Abstract
All-trans-retinoic acid stimulates dendritic growth in hippocampal neurons within minutes by activating mitogen-activated protein kinase and mTOR and increasing dendritic translation of calcium calmodulin-dependent protein kinase II α and the α-amino-3-hydroxyl-5-methyl-4-isoxazole propionate receptor subunit GluR1. Hippocampal neurons express RARα in dendrites, and knocking down RARα prevents all-trans-retinoic acid effects on dendritic growth. Here we show, by liquid chromatography/mass spectrometry analysis of immunoaffinity isolates of hippocampal neurons, that RARα partners with many RNA-binding proteins and translation factors conveyed in dendritic RNA transport granules, including the purine-rich element-binding protein, Pur α. The interaction of RARα with Pur α, an RNA-binding protein required for dendritic RNA transport, and other RNA-binding proteins was confirmed by tandem affinity purification. Confocal microscopy confirmed localization of neuronal RARα in dendritic RNA granules with Pur α and FMRP (the fragile × mental retardation protein). Hippocampal RARα also associates with mRNA, e.g. encoding GluR1 and calcium calmodulin-dependent protein kinase II α. Consistent with a granule function of conveying translationally silenced mRNA, RARα inhibits translation initiation, independent of 7-methylguanylate cap or poly(A) tail, and prompts mRNA redistribution to silencing ribonucleoprotein particles. These data afford a mechanism for rapid stimulation of dendritic growth by all-trans-retinoic acid and reveal that the ligand-dependent transcription factor RARα also regulates translation.
The developed nervous system relies on atRA2 for synaptic plasticity that underlies hippocampus-dependent spatial learning (1, 2). Restricting the dietary atRA precursor retinol (vitamin A) or genetically impairing atRA signaling impairs hippocampus-dependent learning (3–6). Retinoid-compromised rodents err markedly more frequently in tests related to hippocampal function, such as the radial arm and Morris water mazes, and have diminished long term potentiation and abolished long term depression. Conversely, atRA dosing reduces frequency of relational memory errors in older mice relative to young mice (7).
Synaptic plasticity relies on protein synthesis in dendrites (8, 9). Large ribonucleoparticles or RNA granules convey translationally silenced mRNA from neuronal cell bodies to dendrites. These RNA encode diverse classes of postsynaptic proteins, including ionotropic glutamate receptors and kinases such as CaMKII α (10). Synaptic activity triggers translation of dendritic mRNA to support rapid synaptic structure and efficacy modifications (10, 11). For example, the α-amino-3-hydroxyl-5-methyl-4-isoxazole propionate receptor subunits GluR1 and GluR2 insert into synaptic membranes after local translation to enhance the number, strength, and stability of synapses (12).
atRA stimulates dendritic growth in primary mouse hippocampal neurons; siRNA knockdown and small hairpin RNA knockdown indicate a central function for RARα in atRA-induced dendritic growth (13). Responses to atRA occur in less than 30 min and include induction of somatodendritic translation of GluR1 and CaMKII α mRNAs. Intense dendritic expression of RARα in hippocampus in vivo and in primary hippocampal neurons suggests a contribution to translation control, in addition to its established function as a transcription factor. Here we provide direct evidence that RARα clusters in dendritic RNA granules and can repress translation. These data provide a mechanism for the rapid stimulation of synaptic remodeling by atRA and indicate that a nuclear ligand-dependent transcription factor can regulate dendritic translation.
EXPERIMENTAL PROCEDURES
RARα-associated Proteins—To produce RARα bait, COS cells (2 × 107) transfected with the Myc-RARα chimera were lysed in 0.5% Nonidet P-40, 50 mm Tris, pH 7.3, 150 mm NaCl, containing protease inhibitors. Supernatants (12,000 g, 10 min) were incubated 2.5 h at 4 °C with c-Myc beads. The beads were washed with lysis buffer three times.
To identify proteins associated with RARα, the beads were incubated 5 h at 4 °C with lysates (0.5% Triton X-100, 50 mm Tris, pH 7.3, 150 mm NaCl, 2.5 mm MgCl2, 1000 units/ml RNaseOUT, protease inhibitors) from primary hippocampal neurons (2 × 107; DIC24–30). The beads were washed four times, eluted with 2× loading buffer, and resolved by 4–20% gradient SDS-PAGE. The bands were processed in a DigestPro (INTAVIS Bioanalytical Instruments AG), washed, reduced with dithiothreitol, alkylated with iodoacetamide, and trypsin-digested. The digests were loaded onto a C18 cartridge and eluted at 250 nl/min with A (0.5% acetic acid in water) and B (0.5% acetic acid in 80% acetonitrile/20% water) in a 15-min linear gradient from 2% B at 0 min to 80% B, held at 80% B for 5 min, returned to 2% B over 10 min from a reverse phase column (Vydac 238EV5.07515, 75 μm × 150 mm) fitted with a coated spray tip (FS360-50-5-CE; New Objective, Inc.). Nano-liquid chromatography/electrospray ionization/tandem mass spectrometry was done with an AB QStar Pulsar, with a Proxeon Biosystems nano-electrospray source, with resolution >10,000. A 5-s tandem mass spectrometry scan followed a 1-s survey scan. Ions between m/z 400 and 1,000, charges between +2 to +5, and intensities >40 counts were fragmented. Liquid chromatography/tandem mass spectrometry data were submitted to Mascot for analyses.
