Abstract
Hepatocyte growth factor (HGF) and its high affinity receptor, the tyrosine kinase Met, play a key role in embryo development and tumor invasion. Both HGF and Met are established targets for cancer therapy. However, the mechanism of their interaction is complex and remains elusive. HGF is secreted as a monomeric precursor (pro-HGF) that binds to but does not activate Met. Mature HGF is a α/β heterodimer containing a high affinity Met-binding site in the α-chain (HGF-α) and a low affinity Met-binding site in the β-chain (HGF-β). The extracellular portion of Met contains a semaphorin (Sema) domain, a cysteine-rich hinge (plexin-semaphorin-integrin), and four immunoglobulin-like domains (immunoglobulin-like regions in plexins and transcription factors (IPT) 1-4). HGF-β binds to Sema through a low affinity contact. The domain of Met responsible for high affinity binding to HGF-α has not been identified yet. Here we show that this long sought after binding site lies in the immunoglobulin-like region of Met and more precisely in IPT 3 and 4. We also show that IPT 3 and 4 are sufficient to transmit the signal for kinase activation to the cytoplasm, although the lack of Sema makes the receptor equally sensitive to mature HGF and pro-HGF. Finally, we provide evidence that soluble Met-derived proteins containing either the low affinity or high affinity HGF-binding site antagonize HGF-induced invasive growth both in vitro and in xenografts. These data suggest that the immunoglobulin-like region of Met cooperates with the Sema domain in binding to HGF and in controlling Met kinase activity. Although the IPT-HGF-α interaction provides binding strength, the Sema-HGF-β contact confers selective sensitivity to the active form of the ligand.
The Met tyrosine kinase is the product of the c-met proto-oncogene and the high affinity receptor for hepatocyte growth factor (HGF)2 (1, 2). It consists of a 50-kDa α-subunit and a 145-kDa β-subunit, which are linked by a disulfide bond (3, 4). The α-subunit is completely extracellular, whereas the β-subunit includes (from N to C termini) an extracellular region, a transmembrane domain, and a cytoplasmic tyrosine kinase domain. The mature heterodimeric receptor is generated by proteolytic processing and terminal glycosylation from a 170-kDa single-chain precursor (4, 5).
HGF, also known as scatter factor, is a heparin-binding glycoprotein with a broad spectrum of biological activities including cell proliferation, motility, survival, and morphogenesis (6, 7). It is synthesized and secreted as an inactive single chain precursor (pro-HGF) that is stored in the extracellular matrix because of its high affinity for proteoglycans. In the extracellular environment, pro-HGF undergoes proteolytic cleavage at residues Arg494-Val495 to give rise to the biologically active form, a disulfide-linked α/β heterodimer (8, 9). The α-chain consists of an N-terminal domain followed by four kringle domains; the β-chain shares structural homology with the chymotrypsin family of serine proteases but lacks proteolytic activity. In fact, two of the three critical residues that form the catalytic triad typical of serine proteases are not conserved in HGF (10). Despite its inability to signal, pro-HGF binds to Met at high affinity (10) and displaces active HGF (11).
HGF-Met signaling is essential during embryogenesis (12, 13) and tissue regeneration in the adult life (14-17). Importantly, deregulated HGF-Met signaling plays a key role in tumorigenesis and metastasis (6, 18). Inappropriate Met activation by different mechanisms including autocrine HGF stimulation, receptor overexpression, gene amplification, and point mutation is described in a wide variety of human malignancies and correlates with poor prognosis (19). In the last few years, the HGF-Met pathway has been emerging as an appealing target for cancer therapy (20). A variety of Met/HGF inhibitors have been developed, including small molecule compounds targeting Met kinase activity (21-26) or neutralizing anti-Met (27, 28), anti-HGF antibodies (29-31), decoy receptors (32), and HGF-derived factors (33).
Remarkably, despite the great biological and therapeutic importance of this pathway, the mechanism by which HGF activates Met remains poorly understood. Recently, a number of structure-function studies have shed some light onto the interactions between the extracellular portion of Met and HGF. The extracellular region of Met has a modular structure, which encompasses three functional domains. A Sema domain (present also in semaphorins and plexins) spans the first 500 residues at the N terminus of the protein and has a seven-bladed β-propeller structure (34). A PSI domain (also found in plexins, semaphorins, and integrins) covers about 50 residues and contains four conserved disulfide bonds (35). The remaining 400 residues linking the PSI domain to the transmembrane helix are occupied by four IPT domains (36).
HGF is a bivalent ligand containing a high affinity binding site for Met in the α-chain and a low affinity binding site in the β-chain. Cooperation between the α- and the β-chain is required for the biological activity of HGF; whereas the α-chain, and more precisely the N-domain and the first kringle, is sufficient for Met binding, the β-chain is necessary for Met activation (37). Resolution of the crystal structure of the SEMA and PSI domains of Met in complex with the β-chain of HGF revealed that the low affinity binding site for HGF is located in blades 2 and 3 of the β-propeller and that the portion of HGF-β that binds to Met is the same region that serine proteases use to bind their substrates or inhibitors (34). Importantly, determination of HGF β-chain crystal structure at 2.53-Å resolution and specific mutagenesis analysis unveiled that the residues involved in Met binding in the activation pocket of HGF β-chain get exposed only following proteolytic conversion of pro-HGF, thus explaining why pro-HGF binds to Met at high affinity without activating it (38, 39).
Although the low affinity interaction between the β-chain of HGF and the Sema domain of Met is well characterized both structurally and functionally, at the moment it is not clear what region of Met binds to the α-chain of HGF at high affinity. To address this issue, we analyzed the interactions between isolated domains of Met and HGF both in vitro and in living cells. We report that the IPT region of Met, previously thought to merely function as a “stalk” presenting the Sema domain to the ligand (40), actually contains the high affinity binding site for the α-chain of HGF. We also show that engineered proteins derived from the IPT region of Met inhibit HGF-induced invasive growth in vitro and display anti-tumor activity in mice.
EXPERIMENTAL PROCEDURES
Protein Engineering—Soluble or transmembrane receptors and engineered ligands described in this work have been generated by standard PCR and genetic engineering techniques. All of the factors conserve the signal peptide of their parental protein at the N terminus. The amino acid sequences of soluble Met proteins (GenBank™ accession number X54559) correspond to aa 1-24 (signal peptide) plus: Decoy Met, aa 25-932; Sema, aa 25-515; Sema-PSI, aa 25-562; PSI-IPT, aa 516-932; IPT, aa 563-932; IPT Δ1, aa 657-932; IPT Δ1-2, aa 742-932; IPT 3, aa 742-838; and IPT 4, aa 839-932. At the C terminus of each molecule a double FLAG (SDYKDDDDK) or single Myc (EQKLISEEDLN) epitope sequence and a polyhistidine tag (HHHHHHH) were added for protein detection and purification. The engineered, transmembrane MetΔ25-741 is identical to wild-type Met except for the deleted region (aa 25-741). The amino acid sequences of engineered HGF proteins (GenBank™ accession number M73239) correspond to aa 1-31 (signal peptide) plus: HGF, aa 32-728; HGF-α, aa 32-473; HGF NK1, aa 32-205; and HGF-β, aa 495-728. The above Myc or FLAG epitope and polyhistidine tag were added at the C terminus. Uncleavable HGF has been described before (11). NK1-NK1 is a dimeric form of HGF NK1 consisting of the same N-terminal region of HGF repeated in tandem (aa 1-205 directly linked to aa 32-205 without spacer). The amino acid sequence of angiostatin corresponds to aa 1-19 (signal peptide) of human plasminogen (GenBank™ accession number X05199) plus aa 98-459 (kringles 1-4), the above Myc epitope, and a polyhistidine tag. The cDNAs encoding all of the engineered proteins were subcloned into the lentiviral transfer vector pRRLsin.PPT.CMV.eGFP. Wpre (41) in place of the gfp cDNA.
