1 state (29). Cells were then stimulated with 10% FBS and subjected to flow cytometry analysis at various times thereafter (Fig. 3A). Upon serum starvation, the percentages of cells with G1 DNA content in untransfected and siFoxM1-transfected cells accumulated to 62 and 59%, respectively, suggesting that most of these cells were synchronized in G0/G1 (Fig. 3A). Within 14 h after serum stimulation, the percentage of untransfected cells with G1 DNA content dropped to 36% while the S phase population rose from 24 to 44% (Fig. 3, A and B), indicating the reentry of these cells into S phase. By contrast, the percentages of FoxM1-depleted cells in G0/G1 and S phase remained the same after serum stimulation. Thus, depletion of FoxM1 prevented the progression of G0/G1 cells into S phase.
FIGURE 3.
FoxM1 depletion inhibits cell cycle reentry and progression in response to serum stimulation. A, U2OS cells were untreated or transfected with siFoxM1 and maintained in 0.1% FBS for 48 h. Cells were then stimulated with 10% FBS and harvested at various times (0–24 h) thereafter for flow cytometry analysis. Percentages of cells in the G1 phase were counted. B, cells were treated as in A, and percentages of cells in S phase were quantified. C, U2OS cells were either untreated, transfected with siFoxM1 or siJNK1, or treated with SP600125 (10 μm) and cultured in 10% FBS. Numbers of viable cells at indicated days were counted. D, cells were either untreated, transfected with siFoxM1, cotransfected with siFoxM1 and pHA-JNK1, or treated with SP600125. Percentages of cells in G1, S, and G2/M phases were analyzed by flow cytometry 72 h after transfection or 24 h after JNK inhibitor treatment. E, cells were treated as in D and serum-starved for 48 h, followed by the addition of 10% FBS. Cells were pulse-labeled with BrdU for 1 h following 16 h of serum stimulation, and the percentages of cells with BrdU-positive staining were quantified. F, Western blot analysis demonstrated diminished JNK1 protein level in siJNK1-transfected U2OS cells for 72 h. Error bars represent ± S.D. from experiments done in triplicate. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Next we investigated whether JNK1 expression induced by FoxM1 is essential for G1/S progression. U2OS cells were transfected with siFoxM1 or siJNK1, and their growth rates were determined. Depletion of FoxM1 or JNK1 by siRNA (Figs. 1C and 3F), or treatment of cells with SP600125 to inhibit JNK activity all resulted in similar reductions in cell proliferation, indicating that FoxM1 and JNK1 are both critical for cell proliferation (Fig. 3C). To assess the specific role of JNK1, we analyzed the effects of re-expressing JNK1 in FoxM1-depleted cells via transfection of the expression vector pHA-JNK1 (Fig. 3F). FoxM1-depleted cells showed a 50% decrease in S-phase cells compared with untreated U2OS cells as judged by flow cytometry (Fig. 3D). Expression of JNK1 in FoxM1-depleted cells rescued S-phase progression, restoring S phase cells to 85% of the level found in untreated cells (Fig. 3D). Cells treated with the JNK-specific inhibitor SP600125 also exhibited a 40% decrease of S-phase cells, further reinforcing the notion that JNK is required for G1/S entry. To support the interpretation that FoxM1-regulated JNK1 expression is critical for G1/S progression further, we tested the role of JNK1 in DNA synthesis. Knockdown of FoxM1 by siRNA or inhibition of JNK activity by SP600125 both severely inhibited DNA synthesis, whereas re-expression of JNK1 in FoxM1-depleted cells restored DNA synthesis (Fig. 3E). These findings indicate that JNK1 expression in FoxM1-depleted cells functionally rescued a block in DNA synthesis and promoted G1/S progression. Interestingly, JNK1 expression in FoxM1-deficient cells resulted in a 3-fold increase in percentage of G2/M (4N) cells (42.2 ± 2.1%) compared with control cells (14.8 ± 0.8%), and a 50% increase compared with FoxM1-depleted cells (30.3 ± 0.8%)(Fig. 3D). These results suggest that expression of JNK1 in FoxM1-depleted cells was unable to overcome the G2/M block caused by FoxM1 deficiency.