TAP determination of RARα interacting proteins was done with a kit (Stratagene) (14).
RARα-associated mRNA—RARα was immunoprecipitated from lysates of primary hippocampus neurons (DIC14–15) with anti-RARα (Santa Cruz). mRNA was released by boiling twice (5 min each, 10 mm Tris, pH 7.5, 1 mm EDTA, 1% SDS), purified with MegaClear (Ambion), and reverse transcribed with Superscript II (Invitrogen). Primers used following RARα immunoprecipitation were (forward and reverse, respectively): GluR1, 5′-AGTCGAAGCGGATGAAGGG-3′, 5′-GTTGTGGTGGTTGGAGGC-3′; GluR2, 5′-TTCTAACAGCATACAGATAGG-3′, 5′-AAGCATTGGTGACTGCGAAAC-3′; CaMKII α 5′-ATCGCCTATATCCGCATCAC-3′, 5′-GGACAAAGAGCGGATCTCTG-3′; laminin B1, 5′-CATTGAGAACGTGGTCACCAC-3′, 5′-GAACGAGCTCTCACAGTCGTAG-3′; and RARα, 5′-TCACAGACCTTCGGAGCATC-3′, 5′-CCAGTTCTGTCTGAGAGGAC-3′.
Plasmids and Cell Culture—NonO, 14-3-3ζ, hnRNP U, PSF, PABP1, Pur α, RARα, VCP, and REP1 were PCR-amplified from DIC14 hippocampal neuron cDNA and cloned in frame into EGFP-N1 (Clontech). An N-terminal Myc tag was included in PCR primers. DsRed-Pur α and NTAP-RARα were generated by releasing Pur α or RARα cds from EGFP expression vectors and inserting them into DsRed-N1 (Clontech) or NTAP-B (Stratagene) in frame with DsRed and TAP tag, respectively. RARα deletion mutants with N-terminal hemagglutinin were PCR-amplified from EGFP-RARα and inserted into EGFP-N1. An hemagglutinin tag was included in the primer. A dominant negative RARα was made by ligating cds residues 1–403 lacking the AF-2 core in frame with EGFP in EGFP-N1 (15). The FL cds was PCR-amplified from pGL3 (Promega) and inserted into pcDNA3.1(+) (Invitrogen) to generate pcDNA-FL. pSL-MS2 12X and Pol II-MS2-GFP were gifts from Robert H. Singer (Albert Einstein College of Medicine). MS2 12X was subcloned into pcDNA-FL. MS2 cds was subcloned into EGFP-N1 to generate EGFP-MS2. MS2-RARα-EGFP and MS2-RARα/domains-EGFP were generated by PCR amplification of MS2 cds from Pol II-MS2-GFP and cloned into EGFP expression vectors. Nontargeting siRNA and GW182 siRNA were obtained from Dharmacon. COS cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Invitrogen). Primary cultures of hippocampal neurons were prepared from e17 mouse embryos. Neurons and COS cells were transfected with Lipofectamine 2000 (Invitrogen) according to instructions, except Lipofectamine was 0.5 μl/well in 24-well plates for neurons, and the medium was unchanged.
RRL Translation—FL RNA was transcribed with T7 message machine (Ambion) using linearized pcDNA-FL. For the uncapped transcript, the nonfunctional cap analog ApppG was used in place of GpppG (NEB). Fifty-100 base poly(A) tails were added with Escherichia coli poly(A) polymerase (Ambion). Translation was done with a nuclease-treated RRL following instructions (Promega), except reactions used 90 ng of RNA, 0.6 mm MgCl2, and 125 mm KCl to promote cap-poly(A) synergy (16, 17). FL and Renilla luciferase activities were measured with luciferase systems (Promega). Northern blotting was done with Northern-Max (Ambion) using psoralen-biotin-labeled FL or β-actin probes generated by in vitro transcription. The signals were developed with a Bright-Star BioDetect kit (Ambion) and quantified with Image J.