Enzyme-linked Immunosorbant Assays—All of the engineered receptors and factors were collected from the conditioned medium of lentiviral vector-transduced MDA-MB-435 human melanoma cells in the absence of serum. Factor purification was performed by immobilized metal affinity chromatography as previously described (42). Conversion of pro-HGF into active HGF was performed by incubating purified pro-HGF (maximal concentration, 100 ng/μl) with 2-10% FBS (Sigma) at 37 °C for 24 h. Factor conversion was analyzed by Western blotting using anti-HGF antibodies (R & D Systems, Minneapolis, MN). Uncleavable HGF subjected to the same incubation with FBS was used as pro-HGF in all assays that compared active HGF with unprocessed HGF. Binding of engineered ligands to soluble receptors was measured by ELISA using FLAG-tagged soluble receptors in solid phase and Myc-tagged engineered ligands in liquid phase. A fixed concentration of purified soluble receptor (100 ng/well) was adsorbed to 96-well ELISA plates. Protein-coated plates were incubated with increasing concentrations of engineered ligands, and binding was revealed using biotinylated anti-HGF antibodies (R & D) or anti-Myc antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) as indicated. Binding data were analyzed and fit using Prism software (Graph Pad Software, San Diego, CA).
Cell Culture—MDA-MB-435 human melanoma cells were obtained from the Georgetown University Tissue Culture Shared Resource (Washington, DC). The cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS (Sigma). TOV-112D human ovarian carcinoma cells were obtained from ATCC (Manassas, VA) and were cultured using a 1:1 mixture of MCDB 105 medium and medium 199 supplemented with 15% FBS (all from Sigma). A549 human lung carcinoma cells were also obtained from ATCC and maintained in RPMI supplemented with 10% FBS.
Lentiviral Vectors—Vector stocks were produced as previously described (43). Viral p24 antigen concentration was determined by the human immunodeficiency virus, type 1 p24 core profile ELISA kit (PerkinElmer Life Sciences) according to the manufacturer's instructions. The cells were transduced in six-well plates (105 cells/well in 2 ml of medium) using 40 ng/ml of p24 in the presence of 8 μg/ml polybrene (Sigma) as described (43).
Immunoprecipitation and Western Blot Analysis—Cell lysis, immunoprecipitation, and Western blot analysis were performed using extraction buffer as described (44). Signal was detected using ECL system (Amersham Biosciences) according to the manufacturer's instructions. Anti-Met antibodies for immunoprecipitation were obtained as described (45). Anti-Met antibodies for Western blot were purchased from Santa Cruz. Anti-FLAG antibodies were obtained from Sigma. Met phosphorylation analysis in lentiviral vector-transduced MDA-MB-435 cells was performed as previously described (32).
HGF Cross-linking and Met Activation Analysis—Lentiviral vector-transduced TOV-112D cells expressing MetΔ25-741 were subjected to surface biotinylation analysis using an ECL™ surface biotinylation module kit (Amersham Biosciences) according to the manufacturer's instructions. Chemical cross-linking was performed as previously described (11). Briefly, the cells were deprived of serum growth factors for 3 days and then incubated with 1 nm HGF for 3 h. The cell lysates were immunoprecipitated using antibodies directed against the C-terminal portion of Met (45), resolved by SDS-PAGE using a 3-10% polyacrylamide gradient, and analyzed by Western blotting using biotinylated anti-HGF antibodies (R & D). For receptor activation analysis, TOV-112D cells expressing MetΔ25-741 were deprived of serum growth factors for 3 days and then stimulated with 1 nm HGF, uncleavable HGF, HGF NK1, or NK1-NK1 for 10 min. The cells were lysed using extraction buffer as described (44). Cellular proteins were immunoprecipitated with anti-Met antibodies as above and analyzed by Western blotting using anti-phosphotyrosine antibodies (Upstate Biotechnologies, Inc., Lake Placid, New York). The same blots were reprobed with anti-Met antibodies (45).
Biological Assays—Collagen invasion assays using MDA-MB-435 cells were performed using preformed spheroids as described (32). Briefly, the spheroids were generated by incubating cells overnight (700 cells/well) in nonadherent 96-well plates (Greiner, Frickenhausen, Germany) in the presence of 0.24 g/ml methylcellulose (Sigma). The spheroids were embedded into a collagen matrix containing 1.3 mg/ml type I collagen from rat tail (BD Biosciences, Bedford, Massachusetts) and 10% FBS using 96-well plates (40 spheroids/well). Embedded spheroids were cultured at 37 °C for 24 h and then stimulated with 30 ng/ml HGF (R & D) or no factor for additional 24 h. The number of tubules sprouting from each spheroid was scored by microscopy. At least 12 spheroids/experimental point were analyzed.
Tumorigenesis Assays—Lentiviral vector-transduced MDA-MB-435 tumor cells (3 × 106 cells/mouse) in 0.2 ml of Dulbecco's modified Eagle's medium were injected subcutaneously into the right posterior flank of six-week old immunodeficient nu-/- female mice on Swiss CD-1 background (6 mice/group; Charles River Laboratories, Calco, Italy). Tumor size was evaluated every 2 days using a caliper. Tumor volume was calculated using the formula V = 4/3πx2y/2, where x is the minor tumor axis and y is the major tumor axis. A mass of 15 mm3, corresponding approximately to the initial volume occupied by injected cells, was chosen as threshold for tumor positivity. Mice whose tumors were below this threshold were considered tumor-free. After ∼4 weeks, the mice were euthanized, and the tumors were extracted for analysis. The animals were subjected to autopsy. The tumors and lungs were embedded in paraffin and processed for histology. Micrometastasis analysis was performed by microscopy on serial lung sections stained with hematoxylin and eosin. Tumor sections were stained with hematoxylin and eosin and analyzed by an independent pathologist not informed of sample identity. Transgene expression was determined on tumor sections by immunohistochemistry using anti-FLAG antibodies (Sigma). The sections were counterstained with Mayer's hematoxylin (Sigma). Tumor angiogenesis was analyzed by immunohistochemistry using anti-von Willebrand factor antibodies (DAKO, Glostrup, Denmark). The sections were counterstained as above. Vessel density was assessed by microscopy. At least 12 fields/animal were analyzed. All of the animal procedures were approved by the Ethical Commission of the University of Turin (Turin, Italy) and by the Italian Ministry of Health.
Statistical Analysis—Statistical significance was determined using a two-tail homoscedastic Student's t test (array 1, control group; array 2, experimental group). For all of the data analyzed, a significance threshold of p < 0.05 was assumed. In all of the figures, the values are expressed as the means ± standard deviation, and statistical significance is indicated by single (p < 0.05) or double (p < 0.01) asterisks.