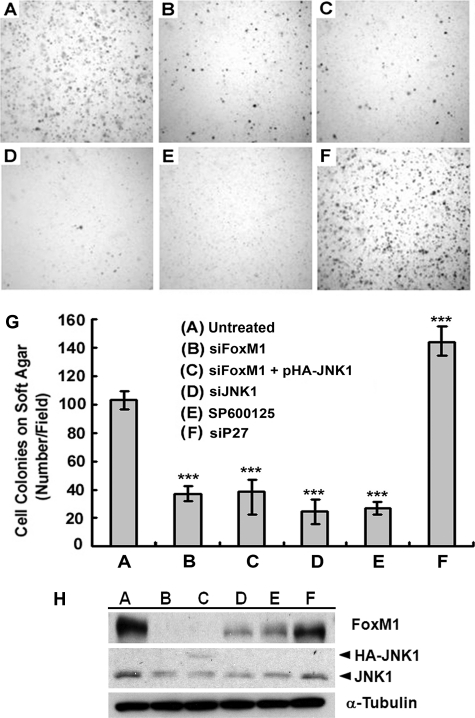
FoxM1 and JNK1 Play Critical Roles in Anchorage-independent Growth and Invasiveness of Cancer Cells—FoxM1 plays a critical role in tumor growth and promotes metastasis (7, 8, 41). To understand how FoxM1 and JNK1 may participate in this process, we examined the ability of U2OS cells to form colonies in soft agar after treatments that perturb their expression of FoxM1 or JNK1. Knockdown of FoxM1 by siRNA reduced colony formation to 36% of control (Fig. 4). Knockdown of FoxM1 is known to result in p27Kip1 accumulation (16), and consistently, transfection of cells with p27Kip1 siRNA resulted in increased colony formation. Ablation of JNK1 activity by either siJNK1 or treatment with the inhibitor SP600125 also inhibited colony formation to ∼25% of control level (Fig. 4). Expression of JNK1 in FoxM1-depleted cells via transfection of the pHA-JNK1 vector failed to rescue colony formation in soft agar. These results indicate that FoxM1 is a critical regulator of anchorage-independent growth, and JNK1 is a necessary but insufficient downstream target of FoxM1 in this process.
FIGURE 4.
FoxM1- or JNK1-depletion inhibits anchorage-independent growth in soft agar. U2OS cells were either left untreated (A), or transfected with siFoxM1 (B), siFoxM1 with pHA-JNK1 (C), siJNK1 (D), sip27 (F), or treated with SP600125 (E) as indicated. One day after treatment, cells were trypsinized and replated on soft agar. Microphotographs showed colonies grown in soft agar for 2 weeks. G, numbers of colonies per microscopic field were counted; error bars represent ± S.D. from experiments done in triplicate. H, total cell lysates harvested in cells treated in A–F as indicated were immunoblotted to determine the levels of expression of FoxM1, JNK1, HA-JNK1, and α-tubulin. ***, p < 0.001.
The abilities of tumor cells to migrate and invade into the neighboring tissue are critical for metastasis. To dissect the specific roles of FoxM1 and its downstream target JNK1 in tumorigenesis further, we tested whether they are essential for tumor cell migration and invasion. U2OS cells were subjected to treatments that altered their expression of FoxM1 or JNK1, and were grown to monolayers before being subjected to an in vitro scratch wound assay. Depletion of either FoxM1 or JNK1 by siRNA, or inhibition of JNK activity by SP600125, significantly diminished the wound closure rate compared with the untreated control or control siRNA-transfected cells (Fig. 5). Expression of JNK1 in FoxM1-depleted cells did not rescue this defect. Next, we investigated the roles of FoxM1 and JNK1 in the invasive properties of U2OS tumor cells using a transwell invasion chamber assay. Depletion of FoxM1 or JNK1 by RNAi, or inhibition of JNK1 activity by SP600125, reduced the invasion of U2OS cells through the Matrigel-coated transwell by 70–85% (Fig. 6). Expression of JNK1 in FoxM1-depleted cells did not rescue this defect. Taken together, these results show that FoxM1 plays an essential regulatory role in tumor cell migration, invasion, and anchorage-independent growth, with JNK1 being a necessary but insufficient downstream mediator of FoxM1 in these processes.