Subcellular Fractionation—Brains from 1-month-old mice were homogenized in a lysis buffer of 10 mm Hepes, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, and a protease inhibitor mixture (Roche Applied Science). The lysates were centrifuged 1000 × g for 10 min, 10,000 × g for 10 min, and 100,000 × g for 1 h to obtain S1, S2 and P2, and S3 and P3, respectively. Equal amounts of protein from each fraction were loaded onto SDS-PAGE for Western analysis with anti-RARα (Santa Cruz, 1:750), anti-FMRP (Chemicon, 1:1000) and anti-S6 (Cell signaling, 1:1000). For solubilization, COS cells (3 × 106) transfected 24 h with pcDNA-FL-MS2BS and MS2-EGFP or MS2-RARα or domains-EGFP were lysed in 200 μl of buffer C (0.25 m sucrose, 10 mm Tris, pH 7.4, 25 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, 30 units/ml RNaseOUT) with digitonin (50 mg/ml; Sigma) on ice for 15 min. The lysates were centrifuged 1000 × g for 5 min. Supernatants were spun 14,000 × g for 5 min to obtain RNA for purification with TRIzol reagent (Invitrogen). The pellets were washed with buffer C twice and resuspended in TRIzol. RNA was isolated according to the TRIzol protocol (Invitrogen). Cytoplasmic and nuclear RNA were isolated with a cytoplasmic and nuclear RNA isolation kit (Norgen). In a separate experiment, brains from 23-day-old mice (littermates) were homogenized in lysis buffer and divided into two groups. One group was supplemented with 80 units/ml RNaseOUT. The other group was not and was used for RNase treatment. The S2 fraction was divided, and one portion was treated with 33 μg/ml RNase A at 37 °C for 30 min and then separated into S3 and P3 fractions.
Sucrose Density Gradients—Polysome profiling was done with 15–45% sucrose gradients (18). Nine hundred-μl fractions were collected from tube bottoms. RNA was isolated by two-round precipitation with 8 m guanidine HCl and ethanol. Protein was precipitated with 2 volumes of acetone.
Immunostaining—DIC12 hippocampal neurons were fixed in 4% paraformaldehyde with 4% sucrose, permeabilized with 0.2% Triton X-100 5 min, and blocked with 5% goat serum and 3% bovine serum albumin in phosphate-buffered saline 1 h. The cells were incubated with anti-RARα (1:100; Santa Cruz Biotechnology sc-551, polyclonal to C terminus epitope) and anti-FMRP (1:100; Chemicon) or anti-Tuj1 (1:100, Chemicon) overnight at 4 °C, followed by Alexa 488- and Alexa 555-conjugated-secondary antibodies (1:100) (Invitrogen) incubation at room temperature for 1 h. MAP2 staining was done using anti-MAP2 (1:100, Chemicon) and Alexa 647-conjugated-secondary antibody (1:20) (Invitrogen) on DIC12 neurons transfected with EGFP-RARα and DsRed-Pur α for 24 h. The neurons were washed with phosphate-buffered saline three times and mounted for microscopy. Confocal microscope images were obtained with a LSM510 confocal microscope in the College of Natural Resources BioImaging facility.
RESULTS AND DISCUSSION
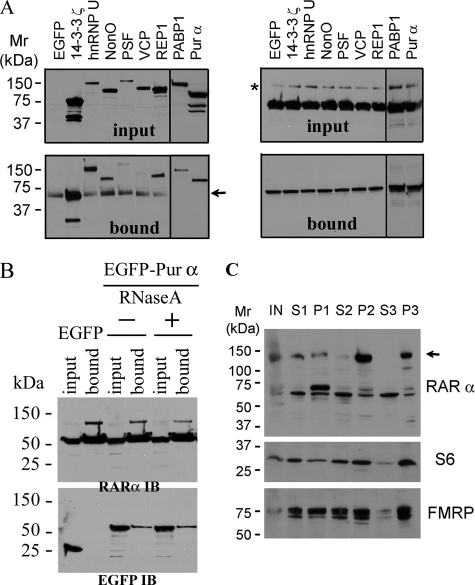
RARα Associates with RNA-regulating Proteins in Neuronal Granules—We used a Myc-RARα chimera bound to anti-Myc-conjugated beads as bait to isolate RARα protein partners from mouse hippocampal neurons and resolved the isolate by gel electrophoresis. We analyzed 22 gel bands by electrospray ionization-quadrupole-time-of-flight mass spectrometry, which identified 58 proteins (Table 1). RARα-associated proteins included many RNA-binding proteins and translation factors associated with dendritic RNA transport granules (19, 20). We selected eight proteins to confirm as RARα-interacting partners by streptavidin/calmodulin tandem affinity purification. This second immunoaffinity technique verified seven of the eight as RARα partners: Pur α, PABP1, REP1, PSF, NonO, hnRNP U, and 14-3-3ζ; the exception was VCP (Fig. 1A). TAP analysis was repeated with EGFP-Pur α and RARα with RNase A treatment to determine whether this interaction required mRNA (Fig. 1B). RNase A treatment did not abolish the interaction between Pur α and RARα, indicating RNA independence.