RESULTS
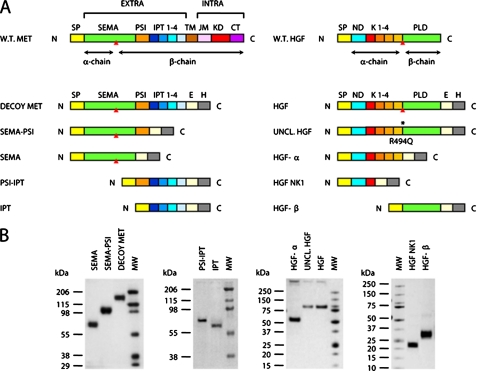
Engineering of HGF/Met Functional Domains—A schematic representation of the functional domains contained in Met and HGF is shown in Fig. 1A. The extracellular portion of Met includes a Sema domain, a PSI hinge, and four IPT modules (left panel). HGF is composed of a α- and a β-chain joined by a disulfide bridge in the mature protein. The α-chain in turn comprises a N-terminal domain and four kringles (right panel). To analyze the interactions between Met and HGF, we expressed all of these functional domains as individual, soluble proteins. Functional domains were engineered to contain the signal peptide of the parental protein at their N terminus, so that they could be properly secreted. At the C terminus, we added an exogenous epitope (FLAG or Myc) for antibody recognition and a polyhistidine tag for protein purification.
FIGURE 1.
Engineering and purification of Met and HGF subdomains. A, schematic representation of the engineered proteins used in this study. Left panel, engineered receptors. W.T. MET, wild-type Met; EXTRA, extracellular portion; INTRA, intracellular portion; SP, signal peptide; SEMA, semaphorin homology domain; PSI, plexin-semaphorin-integrin homology domain; IPT 1-4, immunoglobulin-plexin-transcription factor homology domains 1-4; TM, transmembrane domain; JM, juxta-membrane domain; KD, kinase domain; CT, C-terminal tail; E, FLAG or Myc epitope; H, polyhistidine tag. The red triangle indicates the proteolytic cleavage site between the α- and β-chain. Right panel, engineered ligands. W.T. HGF, wild-type HGF; ND, N-domain; K1-4, kringle 1-4; PLD, protease-like domain; UNCL. HGF, uncleavable HGF. The asterisk indicates the R494Q amino acid substitution in the proteolytic site. B, Coomassie staining of affinity-purified receptors and ligands. Each protein group (Sema, Sema-PSI, Decoy Met; PSI-IPT, IPT; HGF-α, Uncleavable HGF, HGF; HGF NK1, HGF-β) has been resolved by SDS-PAGE in nonreducing conditions. MW, molecular mass marker.
All of the cDNAs encoding the engineered factors were subcloned into the lentiviral vector pRRLsin.PPT.CMV.Wpre (41), and recombinant lentiviral particles were produced as described under “Experimental Procedures.” Recombinant proteins were collected from the conditioned medium of lentiviral vector-transduced MDA-MB-435 human melanoma cells and purified to homogeneity by affinity chromatography. Purified proteins were resolved by SDS-PAGE (Fig. 1B) and quantified against bovine serum albumin standards (not shown).
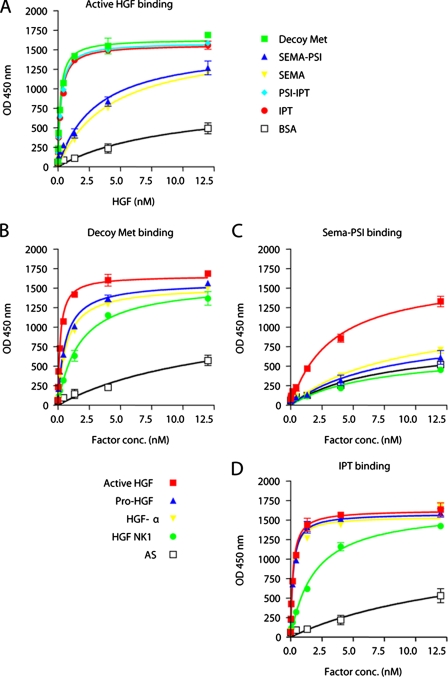
ELISA Analysis of Met-HGF Interactions—The ability of Met ectodomains to interact with HGF was tested in ELISA binding assays. Soluble receptors (Decoy Met, Sema-PSI, Sema, PSI-IPT, IPT) were immobilized in solid phase and exposed to increasing concentrations of active HGF. Binding was revealed using biotinylated anti-HGF antibodies. Nonspecific HGF binding was determined using bovine serum albumin in solid phase instead of soluble Met domains. Binding affinity was determined by nonlinear regression analysis as described under “Experimental Procedures.” In these conditions, decoy Met bound to HGF with a KD value of ∼0.2-0.3 nm. Consistent with previous measurements (40), Sema-PSI and Sema bound to HGF with an affinity at least one log lower compared with decoy Met. Surprisingly, both PSI-IPT and IPT bound to HGF very efficiently, with almost the same affinity as decoy Met (Fig. 2A). The presence or absence of the PSI domain did not affect the binding affinity for HGF of either Sema or IPT. Because almost all of the Sema domains found so far in nature have a PSI module at their C terminus, we therefore continued our binding analysis using decoy Met, Sema-PSI, and IPT.
FIGURE 2.
ELISA analysis of HGF-Met interactions. A, binding of Met subdomains to active HGF. Engineered receptors were immobilized in solid phase and exposed to increasing concentrations of active HGF in liquid phase. Binding was revealed using anti-HGF antibodies. Nonspecific binding was measured by using bovine serum albumin instead of purified receptors in solid phase. B-D, binding of Decoy Met, Sema-PSI, and IPT to different forms of HGF. Engineered receptors were immobilized in solid phase and exposed to increasing concentrations of Myc-tagged active HGF, pro-HGF, HGF-α, or HGF NK1 in liquid phase. Binding was revealed using anti-Myc antibodies. Nonspecific binding was measured by using Myc-tagged angiostatin (AS) in liquid phase.
To determine the affinity of each Met module for pro-HGF, HGF-α, HGF NK1, and HGF-β and to compare it with that for active HGF, engineered receptors were immobilized in solid phase and exposed to increasing concentrations of Myc-tagged ligands. Binding was revealed using anti-Myc antibodies. Nonspecific binding was determined using the kringle-containing protein angiostatin, which is also tagged with a Myc epitope, in liquid phase. Pro-HGF, HGF α-chain, and HGF NK1, which represents the minimal Met-binding module of HGF α-chain, bound to Decoy Met with a 3-, 4-, and 10-fold reduced affinity compared with active HGF, respectively (Fig. 2B). Binding of HGF-β to decoy Met (or to any other Met domain) was too low to be detected in this kind of assay (not shown). Sema-PSI bound at a significant affinity to active HGF only, whereas binding to pro-HGF, HGF-α, or HGF NK1 was indistinguishable from nonspecific binding (Fig. 2C). In contrast, IPT bound to active HGF, pro-HGF, and HGF-α with the same high affinity (Fig. 2D). HGF NK1 bound to IPT 10 times less tightly than active HGF, i.e. with the same affinity as it bound to Decoy Met. These data suggest that the IPT region of Met binds to the α-chain of HGF at high affinity independently of proteolytic processing of the ligand.