FIGURE 5.
FoxM1- or JNK1-depletion compromises in vitro wound closure. U2OS cells were plated and grown to confluence before scratch wounds were inflicted. A–F, phase contrast micrographs of scratch wounds at the time of wounding (t = 0 h) and 5-h postwounding (t = 5 h) are shown. G, wound closure rates were calculated and presented in the bar graph. Error bars represent ± S.D. from experiments done in triplicate. H, total cell lysates harvested in cells treated in A–F as indicated were immunoblotted to determine the levels of expression of FoxM1, JNK1, HA-JNK1, and α-tubulin. **, p < 0.01 and ***, p < 0.001.
FIGURE 6.
Impaired invasion in cells depleted of FoxM1 or JNK1. A–F, cells were treated as above, and their migration through Matrigel invasion chambers toward 5% FBS was monitored. Migrated cells were stained with Giemsa and photographed. G, numbers of migrated cells were counted in five fields per well. The error bar represent results ± S.D. in experiments done in triplicate. H, total cell lysates harvested in cells treated in A–F as indicated were immunoblotted to determine the levels of expression of FoxM1, JNK1, HA-JNK1, and α-tubulin. ***, p < 0.001.
FoxM1 Regulates Expression of MMP-9, but Not MMP-2, through JNK1 in U2OS Cells—Matrix metalloproteinases (MMPs) are implicated in tumor cell invasion and metastasis by degrading the extracellular matrix. In particular, MMP-2 and MMP-9 can degrade type IV collagen, thereby facilitating the breakdown of basal lamina and allowing neoplastic epithelial cells to invade the underlying stroma. Thus, we examined whether FoxM1 can regulate MMP-2 and MMP-9 expression, which may contribute to tumor cell invasion. Depletion of FoxM1 by siRNA curtailed MMP-2 and MMP-9 expression in U2OS cells by 50 and 60%, respectively (Fig. 7), indicating that MMP-2 and MMP-9 are both regulated by FoxM1. However, inhibition of JNK activity by SP600125 decreased MMP-9 expression by 50%, but had no effect on MMP-2 expression. Furthermore, expression of JNK1 in FoxM1-depleted cells resulted in a 3.8-fold increase in MMP-9 expression compared with the untreated control, but had no effect on MMP-2 expression (Fig. 7). These results indicate that MMP-9 and MMP-2 gene expression is regulated by FoxM1 through JNK1-dependent and -independent pathways, respectively.
FIGURE 7.
Diminished expression of MMP-9 and MMP-2 in FoxM1-deficient U2OS cells. A, expression of MMP-9 in U2OS cells transfected with siFoxM1 or treated with SP600125 was compared with that in untreated control cells as determined by qTR-PCR. Re-expression of JNK1 in FoxM1-deficient U2OS cells significantly increased MMP-9 expression. B, expression of MMP-2 cells treated as above was determined by qRT-PCR and calculated as a percentage relative to untreated cells. Error bars represent ± S.D. in triplicate experiments. C, total cell lysates harvested in cells treated in A and B as indicated were immunoblotted to determine the levels of expression of FoxM1, JNK1, HA-JNK1, and α-tubulin. **, p ≤ 0.01 and ***, p ≤ 0.001.
DISCUSSION
The principal findings of this study provide new insights into the mechanism by which FoxM1 regulates cell cycle progression. FoxM1 directly activates the transcription of JNK1, which plays an essential role in mediating FoxM1 functions in the G1/S transition. Both FoxM1 and JNK1 are critical for cell migration, invasion, and anchorage-independent growth in U2OS osteosarcoma cells. These functions may be mediated in part through the JNK1-dependent expression of MMP-9 and the JNK-independent expression of MMP-2, both regulated by FoxM1. Thus, FoxM1 acts as an upstream regulator of the JNK1 signaling pathway to control cell cycle progression and tumor cell migration and invasion (Fig. 8).
FIGURE 8.