TABLE 1.
RARα-interacting proteins
Lysates of DIC14-21 hippocampal neurons were probed with RARα bound to anti-Myc-conjugated beads. The isolate was subjected to gradient gel electrophoresis, and 22 bands were analyzed with electrospray ionization-quadrupole-time-of-flight mass spectrometry. Bold type designates select interactions confirmed by TAP.
| Type (number) | Identity |
|---|---|
| RNA transport (4) | Pur α, Pur β, FXR, Tho complex 4 |
| Protein synthesis (19) | PABP1, eIF3α, eIF3β, ribosomal proteins S3, S4, S5, S6, S12, S13, S16, S17, S19, S25, S26, L23, 40 S ribosomal protein SA (p40), 60 S ribosomal protein, putative ribosomal S4, Hsp70 |
| RNA helicases (3) | DDX1, DDX3, DDX5 |
| hnRNP (4) | hnRNP U, hnRNP A/B, hnRNP D0 (AU-rich element RNA-binding protein), hnRNP Q (SYNCRIP), |
| Other RNA associated (9) | NonO, PSF, GP137 (RNA granule protein 103), EWS, FUS, cold-inducible RBP, RNA-binding motif protein 3, G3BP, single strand DNA-binding protein 1 |
| Endocytotic (4) | AP2A1, AP2A2, AP2B1, clathrin coat assembly AP50 (AP2 ml) |
| Cytoskeletal (5) | vimentin, desmin, tubulin α3, β-actin, tropomodulin |
| Others (10) | REP1, 14-3-3ζ, VCP, 14-3-3β, 14-3-3γ, VG potassium channel α-2 subunit, p100-coactivator, hypothetical protein LOC 68045, nucleotide-diphosphate kinase 2, tumor metastastic process associated protein NM23 |
FIGURE 1.
RARα associates with neuronal RNA granule proteins. A, RARα and Myc-tagged neuronal proteins selected from Table 1 were co-transfected into COS cells to confirm their association. RARα was isolated by TAP. Total (input) and RARα pull-down fractions (bound) were immunoblotted for Myc (left) or RARα (right, C-terminal antibody). The arrow indicates a nonspecific band. The asterisk indicates a high molecular weight form of RARα. B, RARα and either EGFP or EGFP-tagged Pur α were co-transfected into COS cells; RARα was isolated by TAP. Total (input) and RARα pull-down fractions (bound) were immunoblotted for RARα (top, RARα IB) or EGFP (bottom, EGFP IB) with (+) or without (–) RNase A treatment. C, subcellular distribution of RARα in mouse brain homogenates. Equal amounts of protein from each fraction were immunoblotted for RARα, S6, or FMRP: S1, 1000 × g supernatant; S2, 10,000 × g supernatant; P2, 10,000 × g pellet; S3, 100,000 × g supernatant; P3, 100,000 × g pellet. The arrow indicates a modified form of RARα.
Many RARα-associated proteins populate granules that house RNA-binding proteins. Therefore, we used differential centrifugation to determine subcellular loci of RARα in mouse brain. A high molecular weight form of RARα localized in the RNA granule-containing P3 and the membrane containing P2 fractions with two other granule proteins, FMRP and ribosomal protein S6 (Fig. 1B). This experiment was repeated, except the S2 fraction was treated with RNase A. RARα again isolated with the P3 fraction, indicating that mRNA is not obligatory to its localization in the RNA granule fraction. This RARα migrated predominantly as an SDS-resistant band at ∼140 kDa, suggesting covalent modification and/or a multiprotein complex. We are in the process of analyzing the precise composition of this complex but have confirmed that its major constituent is RARα by redoing the Western blot with a monoclonal antibody raised against the N terminus of RARα (Chemicon MAB5346)[em]the first antibody used was a polyclonal raised against the C terminus[em]and proteomics analysis with liquid chromatography/mass spectrometry (data not shown).