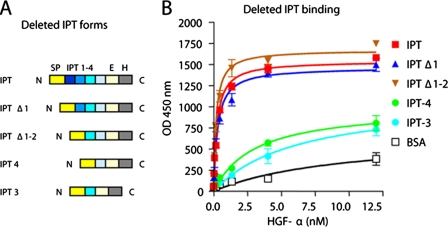
The α-Chain of HGF Binds to IPT Domains 3 and 4 with High Affinity—The IPT region of Met extends for about 400 amino acids and contains four IPT domains. To more finely map the IPT-HGF interface, we engineered a series of IPT variants that were deleted in one or more domains (Fig. 3A). IPT Δ1 and IPT Δ1-2 are two N-terminal deleted forms of IPT lacking the first or the first two immunoglobulin-like domains, respectively. IPT 3 and IPT 4 correspond to the two C-terminal immunoglobulin-like domains expressed as single proteins. Protein production and purification were performed as described above. The ability of the engineered IPTs to interact with HGF α-chain was investigated in ELISA binding assays using the whole IPT region as a control. IPT, IPT Δ1, IPT Δ1-2, IPT 3, and IPT 4 were immobilized in solid phase and exposed to increasing concentrations of HGF-α. Binding was revealed using anti-HGF antibodies. Nonspecific binding was measured using bovine serum albumin as above. As shown in Fig. 3B, deletion of the first two immunoglobulin-like domains did not substantially affect HGF binding. In fact, IPT Δ1-2, a protein corresponding to the last two immunoglobulin-like domains of Met, bound to the α-chain of HGF with equal if not higher strength than IPT. However, further deletion of either the third or fourth immunoglobulin-like domain did almost completely impair HGF-α binding. Similar results were obtained using active HGF or pro-HGF instead of HGF-α (not shown). These data suggest that the last two immunoglobulin-like domains of Met, which lie close to the transmembrane helix in the context of a bona fide Met, are sufficient for binding the α-chain of HGF at high affinity.
FIGURE 3.
IPT domains 3 and 4 are sufficient to binding to HGF-α at high affinity. A, schematic representation of deleted IPT variants. Color code and legend as in Fig. 1A. B, ELISA analysis of interactions between IPT variants and HGF-α. Engineered IPTs were immobilized in solid phase and exposed to increasing concentrations of HGF-α in liquid phase. Binding was revealed using biotinylated anti-HGF antibodies.
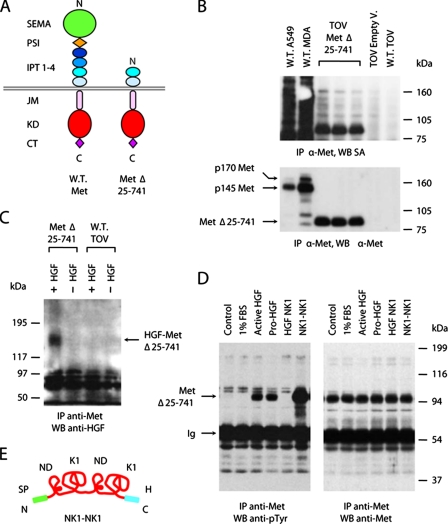
IPT Domains 3 and 4 Are Sufficient for Binding to HGF in Living Cells—To determine whether HGF could bind to IPT 3 and 4 in the context of a membrane-anchored receptor, we engineered a Met protein carrying a large deletion in the extracellular region. We deleted amino acids 25-741, corresponding to the Sema domain (aa 25-515), the PSI domain (aa 516-562), and the first two IPT domains (IPT 1 and 2, aa 563-741), generating a recombinant receptor containing IPT domains 3 and 4, the transmembrane helix, and the full cytoplasmic region (Fig. 4A). The cDNA encoding the engineered receptor MetΔ25-741 was subcloned into the same lentiviral vector described above (41). Recombinant lentiviral particles were used to transduce the human ovary carcinoma cell line TOV-112D, which lacks endogenous Met expression as determined by reverse transcription-PCR analysis (32). Surface biotinylation analysis revealed that MetΔ25-741 was properly expressed and exposed on the membrane of TOV-112D cells (Fig. 4B).
FIGURE 4.
IPT domains 3 and 4 are sufficient for binding to HGF in living cells. A, schematic representation of the deleted MetΔ25-741 receptor. The color code and legend are as in Fig. 1A. B, surface biotinylation analysis. Cellular proteins were immunoprecipitated (IP) using antibodies directed against the C-terminal portion of Met and analyzed by Western blotting (WB) using horseradish peroxidase-conjugated streptavidin (SA). The same blots were reprobed with anti-Met antibodies. W.T., wild type; A549, A549 human lung carcinoma cells; MDA, MDA-MB-435 human melanoma cells; TOV, TOV-112D human ovary carcinoma cells; Empty V., empty vector. The p170 band corresponds to unprocessed Met; p145 is the mature form of the receptor. C, chemical cross-linking analysis. TOV-112D cells expressing MetΔ25-741 (Met Δ25-741) and wild-type TOV-112D cells (W.T. TOV) were incubated with HGF and then subjected to chemical cross-linking. The cell lysates were immunoprecipitated using anti-Met antibodies and analyzed by Western blotting using anti-HGF antibodies. The arrow indicates HGF-MetΔ25-741 complexes. D, Met phosphorylation analysis. TOV-112D cells expressing MetΔ25-741 were stimulated with 1% FBS as a negative control and with equal amounts of HGF, pro-HGF, HGF NK1, or NK1-NK1. Receptor phosphorylation was determined by immunoprecipitation with anti-Met antibodies and Western blotting with anti-phosphotyrosine (anti-pTyr) antibodies. The same blots were reprobed using anti-Met antibodies. The arrows indicate bands corresponding to MetΔ25-741 or immunoglobulins (Ig). E, schematic representation of NK1-NK1. From N to C termini: SP, signal peptide; ND, N-domain; K1, kringle 1; H, polyhistidine tag.
To examine whether MetΔ25-741 could bind to HGF, we incubated lentiviral vector-transduced cells in the presence or absence of recombinant HGF and subsequently treated them with the cross-linking agent BS3. Cell lysates were immunoprecipitated with an antibody raised against the C-terminal portion of Met, resolved by SDS-PAGE, and analyzed by Western blotting using anti-HGF biotinylated antibodies. As a control, the same analysis was performed on wild-type TOV-112D cells. Immunoblots showed a distinct band with a molecular mass of ∼180 kDa in the lane corresponding to cells expressing MetΔ25-741 treated with HGF but not in lanes corresponding to the same cells without HGF or to wild-type TOV-112D cells, either in the presence or absence of the ligand (Fig. 4C). Considering that both MetΔ25-741 and HGF have a molecular mass of ∼90 kDa, the immunoprecipitated cross-linked protein is compatible with a 1:1 complex formed by HGF plus MetΔ25-741.
We next tested whether HGF binding to MetΔ25-741 could induce Met kinase activation. To this end, we stimulated lentiviral vector-transduced TOV-112D cells with pro-HGF or active HGF, and cell lysates were immunoprecipitated with anti-Met antibodies as above. Receptor activation was determined by Western blot analysis using anti-phosphotyrosine antibodies. The same blots were reprobed with anti-Met antibodies to normalize the amount of receptor immunoprecipitated. Remarkably, both pro-HGF and active HGF were capable of inducing robust phosphorylation of MetΔ25-741 (Fig. 4D). This may be due to the ability of pro-HGF and HGF to form homodimers.3 In any case, because pro-HGF binding to full size Met does not induce kinase activation (10, 11), this suggests that the Sema domain somehow exerts an auto-inhibitory effect on Met catalytic activity that is released upon binding to active HGF. Receptor stimulation was also performed using HGF NK1 and an engineered dimeric ligand consisting of two NK1 fragments repeated in tandem (NK1-NK1; Fig. 4E). As shown in Fig. 4D, NK1-NK1 stimulation of lentiviral vector-transduced TOV-112D cells resulted in potent phosphorylation of MetΔ25-741, whereas stimulation with monomeric NK1 had no effect. These results suggest that the two C-terminal IPT domains of Met (IPT 3 and 4) are sufficient to bind to HGF (and more precisely to HGF NK1 that represents the minimal Met-binding module in the α-chain of HGF) and to transmit the signal for receptor activation to the cytoplasmic kinase domain, presumably following ligand-induced receptor dimerization. However, they also suggest that IPT 3 and 4 alone are not sufficient for distinguishing the biologically active form of HGF from its inactive precursor, pro-HGF.