Model for the roles of FoxM1 and JNK1 in cell proliferation, migration, and invasion. FoxM1 transcriptionally activates JNK1, which regulates the expression of a set of genes critical for G1/S progression, including cyclin A. JNK1 also mediates the regulation of MMP-9 by FoxM1, thus contributing to cell migration and invasion. FoxM1 directly regulates MMP2 expression (59) to enhance cell migration and invasion.
We show that inhibition of JNK activity alone is sufficient to recapitulate the G1/S block elicited by FoxM1 deficiency, whereas reconstitution of JNK1 efficiently rescues this defect in FoxM1-depleted cells. These findings imply that the JNK1-dependent signaling pathway is necessary and sufficient for mediating the FoxM1 effects in promoting G1/S progression. Diminished JNK1 protein level in FoxM1-depleted cells resulted in reduced phosphorylation and activation of c-Jun (42) and ATF-2 (Fig. 1, B and C). Activated c-Jun and ATF-2 form AP-1 transcription factors that induce the expression of multiple genes essential for G1/S progression, including cyclin D1, cyclin E, and cyclin A (24, 26, 27, 34, 44–47). Indeed, the expression of cyclin A2 was diminished in FoxM1-depeleted cells (Fig. 1C). Consequently, down-regulation of FoxM1 or inhibition of JNK resulted in reduced DNA synthesis, a defect that was rescued by the expression of JNK1 (Fig. 3E). Therefore, JNK1 deficiency can directly explain the G1/S cell cycle block observed in FoxM1-depleted cells (Fig. 8). These findings are consistent with the cell proliferation defects observed in JNK1–/– fibroblasts (37). While FoxM1 also transcriptionally activates Skp2 and Cks1 to facilitate the degradation of CDK inhibitors p21Cip1 and p27Kip1 (12, 15, 16), overexpression of JNK1 in FoxM1-depleted cells apparently overrides the defects stemming from deficiency in Skp2 and Cks1 that may inhibit the G1/S transition. How JNK1 is able to override these defects mechanistically is currently under investigation.
FoxM1 directly binds and activates the JNK1 promoter as judged by ChIP and transcription assays (Fig. 2). However, FoxM1 has no effect on JNK2 expression. Recent studies show that JNK1 and JNK2 play opposing roles in cell proliferation. Whereas JNK1 phosphorylates and activates the transcriptional activity of c-Jun, JNK2 is preferentially bound to c-Jun in unstimulated cells and contributes to the degradation of c-Jun (37). Thus, JNK1 is a positive regulator of cell proliferation, whereas JNK2 plays a negative role in this process (37, 49). Our finding that FoxM1 specifically activates JNK1 but not JNK2 is consistent with the role of FoxM1 as a positive regulator of cell proliferation.
Inhibition of JNK activity by SP600125 appears to block cells from mitotic progression as well, resulting in an accumulation of cells in G2/M as previously reported (Fig. 3D) (50). However, in this instance it is unclear which JNK isoform plays the dominant role in G2/M transition. It has been shown that inhibition of JNK2, but not JNK1, caused an accumulation of cells with 4N DNA content (51), suggesting that JNK2 may play a role in mitotic exit. Expression of JNK1 is unable to overcome the blockade in mitotic exit in FoxM1-depleted cells (Fig. 3D). In this regard, FoxM1 has been shown to directly regulate the expression of a large number of genes critical for mitotic progression, including Cdc25B, aurora B, PLK-1, Cenp-A, and Cenp-B (12–14). The inability of JNK1 to compensate for FoxM1 deficiency indicates that JNK1 expression is unable to reverse the deficit in expression of these genes for mitotic progression.
Recently, Das et al. (28) showed that JNK-deficient cells exhibit early senescence through up-regulation of p53. Interestingly, FoxM1–/– MEFs also display phenotypes of premature senescence with the expression of senescence-associated β-galactosidase, p16Ink4A, and p19ARF, resulting in a G2/M block (12). These phenotypes are similar to those of MEFs deficient in MKK7, the upstream activating kinase of JNK, which also exhibit a premature senescence phenotype associated with a G2/M block (52). Moreover, cells that are deficient in the JNK substrate c-Jun or its binding partner JunD also show elevated p53 expression and early senescence (53–55). These results establish the critical role of the JNK pathway in protecting cells from senescence, and suggest that down-regulation of JNK contributes to premature senescence induced by FoxM1 deficiency.