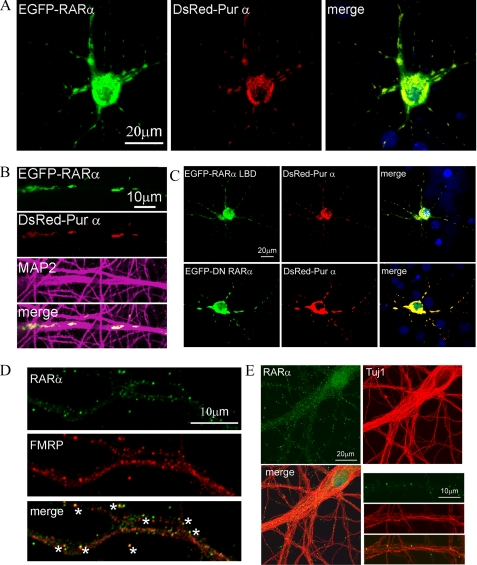
Pur α, a granule RNA-binding protein required for dendritic RNA transport, was confirmed as an RARα-interacting partner in both immunoaffinity analyses. We therefore applied fluorescence microscopy to determine whether RARα localized with Pur α in neurons. Soma and dendrites of neurons co-expressed RARα and Pur α (Fig. 2, A and B). Dendritic expression of both was punctated. RARα also co-localized with the dendritic marker MAP2. Extranuclear localization of RARα required only the LBD (residues 200–390) and not the transcription activation domain or transcription activity, as shown by co-localization of both the RARα LBD and a dominant negative RARα mutant with Pur α (Fig. 2C). Immunofluorescence also confirmed that ∼50% of endogenous RARα co-localized with FMRP (Fig. 2D). RARα puncta interspersed among neuronal microtubules with neuron-specific α-tubulin III (Fig. 2E).
FIGURE 2.
Localization of RARα in neuronal granules. A, co-localization in DIC12 hippocampal neurons of RARα (green) with Pur α (red). B, enlargement of dendrites from A, and co-localization of RARα with MAP2 (purple). C, localization of the RARα LBD (EGFP-RARα-LBD) and a dominant negative RARα mutant (EGFP-DN-RARα) in DIC12 hippocampal neurons with DsRed-Pur α. D, a representative dendrite showing partial co-localization of endogenous RARα (green) and FMRP (red) in DIC12 hippocampal neurons. Asterisks denote co-localizing puncta. E, RARα puncta intersperse among α-tubulin III-labeled (Tuj1) neuronal microtubules. The larger panels show a cell, whereas the smaller panels show a magnified dendritic region.
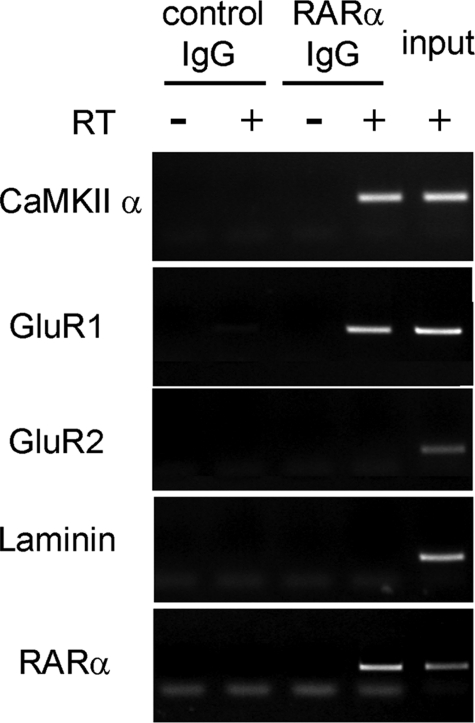
RARα Represses Translation—Synaptic modification relies on dendritic protein synthesis supported by translation of CaMKII α and the α-amino-3-hydroxyl-5-methyl-4-isoxazole propionate receptor subunits GluR1 and GluR2 (21, 22). atRA induces dendritic translation of CaMKIIα and GluR1 mRNA but not of GluR2 mRNA (13). Therefore, we immunoprecipitated RARα from mouse hippocampus and analyzed associated mRNA (Fig. 3). The precipitate included mRNA encoding CaMKII α, GluR1, and RARα but not GluR2 or the nondendritic protein laminin B1, consistent with RARα modifying translation of CaMKII α and GluR1, but not of GluR2.
FIGURE 3.
RARα associates specifically with CaMKII α and GluR1 mRNA in neurons. Lysates of DIC14–15 hippocampal neurons were immunoprecipitated with IgG or anti-RARα and analyzed by reverse transcription-PCR without (–RT) or with (+RT) reverse transcriptase with primers for mRNA indicated at the left.