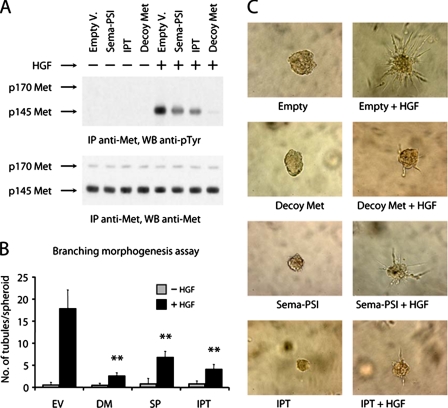
Soluble IPT Inhibits HGF-induced Invasive Growth in Vitro—In a previous study, we demonstrated that the extracellular portion of Met expressed as a soluble protein (Decoy Met) inhibits HGF-induced invasive growth both in vitro and in mouse models of cancer (32). Recombinant soluble Sema-PSI was also shown to inhibit both ligand-dependent and -independent Met phosphorylation (46). Based on these results, we tested whether soluble IPT displayed HGF/Met antagonistic activity in living cells. MDA-MB-435 human melanoma cells, which express Met and are an established model system for analysis of HGF-mediated invasive growth (32), were transduced with lentiviral vectors encoding soluble Decoy Met, Sema-PSI, or IPT. Cells transduced with an empty vector were used as control. Lentiviral vector-transduced cells secreting comparable levels of soluble factors (∼50 pmol/106 cells/24 h) were serum-starved for several days, allowing the recombinant factors to accumulate in the medium, and then stimulated with recombinant HGF. Met tyrosine phosphorylation was determined by immunoblotting with anti-phosphotyrosine antibodies as described above. As shown in Fig. 5A, both IPT and Sema-PSI partially inhibited HGF-induced Met phosphorylation, whereas Decoy Met completely neutralized the ability of HGF to induce Met activation. Reprobing of the same immunoblots with antibodies directed against the C-terminal tail of Met revealed no substantial differences in the amounts of immunoprecipitated protein.
FIGURE 5.
Soluble IPT inhibits HGF-induced invasive growth in vitro. A, lentiviral vector-transduced MDA-MB-435 cells were stimulated with recombinant HGF, and Met phosphorylation was determined by immunoblotting using anti-phosphotyrosine antibodies (upper panel). The same blot was reprobed using anti-Met antibodies (lower panel). Empty V., empty vector. B, branching morphogenesis assay. Preformed spheroids of lentiviral vector-transduced MDA-MB-435 cells were embedded in collagen and then stimulated with recombinant HGF to form branched tubules. Collagen invasion was quantified by scoring the mean number of tubules sprouting from each spheroid. EV, empty vector; DM, Decoy Met; SP, Sema-PSI. C, representative images from the experiment described in B. Magnification, 200×. IP, immunoprecipitation; WB, Western blotting.
To test the inhibitory potential of Met ectodomains in a more biological setting, we employed the same cells to perform a HGF-dependent branching morphogenesis assay. Preformed cell spheroids were seeded in a three-dimensional collagen matrix and then stimulated with recombinant HGF to form tubular structures. Branching was quantified by scoring the mean number of tubules sprouting from each colony. As shown in Fig. 5B, both soluble IPT and Sema-PSI inhibited HGF-induced colony branching (empty vector, 17.5 tubules/colony; IPT, 4.0 tubules/colony; Sema-PSI, 6.7 tubules/colony). However, consistent with the results obtained in phosphorylation experiments, decoy Met was a more potent HGF inhibitor than either of its subdomains (2.5 tubules/colony). Representative images of colony morphology are shown in Fig. 5C.
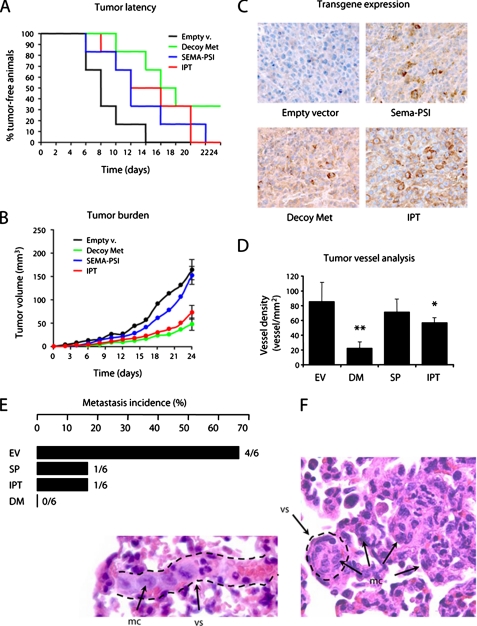
Soluble IPT Displays Anti-tumor and Anti-metastatic Activity in Mice—The above results prompted us to explore the therapeutic potential of soluble IPT in mouse models of cancer. Lentiviral vector-transduced MDA-MB-435 melanoma cells were injected subcutaneously into CD-1 nu-/- mice (six mice/group), and tumor growth was monitored over time. After approximately 3 weeks, the tumors were extracted for analysis, and the mice were subjected to autopsy. In a Kaplan-Meier-like analysis, where the percentage of tumor-free animals is plotted against time, and tumor latency is quantified calculating the median in days, all of the engineered soluble receptors delayed the appearance of experimental tumors. However, IPT was slightly more effective than Sema-PSI, and decoy Met was more potent than either IPT or Sema-PSI (Fig. 6A). Analysis of tumor burden over time revealed that IPT was only slightly less effective than decoy Met, whereas Sema-PSI inhibited neoplastic growth only during the very early stages of the experiment (Fig. 6B). Immunohistochemical analysis of transgene expression showed that Decoy Met, Sema-PSI, and IPT reached similar levels and distribution in tumors (Fig. 6C).
FIGURE 6.
Soluble IPT displays anti-tumor and anti-metastatic activity in mice. CD-1 nu-/- mice were injected subcutaneously with lentiviral vector-transduced MDA-MB-435 cells, and tumor growth was monitored over time. A, Kaplan-Meier-like plots of tumor latency (x axis, time in days; y axis, percent of tumor free-animals). Empty v., empty vector. B, mean tumor volume over time. C, immunohistochemical analysis of tumor sections using anti-FLAG antibodies. Magnification, 400×. D, tumor vessel analysis. Tumor sections were stained with anti-von Willebrand factor antibodies. The number of vessels per square mm of tumor section was determined by microscopy. EV, empty vector; DM, Decoy Met; SP, Sema-PSI. E, metastasis incidence analysis. Upon autopsy, serial lung sections were analyzed by microscopy to determine the presence of micrometastases. Metastasis incidence, i.e. the number of mice with metastasis over the total, is indicated in both percentage (bars) and fraction (at the ends of the bars). F, representative images of micrometastases from the empty vector group. Lung sections were stained with hematoxylin and eosin. The dotted lines identify the walls of blood vessels (vs). Metastatic cells (mc) can be found inside vessels as an embolus or in the parenchyma. Magnification, 400×.