High levels of FoxM1 expression has been associated with a variety of human cancers, including hepatocellular carcinoma, intrahepatic cholangiocarcinomas, basal cell carcinomas, infiltrating ductal breast carcinomas, anaplastic astrocytomas, and glioblastomas (56). In established animal models, FoxM1 is critical for the development and metastasis of chemically induced hepatocellular carcinomas, prostate cancer, lung cancer, and colorectal cancer (7–10). These results establish that FoxM1 plays a critical role in tumor development and suggest that FoxM1 may promote tumor invasion (41). Expression of matrix metalloproteinases, particularly MMP-2 and MMP-9, have been strongly implicated in tumor angiogenesis, invasion and metastasis (57, 58). Indeed, very recent studies published while this manuscript was being prepared reported that FoxM1 can upregulate the expression of MMP-2 and MMP-9, and this expression promotes the invasiveness of glioma cells and pancreatic cancer cells (59, 60). These findings are in agreement with our observation that FoxM1 up-regulates MMP-2 and MMP-9 in U2OS osteosarcoma cells, and support the notion that MMP-2 and MMP-9 play a role in FoxM1-dependent tumor invasion. Our studies further show that FoxM1 regulates MMP-9, but not MMP-2, through a JNK-dependent pathway (Fig. 7). It is known that c-Jun, a JNK substrate and a component of the AP-1 transcriptional factor, can activate MMP-9 transcription through multiple AP-1 binding sites in the MMP-9 promoter (61). By contrast, FoxM1 can bind to the MMP-2 promoter directly and activate its transcription in glioma cells (59). Thus, FoxM1 may induce MMP-9 through JNK1-mediated activation of c-Jun, whereas MMP-2 is likely activated by FoxM1 directly in U2OS cells (Fig. 8). However, it is noteworthy that the activation of MMP-2 has been reported to be JNK-dependent in some cell types but not in others (36, 48, 62, 63), suggesting that the specific mechanism of FoxM1-induced MMP-2 expression may be cell type-dependent.
Our current model for how FoxM1 regulates cell cycle progression and cell migration and invasion through the activation of JNK1 is summarized in Fig. 8. Given the importance of the FoxM1-JNK1 axis in cell cycle progression and tumor cell invasion, targeting its function holds promise as an attractive cancer therapeutic approach. Indeed, treatment of mice with a cell-penetrating peptide that blocks FoxM1 activity significantly reduces tumor growth in chemically induced hepatocellular carcinoma (43). The possibility that intervention of FoxM1 signaling as a therapeutic strategy in cancers associated with FoxM1 overexpression clearly warrants further investigation.
Acknowledgments
We thank Dr. A. N. Kong (Rutgers) for providing the pcDNA3-HA-JNK1 plasmid, Dr. K. L. Hagen for flow cytometry analysis, and H. Yoder, A. Monson, and W. N. Yu for help and assistance.
This work was supported by National Institutes of Health Grants AG21842, DK54687, CA124488 (to R. H. C.), CA46565 and HL081390 (to L. F. L.), CA100035, and CA124488 (to P. R.), and DK44525 and DK68503 (to A. L. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This report is dedicated to the memory of Dr. Robert H. Costa, who unfortunately passed away after a heroic fight with cancer.
Footnotes
The abbreviations used are: Fox, Forkhead box; EMSA, electrophoretic mobility shift assay; JNK, c-Jun N-terminal kinase; ATF-2, activating transcription factor-2; CDKI, cyclin-dependent kinase inhibitors; MEFs, mouse embryonic fibroblasts; MMPs, matrix metalloproteinases; FBS, fetal bovine serum; BrdU, bromodeoxyuridine; siRNA, short interfering RNA; ChIP, chromatin immunoprecipitation assay; qRT-PCR, quantitative real-time RT-PCR; WT, wild type.