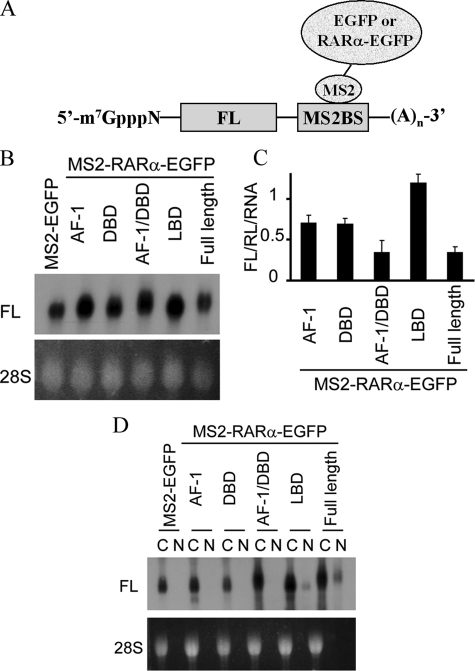
To determine whether RARα can affect translation directly, we applied a tethering assay that has established translation regulation by exon junction complex proteins, DEAD box helicases, etc. (23–25). Translation was evaluated from an FL mRNA harboring tandem repeats of the MS2BS in its 3′-untranslated region (Fig. 4A). This construct was co-expressed with constructs expressing chimera of the bacteriophage protein MS2 with EGFP or MS2-RARα-EGFP. Renilla luciferase also was co-transfected to normalize for variations in FL mRNA expression. The RARα construct reduced FL translation (FL/Renilla luciferase signal) 86 ± 2% (mean ± S.E., n = 9, 3 replicates in each of three experiments) relative to MS2-EGFP. A second experiment compared RARα to each of its domains. In this case, to exclude mRNA expression differences as underlying differences in FL activity, the ratios FL/Renilla luciferase activity were normalized to amounts of FL mRNA (Fig. 4B). The degree of suppression by full-length RARα remained the same (Fig. 4C). The combined ligand-independent activation function-1 (AF-1) and DNA-binding domains of RARα (residues 1–199) reduced translation as well as full-length RARα, but the individual AF-1 (residues 1–87) or DNA-binding domain (residues 87–199) were less potent. The LBD did not repress translation. MS2-RARα-EGFP did not reduce translation from FL mRNA lacking MS2BS (not shown). Insertion of MS2BS into FL mRNA did not hinder its nuclear export (Fig. 4D). These data indicate that RARα suppresses FL activity through changes in translation effected by proximity of the first 199 RARα residues to FL mRNA.
FIGURE 4.
RARα suppresses translation in intact cells. A, a tethering assay for evaluating translation suppression relies on co-expressing FL mRNA, harboring tandem repeats of MS2BS in its 3′-untranslated region, with a chimera of MS2 and a query protein. B, Northern blotting of FL-MS2BS mRNA expressed during tethering assays. RNA was isolated from COS cells 24 h after transfection with FL-MS2BS and MS2-EGFP or MS2-(RARα or domains)-EGFP. The 28 S rRNA was ethidium bromide-stained. C, FL/Renilla luciferase (RL) activities in the presence of MS2-RARα or domains-EGFP relative to control (FL/Renilla luciferase of MS2-EGFP, set as 1), normalized to the mRNA signals of FL-MS2BS mRNA shown in B; the data are the means ± S.E., n = 9 (three replicates in each of three experiments). Renilla luciferase was co-transfected to normalize for variations in FL activity caused by variations in FL mRNA. D, Northern blotting of FL-MS2BS mRNA expressed with MS2-EGFP or MS2-RARα or domains-EGFP indicating nuclear export. Lanes C, cytoplasm; lanes N, nuclei.
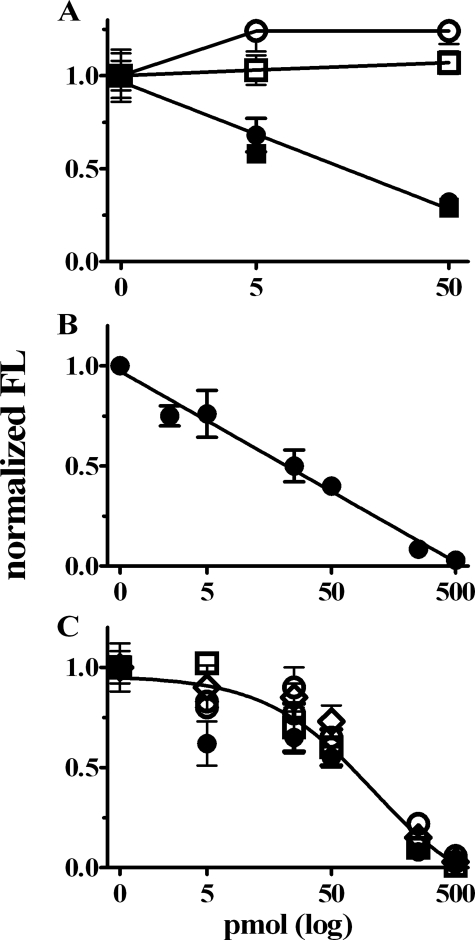