Because HGF is a potent pro-angiogenic factor (47), we determined whether inhibition of HGF/Met in tumors resulted in impairment of angiogenesis. Tumor sections were analyzed by immunohistochemistry using antibodies against von Willebrand factor, and vessel density was assessed by microscopy (Fig. 6D). IPT decreased tumor vessel density by 1.5 times, whereas decoy Met achieved a much stronger inhibition (approximately four times); Sema-PSI did not significantly affect tumor angiogenesis.
Upon autopsy, lungs from the mice described above were extracted and processed for histology. Serial lung sections were stained with hematoxylin and eosin and analyzed by microscopy to determine the presence of micrometastases. The results are shown in Fig. 6E. In the control group, four of six mice (67%) were bearing micrometastases. In the IPT and Sema-PSI group, micrometastases could be found in only one of six mice (17%), whereas no metastasis could be found in the decoy Met group. Metastatic lesions were both parenchymal (extravascular) and embolic (intravascular; see Fig. 6F for representative images).
In conclusion, both soluble Sema-PSI and IPT, capable of binding to HGF autonomously, interfere with tumor growth and invasion in animal models. However, the full extracellular portion of Met achieves a more potent inhibition, suggesting that the two HGF-binding sites contained in Sema and IPT cooperate in neutralizing the ligand.
DISCUSSION
The data presented in this study suggest that the α-chain of HGF binds to the IPT region of Met at high affinity and that it does so independently of proteolytic processing of the ligand. They also suggest that HGF binding to IPT in the context of a transmembrane Met lacking the Sema domain is sufficient for transmitting the signal for receptor activation to the cytoplasmic kinase domain, although without distinction between the inactive and active form of the ligand. Finally, they provide evidence that engineered proteins derived from the IPT region and Sema domain of Met are capable of neutralizing the pro-invasive activity of HGF both in vitro and in vivo.
It has been known for long time that HGF is a bivalent factor. Early protein engineering studies identified a high affinity Met-binding site in the N-domain and first kringle (10). Subsequently, combined biochemical and biological analysis demonstrated that the serine protease-like domain (β-chain), although not necessary for binding, plays a key role in mediating receptor activation (37). More recently, detailed crystallographic and mutagenesis data have thoroughly characterized both structurally and functionally the low affinity Met-binding site on the β-chain of HGF and its interaction with the Sema domain of Met (34, 38, 39). The interface between the α-chain of HGF and Met remains, however, elusive. Small angle x-ray scattering and cryo-electron microscopy studies suggested the presence of contacts among the N-terminal and first kringle domain of HGF and the Sema domain of Met (48). However, plasmon resonance analysis revealed that this interaction has a very low affinity (about two times lower than that of HGF-β for Sema and 100 times lower than that of HGF-α for the intact receptor) (49). Because this weak interaction cannot account per se for the tight bond between HGF and Met, the high affinity HGF-binding site on Met has still to be identified.
The results presented here contribute to fill this gap and suggest that this long sought after HGF-binding site lies in the IPT region of Met and more precisely in the last two immunoglobulin-like domains close to the cell membrane. Several distinct experimental results suggest that this is the case. First, a soluble, deleted Met receptor containing nothing but the four IPT domains binds to HGF with substantially the same affinity as the entire extracellular portion of Met. Conversely, Sema displays very low affinity toward HGF. Second, IPT binds to active HGF, pro-HGF, or HGF-α with unchanged strength. Third, deletion of IPT 1 and IPT 2 does not affect the affinity of IPT for any form of HGF. Fourth, an engineered Met receptor carrying a large deletion in its ectodomain corresponding to the Sema domain, the PSI module, and the first two immunoglobulin-like domains (MetΔ25-741) retains the ability to bind to HGF and to transduce the signal for kinase activation to the inside of the cell, although it cannot distinguish between active HGF and Pro-HGF. Finally, a dimeric form of HGF NK1, which is known to contain the minimal Met-binding domain of HGF-α, is capable of eliciting activation of MetΔ25-741 as efficiently as if not more powerfully than HGF, thus identifying in IPT 3-4 the HGF NK1-binding site.
Although these data point at a key role for IPT in HGF binding, it is noteworthy that two previous structure/function studies on the extracellular portion of Met failed to identify any ligand-binding site in this region. A first draft of the Met ectodomain map suggested that the Sema domain is necessary and sufficient for HGF binding based on ELISAs (40). However, this analysis was conducted under different conditions. First, these experiments employed fusion proteins between Met subdomains and the Fc portion of a human immunoglobulin, thus forcing receptor modules to be dimeric. Second, the assays were performed using the ligand in solid phase and the engineered receptors in liquid phase, somehow reversing the physiological scenario in which Met is anchored to the membrane and HGF is in solution. Furthermore, the affinity of HGF for the Sema-Fc or Sema-PSI-Fc chimeras was dramatically reduced compared with that of the control chimera containing the entire Met ectodomain, thus implying that the remaining portion of Met (i.e. the IPT region) also contained a HGF binding activity. A second study analyzed the role of the Sema domain in receptor dimerization and suggested that an engineered form of the Met extracellular portion containing a deletion in the Sema domain was not capable of co-precipitating HGF (46). However, HGF-Met interactions were not measured in living cells but in crude cell lysates in the presence of detergent, and no chemical cross-linking was performed. It is quite possible that the different experimental conditions may have affected the sensitivity of the assays, thus leading to underestimation of the IPT function.
In contrast, a third and more recent independent study on the extracellular portion of Met provides support for the involvement of the IPT region in HGF binding. It is well known that Met is also a receptor for Internalin B (InlB), a surface protein of the human bacterial pathogen Listeria monocytogenes (50). ELISA analysis unveiled that the Sema domain of Met is dispensable for high affinity InlB binding. Crystallographic data confirmed that InlB binds tightly to the IPT region of the receptor, whereas it forms a secondary contact with a portion of the Sema domain not involved in HGF binding. In displacement experiments, the authors showed that InlB and HGF partially compete for binding to the Met ectodomain, thus suggesting that Sema does not represent the sole HGF-binding module on the Met receptor (51).
The bivalent nature of both HGF and Met poses a fundamental biological question. Why did evolution select for this unusual, complex mechanism of ligand-receptor interaction? Why does Met need two distinct HGF-binding sites, each with a different affinity? Comprehensive analysis of our results together with the literature leads to a putative functional model.
Because of its high affinity for glycosamino-glycans, HGF diffuses through the extracellular matrix of tissues at a very low rate. Because HGF is provided paracrinally by the stroma, it would not be feasible to control it at a transcriptional level. In case of organ injury, HGF must be ready on site to provide its survival and trophic signals for tissue regeneration. To comply with this requirement, HGF is secreted as an inactive precursor (pro-HGF) that accumulates in the extracellular matrix of tissues. Pro-HGF binds at high affinity to Met via the IPT site. This interaction does not lead to receptor activation in the context of a full size Met but prepares the cell to respond readily to environmental challenges. Upon a variety of insults including wounds, chemical agents, radiation, oxidative stress, or hypoxia, pro-HGF convertase activity is induced locally and promptly converts pro-HGF into active HGF. This catalytic reaction can occur when pro-HGF is receptor-bound as demonstrated in the case of urokinase-type plasminogen activator (52). Following proteolytic processing, the second, low affinity Met-binding site on the β-chain of HGF becomes exposed, and binding to the Sema domain takes place. This interaction results in kinase activation and in launch of the invasive growth program that promotes cell survival, proliferation, motility, and invasion. If only the IPT site were present, both HGF and pro-HGF would elicit receptor activation, thus losing a fundamental degree of regulation. Conversely, if only the Sema domain were present, the strength of interaction between Met and its ligand would be too weak to compete with high capacity, nonspecific interfaces offered by many other components of the cell membrane including proteoglycans.