References
- 1.Kaestner, K. H., Knochel, W., and Martinez, D. E. (2000) Genes Dev. 14 142–146 [PubMed] [Google Scholar]
- 2.Costa, R. H. (2005) Nat. Cell Biol. 7 108–110 [DOI] [PubMed] [Google Scholar]
- 3.Wang, X., Kiyokawa, H., Dennewitz, M. B., and Costa, R. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16881–16886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye, H., Holterman, A. X., Yoo, K. W., Franks, R. R., and Costa, R. H. (1999) Mol. Cell. Biol. 19 8570–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalinichenko, V. V., Gusarova, G. A., Tan, Y., Wang, I. C., Major, M. L., Wang, X., Yoder, H. M., and Costa, R. H. (2003) J. Biol. Chem. 278 37888–37894 [DOI] [PubMed] [Google Scholar]
- 6.Krupczak-Hollis, K., Wang, X., Kalinichenko, V. V., Gusarova, G. A., Wang, I. C., Dennewitz, M. B., Yoder, H. M., Kiyokawa, H., Kaestner, K. H., and Costa, R. H. (2004) Dev. Biol. 276 74–88 [DOI] [PubMed] [Google Scholar]
- 7.Kalinichenko, V. V., Major, M. L., Wang, X., Petrovic, V., Kuechle, J., Yoder, H. M., Dennewitz, M. B., Shin, B., Datta, A., Raychaudhuri, P., and Costa, R. H. (2004) Genes Dev. 18 830–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, I. M., Ackerson, T., Ramakrishna, S., Tretiakova, M., Wang, I. C., Kalin, T. V., Major, M. L., Gusarova, G. A., Yoder, H. M., Costa, R. H., and Kalinichenko, V. V. (2006) Cancer Res. 66 2153–2161 [DOI] [PubMed] [Google Scholar]
- 9.Kalin, T. V., Wang, I. C., Ackerson, T. J., Major, M. L., Detrisac, C. J., Kalinichenko, V. V., Lyubimov, A., and Costa, R. H. (2006) Cancer Res. 66 1712–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida, Y., Wang, I. C., Yoder, H. M., Davidson, N. O., and Costa, R. H. (2007) Gastroenterology 132 1420–1431 [DOI] [PubMed] [Google Scholar]
- 11.Wang, I. C., Meliton, L., Tretiakova, M., Costa, R. H., Kalinichenko, V. V., and Kalin, T. V. (2008) Oncogene [DOI] [PubMed]
- 12.Wang, I. C., Chen, Y. J., Hughes, D., Petrovic, V., Major, M. L., Park, H. J., Tan, Y., Ackerson, T., and Costa, R. H. (2005) Mol. Cell. Biol. 25 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laoukili, J., Kooistra, M. R., Bras, A., Kauw, J., Kerkhoven, R. M., Morrison, A., Clevers, H., and Medema, R. H. (2005) Nat. Cell Biol. 7 126–136 [DOI] [PubMed] [Google Scholar]
- 14.Wonsey, D. R., and Follettie, M. T. (2005) Cancer Res. 65 5181–5189 [DOI] [PubMed] [Google Scholar]
- 15.Liu, M., Dai, B., Kang, S. H., Ban, K., Huang, F. J., Lang, F. F., Aldape, K. D., Xie, T. X., Pelloski, C. E., Xie, K., Sawaya, R., and Huang, S. (2006) Cancer Res. 66 3593–3602 [DOI] [PubMed] [Google Scholar]
- 16.Petrovic, V., Costa, R. H., Lau, L. F., Raychaudhuri, P., and Tyner, A. L. (2008) J. Biol. Chem. 283 453–460 [DOI] [PubMed] [Google Scholar]
- 17.Minden, A., and Karin, M. (1997) Biochim. Biophys. Acta 1333 F85–F104 [DOI] [PubMed] [Google Scholar]
- 18.Ip, Y. T., and Davis, R. J. (1998) Curr. Opin. Cell Biol. 10 205–219 [DOI] [PubMed] [Google Scholar]
- 19.Hall, J. P., Merithew, E., and Davis, R. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14022–14024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy, N. J., and Davis, R. J. (2003) Cell Cycle 2 199–201 [PubMed] [Google Scholar]