RARα Functions as a Sequence-independent Translation Inhibitor—To determine whether RARα represses translation directly, we added purified RARα to a RRL programmed with FL mRNA or with an FL mRNA that included the entire 5′- and 3′-untranslated regions of GluR1. RARα suppressed translation of both FL constructs in a dose-dependent manner, with 50 pmol causing ∼70% inhibition. Control GST did not suppress translation (Fig. 5A). To exclude mRNA degradation, quantitative PCR was done with the FL mRNA in each of the three replicates for the six groups exposed to the three amounts of GST or GST-RARα. The quantities of FL mRNA did not vary, excluding mRNA degradation as the cause of decreased translation (data not shown). Extended dose-dependent analysis showed ∼80–90% repression by 250 pmol RARα (Fig. 5B). To examine whether suppression requires mRNA capping and/or tailing, we tested the effect of RARα on FL mRNA capped with a functional m7GpppN or a nonfunctional analog, ApppG, in the presence or absence of a poly(A) tail. Low levels of mRNA were used in a RRL assay modified so that translation was susceptible to 5′-cap and 3′-poly(A) synergy (16, 17). Similar concentrations of RARα inhibited translation regardless of capping or tailing (Fig. 5C). RARα suppressed FL expression from capped and untailed, capped and tailed, and uncapped and tailed from the beginnings of the assays. The functional half-lives of FL RNA did not change with GST versus RARα treatment, indicating that the stabilities of the mRNAs had not changed during the assays of the various groups (Fig. 6). These data demonstrate that RARα functions independently of mRNA sequence and does not affect mRNA turnover.
FIGURE 5.
RARα suppresses translation in RRL. A, sequence independence of RARα translation suppression. Effect of GST (open symbols) or GST-RARα (filled symbols) on FL mRNA translation by a RRL programmed with FL (squares) or GluR1-FL mRNA (circles). B, dose-dependent inhibition of FL mRNA translation by RARα. C, Cap- and poly(A)-independent inhibition of FL mRNA translation by RARα. Open circles, uncapped and untailed; open triangles, capped and untailed; open squares, uncapped and tailed; filled circles, capped and tailed. The data in A–C were normalized to FL activity in the absence of GST and RARα and represent the means ± S.D., n = 3. FL measurements were made in lysates after 60 min of incubation at 37 °C.
FIGURE 6.
Time courses of RARα effects on FL mRNA translation. RARα (filled circles) and GST (open circles) were compared for effects on FL mRNA translation. A, capped and untailed; B, capped and tailed; C, uncapped and tailed. D–F, each data set in A–C, respectively, was renormalized to its own highest signal to assess mRNA functional half-life. The data are the means ± S.D., n = 3. FL measurements were made in lysates as described in the legend to Fig. 5 with 500 pmol of either GST or RARα-GST.
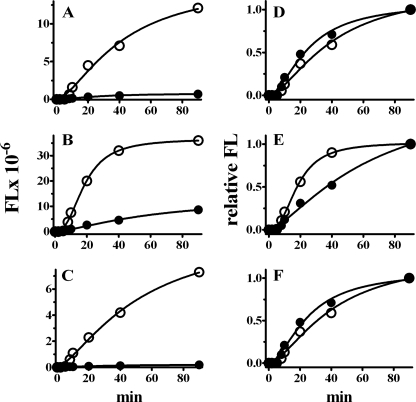
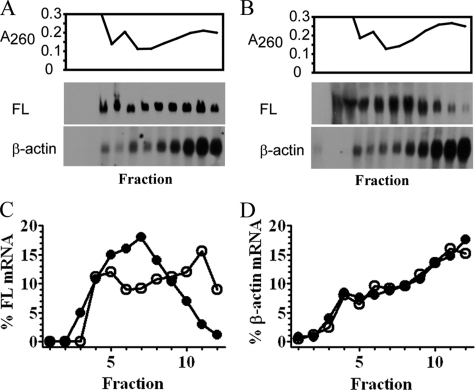
RARα Inhibits Initiation of Translation—We used polysomal profiling to provide insight into the mechanism of translation suppression (18, 29). FL-mRNA shifted toward the top of the gradient, away from polysomes, only when tethered with RARα (Fig. 7). Neither MS2-RARα-EGFP nor MS2-EGFP affected β-actin mRNA. This indicates that RARα inhibits polysome loading of associated mRNA, impeding initiation.
FIGURE 7.