Perhaps not accidentally, the model proposed above shares striking similarities with the L. monocytogenes system (50, 51). Met-mediated bacterial invasion of the host cell occurs in three consecutive steps. First, InlB binds at high affinity to the IPT region of Met. Second, a different portion of InlB binds to the Sema domain at low affinity. This results in receptor activation. Third, heparin-dependent receptor oligomerization amplifies Met signaling and leads to internalization through a yet uncharacterized mechanism. Although Met activation by InlB and Met activation by HGF have different biological goals (bacterial uptake for the former and activation of invasive growth for the latter), the overall strategy is the same. The IPT region is used as a high affinity anchoring site, and a low affinity interaction with the Sema domain provides the signal for receptor dimerization and activation. The fact that two unrelated systems utilizing different ligands employ analogous mechanisms to achieve tyrosine kinase activation supports a critical role of both IPT and Sema in the regulation and control of Met activity.
Consistent with this notion, cooperation between Sema and IPT is observed also when the extracellular portion of Met is used as a biotechnological tool to inhibit HGF-induced invasive growth. In our in vitro analysis and in mouse xenografts, both the IPT and Sema-PSI soluble proteins displayed a significant inhibitory effect. However, none of them could achieve the powerful inhibition displayed by the full Met ectodomain, which contains both the low affinity and high affinity HGF-binding site. This implies that both of these interactions contribute to controlling Met activity. Although the HGF-β-Sema contact had already been identified as a target for therapy (38, 46), the results presented here unveil a second interface that can potentially offer opportunities for pharmacological intervention. Recombinant proteins or antibodies that bind to the IPT region in place of bona fide HGF have a potential application as competitive inhibitors. Paradoxically, one of these proteins may well be HGF NK4, a recombinant HGF inhibitor substantially corresponding to the α-chain of HGF that has been used for some years with success (33) without knowing where on Met it bound. Because we now know that HGF-α binds with high affinity to IPT and not to Sema, the fact that HGF NK4 is an effective HGF antagonist provides support to the idea that IPT is a valid target for therapy. Further development of neutralizing antibodies against this region of Met will substantiate this hypothesis.
Acknowledgments
We thank Mauro Risio for histological analysis and Luigi Naldini for lentiviral technology expertise.
Author's Choice—Final version full access.
This work has been supported in part by the Italian Association for Cancer Research, the Compagnia San Paolo di Torino Foundation, the Cassa di Risparmio di Torino Foundation, the Italian Ministry of Health (Ricerca Finalizzata 2004), and the Italian Ministry of University and Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HGF, hepatocyte growth factor; PSI, plexin-semaphorin-integrin; IPT, immunoglobulin-like regions in plexins and transcription factors; aa, amino acid(s); ELISA, enzyme-linked immunosorbant assay; FBS, fetal bovine serum; InlB, internalin B; Sema, semaphorin.
C. Basilico, A. Arnesano, M. Galluzzo, P. M. Comoglio, and P. Michieli, unpublished results.
References
- 1.Bottaro, D. P., Rubin, J. S., Faletto, D. L., Chan, A. M., Kmiecik, T. E., Vande Woude, G. F., and Aaronson, S. A. (1991) Science 251 802-804 [DOI] [PubMed] [Google Scholar]
- 2.Naldini, L., Vigna, E., Narsimhan, R. P., Gaudino, G., Zarnegar, R., Michalopoulos, G. K., and Comoglio, P. M. (1991) Oncogene 6 501-504 [PubMed] [Google Scholar]
- 3.Tempest, P. R., Stratton, M. R., and Cooper, C. S. (1988) Br. J. Cancer 58 3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano, S., Ponzetto, C., Di Renzo, M. F., Cooper, C. S., and Comoglio, P. M. (1989) Nature 339 155-156 [DOI] [PubMed] [Google Scholar]
- 5.Komada, M., Hatsuzawa, K., Shibamoto, S., Ito, F., Nakayama, K., and Kitamura, N. (1993) FEBS Lett. 328 25-29 [DOI] [PubMed] [Google Scholar]
- 6.Trusolino, L., and Comoglio, P. M. (2002) Nat. Rev. Cancer 2 289-300 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura, T. (1994) Princess Takamatsu. Symp. 24 195-213 [PubMed] [Google Scholar]
- 8.Nakamura, T., Nishizawa, T., Hagiya, M., Seki, T., Shimonishi, M., Sugimura, A., Tashiro, K., and Shimizu, S. (1989) Nature 342 440-443 [DOI] [PubMed] [Google Scholar]
- 9.Hartmann, G., Naldini, L., Weidner, K. M., Sachs, M., Vigna, E., Comoglio, P. M., and Birchmeier, W. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11574-11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lokker, N. A., Mark, M. R., Luis, E. A., Bennett, G. L., Robbins, K. A., Baker, J. B., and Godowski, P. J. (1992) EMBO J. 11 2503-2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzone, M., Basilico, C., Cavassa, S., Pennacchietti, S., Risio, M., Naldini, L., Comoglio, P. M., and Michieli, P. (2004) J. Clin. Investig. 114 1418-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt, C., Bladt, F., Goedecke, S., Brinkmann, V., Zschiesche, W., Sharpe, M., Gherardi, E., and Birchmeier, C. (1995) Nature 373 699-702 [DOI] [PubMed] [Google Scholar]
- 13.Bladt, F., Riethmacher, D., Isenmann, S., Aguzzi, A., and Birchmeier, C. (1995) Nature 376 768-771 [DOI] [PubMed] [Google Scholar]
- 14.Borowiak, M., Garratt, A. N., Wüstefeld, T., Strehle, M., Trautwein, C., and Birchmeier, C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10608-10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh, C. G., Factor, V. M., Sánchez, A., Uchida, K., Conner, E. A., and Thorgeirsson, S. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4477-4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto, K., and Nakamura, T. (2001) Kidney Int. 59 2023-2038 [DOI] [PubMed] [Google Scholar]
- 17.Ohmichi, H., Matsumoto, K., and Nakamura, T. (1996) Am. J. Physiol. 270 L1031-L1039 [DOI] [PubMed] [Google Scholar]
- 18.Birchmeier, C., Birchmeier, W., Gherardi, E., and Vande Woude, G. F. (2003) Nat. Rev. Mol. Cell Biol. 4 915-925 [DOI] [PubMed] [Google Scholar]
- 19.Knudsen, B. S., and Vande Woude, G. (2008) Curr. Opin. Genet. Dev. 18 87-96 [DOI] [PubMed] [Google Scholar]