- 21.Gupta, S., Barrett, T., Whitmarsh, A. J., Cavanagh, J., Sluss, H. K., Derijard, B., and Davis, R. J. (1996) EMBO J. 15 2760–2770 [PMC free article] [PubMed] [Google Scholar]
- 22.Weston, C. R., and Davis, R. J. (2007) Curr. Opin. Cell Biol. 19 142–149 [DOI] [PubMed] [Google Scholar]
- 23.Leaner, V. D., Kinoshita, I., and Birrer, M. J. (2003) Oncogene 22 5619–5629 [DOI] [PubMed] [Google Scholar]
- 24.Wisdom, R., Johnson, R. S., and Moore, C. (1999) EMBO J. 18 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beier, F., Taylor, A. C., and LuValle, P. (2000) J. Biol. Chem. 275 12948–12953 [DOI] [PubMed] [Google Scholar]
- 26.Schwabe, R. F., Bradham, C. A., Uehara, T., Hatano, E., Bennett, B. L., Schoonhoven, R., and Brenner, D. A. (2003) Hepatology 37 824–832 [DOI] [PubMed] [Google Scholar]
- 27.Katabami, M., Donninger, H., Hommura, F., Leaner, V. D., Kinoshita, I., Chick, J. F., and Birrer, M. J. (2005) J. Biol. Chem. 280 16728–16738 [DOI] [PubMed] [Google Scholar]
- 28.Das, M., Jiang, F., Sluss, H. K., Zhang, C., Shokat, K. M., Flavell, R. A., and Davis, R. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15759–15764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Major, M. L., Lepe, R., and Costa, R. H. (2004) Mol. Cell. Biol. 24 2649–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keum, Y. S., Owuor, E. D., Kim, B. R., Hu, R., and Kong, A. N. (2003) Pharm. Res. 20 1351–1356 [DOI] [PubMed] [Google Scholar]
- 31.Dai, Y., Rahmani, M., Pei, X. Y., Khanna, P., Han, S. I., Mitchell, C., Dent, P., and Grant, S. (2005) Blood 105 1706–1716 [DOI] [PubMed] [Google Scholar]
- 32.Zou, X., Ray, D., Aziyu, A., Christov, K., Boiko, A. D., Gudkov, A. V., and Kiyokawa, H. (2002) Genes Dev. 16 2923–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skeen, J. E., Bhaskar, P. T., Chen, C. C., Chen, W. S., Peng, X. D., Nogueira, V., Hahn-Windgassen, A., Kiyokawa, H., and Hay, N. (2006) Cancer Cell 10 269–280 [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, M., Nomura, Y., Suzuki, H., Ichikawa, E., Takeuchi, A., Suzuki, M., Nakamura, T., Nakajima, T., and Oda, K. (1998) Exp. Cell Res. 239 93–103 [DOI] [PubMed] [Google Scholar]
- 35.Beier, F., Lee, R. J., Taylor, A. C., Pestell, R. G., and LuValle, P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1433–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, W. P., III, Douillet, C. D., Milano, P. M., Boucher, R. C., Patterson, C., and Rich, P. B. (2006) Am. J. Physiol. Heart Circ. Physiol. 290 H1988–H1996 [DOI] [PubMed] [Google Scholar]
- 37.Sabapathy, K., Hochedlinger, K., Nam, S. Y., Bauer, A., Karin, M., and Wagner, E. F. (2004) Mol. Cell 15 713–725 [DOI] [PubMed] [Google Scholar]
- 38.Ye, H., Kelly, T. F., Samadani, U., Lim, L., Rubio, S., Overdier, D. G., Roebuck, K. A., and Costa, R. H. (1997) Mol. Cell. Biol. 17 1626–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korver, W., Roose, J., and Clevers, H. (1997) Nucleic Acids Res. 25 1715–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao, K. M., Sha, M., Lu, Z., and Wong, G. G. (1997) J. Biol. Chem. 272 19827–19836 [DOI] [PubMed] [Google Scholar]