- 20.Christensen, J. G., Burrows, J., and Salgia, R. (2005) Cancer Lett. 225 1-26 [DOI] [PubMed] [Google Scholar]
- 21.Puri, N., Khramtsov, A., Ahmed, S., Nallasura, V., Hetzel, J. T., Jagadeeswaran, R., Karczmar, G., and Salgia, R. (2007) Cancer Res. 67 3529-3534 [DOI] [PubMed] [Google Scholar]
- 22.Zou, H. Y., Li, Q., Lee, J. H., Arango, M. E., McDonnell, S. R., Yamazaki, S., Koudriakova, T. B., Alton, G., Cui, J. J., Kung, P. P., Nambu, M. D., Los, G., Bender, S. L., Mroczkowski, B., and Christensen, J. G. (2007) Cancer Res. 67 4408-4417 [DOI] [PubMed] [Google Scholar]
- 23.Ma, P. C., Jagadeeswaran, R., Jagadeesh, S., Tretiakova, M. S., Nallasura, V., Fox, E. A., Hansen, M., Schaefer, E., Naoki, K., Lader, A., Richards, W., Sugarbaker, D., Husain, A. N., Christensen, J. G., and Salgia, R. (2005) Cancer Res. 65 1479-1488 [DOI] [PubMed] [Google Scholar]
- 24.Berthou, S., Aebersold, D. M., Schmidt, L. S., Stroka, D., Heigl, C., Streit, B., Stalder, D., Gruber, G., Liang, C., Howlett, A. R., Candinas, D., Greiner, R. H., Lipson, K. E., and Zimmer, Y. (2004) Oncogene. 23 5387-5393 [DOI] [PubMed] [Google Scholar]
- 25.Christensen, J. G., Schreck, R., Burrows, J., Kuruganti, P., Chan, E., Le, P., Chen, J., Wang, X., Ruslim, L., Blake, R., Lipson, K. E., Ramphal, J., Do, S., Cui, J. J., Cherrington, J. M., and Mendel, D. B. (2003) Cancer Res. 63 7345-7355 [PubMed] [Google Scholar]
- 26.Sattler, M., Pride, Y. B., Ma, P., Gramlich, J. L., Chu, S. C., Quinnan, L. A., Shirazian, S., Liang, C., Podar, K., Christensen, J. G., and Salgia, R. (2003) Cancer Res. 63 5462-5469 [PubMed] [Google Scholar]
- 27.Martens, T., Schmidt, N. O., Eckerich, C., Fillbrandt, R., Merchant, M., Schwall, R., Westphal, M., and Lamszus, K. (2006) Clin. Cancer Res. 12 6144-6152 [DOI] [PubMed] [Google Scholar]
- 28.Petrelli, A., Circosta, P., Granziero, L., Mazzone, M., Pisacane, A., Fenoglio, S., Comoglio, P. M., and Giordano, S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5090-5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao, B., Su, Y., Oskarsson, M., Zhao, P., Kort, E. J., Fisher, R. J., Wang, L. M., and Vande Woude, G. F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7443-7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess, T., Coxon, A., Meyer, S., Sun, J., Rex, K., Tsuruda, T., Chen, Q., Ho, S. Y., Li, L., Kaufman, S., McDorman, K., Cattley, R. C., Sun, J., Elliott, G., Zhang, K., Feng, X., Jia, X. C., Green, L., Radinsky, R., and Kendall, R. (2006) Cancer Res. 66 1721-1729 [DOI] [PubMed] [Google Scholar]
- 31.Kim, K. J., Wang, L., Su, Y. C., Gillespie, G. Y., Salhotra, A., Lal, B., and Laterra, J. (2006) Clin. Cancer Res. 12 1292-1298 [DOI] [PubMed] [Google Scholar]
- 32.Michieli, P., Mazzone, M., Basilico, C., Cavassa, S., Sottile, A., Naldini, L., and Comoglio, P. M. (2004) Cancer Cell 6 61-73 [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto, K., and Nakamura, T. (2003) Cancer Sci. 94 321-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamos, J., Lazarus, R. A., Yao, X., Kirchhofer, D., and Wiesmann, C. (2004) EMBO J. 23 2325-2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlov, G., Perreault, A., Schrag, J. D., Park, M., Cygler, M., Gehring, K., and Ekiel, I. (2004) Biochem. Biophys. Res. Commun. 321 234-240 [DOI] [PubMed] [Google Scholar]
- 36.Bork, P., Doerks, T., Springer, T. A., and Snel, B. (1999) Trends Biochem. Sci. 24 261-263 [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto, K., Kataoka, H., Date, K., and Nakamura, T. (1998) J. Biol. Chem. 273 22913-22920 [DOI] [PubMed] [Google Scholar]
- 38.Kirchhofer, D., Yao, X., Peek, M., Eigenbrot, C., Lipari, M. T., Billeci, K. L., Maun, H. R., Moran, P., Santell, L., Wiesmann, C., and Lazarus, R. A. (2004) J. Biol. Chem. 279 39915-39924 [DOI] [PubMed] [Google Scholar]
- 39.Kirchhofer, D., Lipari, M. T., Santell, L., Billeci, K. L., Maun, H. R., Sandoval, W. N., Moran, P., Ridgway, J., Eigenbrot, C., and Lazarus, R. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5306-5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gherardi, E., Youles, M. E., Miguel, R. N., Blundell, T. L., Iamele, L., Gough, J., Bandyopadhyay, A., Hartmann, G., and Butler, P. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12039-12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Follenzi, A., Ailles, L. E., Bakovic, S., Geuna, M., and Naldini, L. (2000) Nat. Genet. 25 217-222 [DOI] [PubMed] [Google Scholar]
- 42.Michieli, P., Cavassa, S., Basilico, C., De Luca, A., Mazzone, M., Asti, C., Chiusaroli, R., Guglielmi, M., Bossù, P., Colotta, F., Caselli, G., and Comoglio, P. M. (2002) Nat. Biotechnol. 20 488-495 [DOI] [PubMed] [Google Scholar]
- 43.Vigna, E., and Naldini, L. (2000) J. Gene Med. 2 308-316 [DOI] [PubMed] [Google Scholar]
- 44.Longati, P., Bardelli, A., Ponzetto, C., Naldini, L., and Comoglio, P. M. (1994) Oncogene. 9 49-57 [PubMed] [Google Scholar]
- 45.Ruco, L. P., Ranalli, T., Marzullo, A., Bianco, P., Prat, M., Comoglio, P. M., and Baroni, C. D. (1996) J. Pathol. 180 266-270 [DOI] [PubMed] [Google Scholar]
- 46.Kong-Beltran, M., Stamos, J., and Wickramasinghe, D. (2004) Cancer Cell 6 75-84 [DOI] [PubMed] [Google Scholar]
- 47.Bussolino, F., Di Renzo, M. F., Ziche, M., Bocchietto, E., Olivero, M., Naldini, L., Gaudino, G., Tamagnone, L., Coffer, A., and Comoglio, P. M. (1992) J. Cell Biol. 119 629-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gherardi, E., Sandin, S., Petoukhov, M. V., Finch, J., Youles, M. E., Ofverstedt, L. G., Miguel, R. N., Blundell, T. L., Vande Woude, G. F., Skoglund, U., and Svergun, D. I. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 4046-4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes, O., Pillozzi, S., Deakin, J. A., Carafoli, F., Kemp, L., Butler, P. J., Lyon, M., and Gherardi, E. (2007) J. Mol. Biol. 367 395-408 [DOI] [PubMed] [Google Scholar]
- 50.Shen, Y., Naujokas, M., Park, M., and Ireton, K. (2000) Cell 103 501-510 [DOI] [PubMed] [Google Scholar]
- 51.Niemann, H. H., Jäger, V., Butler, P. J., van den Heuvel, J., Schmidt, S., Ferraris, D., Gherardi, E., and Heinz, D. W. (2007) Cell 130 235-246 [DOI] [PubMed] [Google Scholar]
- 52.Naldini, L., Vigna, E., Bardelli, A., Follenzi, A., Galimi, F., and Comoglio, P. M. (1995) J. Biol. Chem. 270 603-611 [DOI] [PubMed] [Google Scholar]