- 41.Chandran, U. R., Ma, C., Dhir, R., Bisceglia, M., Lyons-Weiler, M., Liang, W., Michalopoulos, G., Becich, M., and Monzon, F. A. (2007) BMC. Cancer 7 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derijard, B., Hibi, M., Wu, I. H., Barrett, T., Su, B., Deng, T., Karin, M., and Davis, R. J. (1994) Cell 76 1025–1037 [DOI] [PubMed] [Google Scholar]
- 43.Gusarova, G. A., Wang, I. C., Major, M. L., Kalinichenko, V. V., Ackerson, T., Petrovic, V., and Costa, R. H. (2007) J. Clin. Investig. 117 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakiri, L., Lallemand, D., Bossy-Wetzel, E., and Yaniv, M. (2000) EMBO J. 19 2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hennigan, R. F., and Stambrook, P. J. (2001) Mol. Biol. Cell 12 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albanese, C., D'Amico, M., Reutens, A. T., Fu, M., Watanabe, G., Lee, R. J., Kitsis, R. N., Henglein, B., Avantaggiati, M., Somasundaram, K., Thimmapaya, B., and Pestell, R. G. (1999) J. Biol. Chem. 274 34186–34195 [DOI] [PubMed] [Google Scholar]
- 47.Bakiri, L., Matsuo, K., Wisniewska, M., Wagner, E. F., and Yaniv, M. (2002) Mol. Cell. Biol. 22 4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung, L. W., Leung, P. C., and Wong, A. S. (2006) Cancer Res. 66 10902–10910 [DOI] [PubMed] [Google Scholar]
- 49.Sabapathy, K., and Wagner, E. F. (2004) Cell Cycle 3 1520–1523 [DOI] [PubMed] [Google Scholar]
- 50.Kuntzen, C., Sonuc, N., De Toni, E. N., Opelz, C., Mucha, S. R., Gerbes, A. L., and Eichhorst, S. T. (2005) Cancer Res. 65 6780–6788 [DOI] [PubMed] [Google Scholar]
- 51.MacCorkle, R. A., and Tan, T. H. (2004) J. Biol. Chem. 279 40112–40121 [DOI] [PubMed] [Google Scholar]
- 52.Wada, T., Joza, N., Cheng, H. Y., Sasaki, T., Kozieradzki, I., Bachmaier, K., Katada, T., Schreiber, M., Wagner, E. F., Nishina, H., and Penninger, J. M. (2004) Nat. Cell Biol. 6 215–226 [DOI] [PubMed] [Google Scholar]
- 53.Schreiber, M., Kolbus, A., Piu, F., Szabowski, A., Mohle-Steinlein, U., Tian, J., Karin, M., Angel, P., and Wagner, E. F. (1999) Genes Dev. 13 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weitzman, J. B., Fiette, L., Matsuo, K., and Yaniv, M. (2000) Mol. Cell 6 1109–1119 [DOI] [PubMed] [Google Scholar]
- 55.MacLaren, A., Black, E. J., Clark, W., and Gillespie, D. A. (2004) Mol. Cell. Biol. 24 9006–9018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teh, M. T., Wong, S. T., Neill, G. W., Ghali, L. R., Philpott, M. P., and Quinn, A. G. (2002) Cancer Res. 62 4773–4780 [PubMed] [Google Scholar]
- 57.Egeblad, M., and Werb, Z. (2002) Nat. Rev. Cancer 2 161–174 [DOI] [PubMed] [Google Scholar]
- 58.Deryugina, E. I., and Quigley, J. P. (2006) Cancer Metastasis Rev. 25 9–34 [DOI] [PubMed] [Google Scholar]
- 59.Dai, B., Kang, S. H., Gong, W., Liu, M., Aldape, K. D., Sawaya, R., and Huang, S. (2007) Oncogene 26 6212–6219 [DOI] [PubMed] [Google Scholar]
- 60.Wang, Z., Banerjee, S., Kong, D., Li, Y., and Sarkar, F. H. (2007) Cancer Res. 67 8293–8300 [DOI] [PubMed] [Google Scholar]
- 61.Gum, R., Wang, H., Lengyel, E., Juarez, J., and Boyd, D. (1997) Oncogene 14 1481–1493 [DOI] [PubMed] [Google Scholar]
- 62.Xie, Z., Singh, M., and Singh, K. (2004) J. Biol. Chem. 279 39513–39519 [DOI] [PubMed] [Google Scholar]
- 63.Arai, K., Lee, S. R., and Lo, E. H. (2003) Glia 43 254–264 [DOI] [PubMed] [Google Scholar]