RARα inhibits translation initiation. A and B, polysome distribution of FL-MS2BS mRNA versus endogenous β-actin in COS cells co-transfected with MS2-EGFP (A) or MS2-RARα-EGFP (B). RNA was monitored at A260 (top). mRNA precipitated from each fraction was analyzed by Northern blot with FL or β-actin probes (bottom). C and D, distribution of FL (C) or β-actin mRNA (D) tethered with MS2-EGFP (open circles) or MS2-RARα-EGFP (filled circles) in each fraction, expressed as a percentage of the sum of Northern blot signals in all fractions.
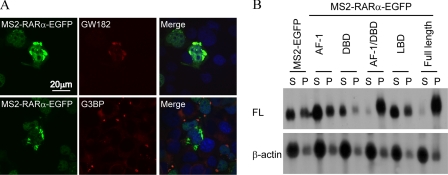
RARα Localizes in Neuronal Ribonucleoprotein Particles—Neuronal ribonucleoprotein particles are structurally and functionally related to PB, which store and/or degrade translationally repressed mRNAs in conjunction with stress granules (26, 27). To determine whether these silencing structures include RARα-suppressed transcripts, we co-transfected COS cells with FL-MS2BS and MS2-RARα-EGFP and probed for the PB marker GW182 or the stress granule marker G3BP (Fig. 8A). The majority of RARα isolated in large aggregates with GW182. In contrast, MS2-RARα-EGFP did not co-localize with G3BP. To confirm association of repressed transcripts with PB, COS cells were co-transfected with FL-MS2BS and MS2-RARα or domains-EGFP or MS2-EGFP and were solubilized with digitonin, which preserves PB (18). FL mRNA tethered with EGFP, or the RARα AF-1, DNA-binding domain or LBD partitioned primarily to soluble fractions, whereas FL mRNA tethered with the combined AF-1/DNA-binding domain or with full-length RARα partitioned primarily to PB (Fig. 8B). We next dissolved PB by siRNA knockdown of GW182 (28). This did not prevent translation suppression by RARα (not shown). These data indicate that RARα silences mRNA independently of PB but sorts silenced mRNA to PB.
FIGURE 8.
RARα redistributes mRNA to PB. A, immunocytochemistry with anti-GW182 or G3BP antibodies (red) of COS cells co-transfected with FL-MS2BS and MS2-RARα-EGFP (green). The nuclei were counterstained with 4′,6′-diamino-2-phenylindole (blue). B, Northern blots of supernatants (lanes S) and pellets (lanes P) from digitonin-solubilized COS cells transfected with FL-MS2BS and MS2-RARα or domains-EGFP. DBD, DNA-binding domain.
Conclusions—Previously, we showed that atRA stimulates dendritic growth within minutes, by activating mitogen-activated protein kinase and mTOR regulation of neuronal translation and increasing dendritic translation of CaMKII α and GluR1 mRNA, without affecting GluR2 (13). We also showed that the atRA effects on dendritic growth were prevented by knocking down RARα with siRNA or small hairpin RNA and hippocampal neurons expressed RARα in dendrites, suggesting a function in translation, in addition to transcription.
Here we have shown by immunoaffinity and proteomic analyses that RARα forms partnerships in neurons with multiple RNA-binding proteins and translation factors and confirmed select RARα partnerships by TAP. Consistent with a function in translation, mouse brain RARα localized in the RNA granule-containing fraction. RARα partnered with the granule RNA-binding protein Pur α and RARα co-localizes in neuronal somatic PB with Pur α and FMRP. Neuronal RARα associated specifically with mRNA encoding CaMKII α and GluR1. Both a tethering assay and a RRL translation assay showed that RARα can repress translation, and polysome profiling showed that RARα moves mRNA away from polysomes, consistent with prevention of translation initiation. These data broaden insight into the mechanisms of plasticity, translation regulation, retinoid action, and the action of RARα, well recognized as a ligand-activated transcription factor but not known as a regulator of translation (30–32).
We propose that RARα silences translation and facilitates delivery of specific mRNA to neuronal dendrites. atRA would stimulate transcription and translation in neurons, providing for rapid insertion of receptors into synapses and generating mRNA in preparation for a new cycle. This mechanism could also modulate homeostatic scaling, a form of metaplasticity that engages local synthesis of GluR1 and other synaptic proteins (8, 12).
Acknowledgments
We thank Dr. Robert H. Singer for providing MS2 plasmids. We thank Dr. Jennifer Doudna for advice.
This work was supported, in whole or in part, by National Institutes of Health Grants DK36870 and AG13566. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: atRA, all-trans-retinoic acid; cds, coding sequences; DIC, days in culture; FL, firefly luciferase; MAP2, microtubule-associated protein 2; MS2BS, binding sites for the bacteriophage protein MS2; PB, somatic processing bodies; RRL, rabbit reticulocyte lysate; TAP, tandem affinity purification; CaMKII, calmodulin kinase II; siRNA, small interfering RNA; EGFP, enhanced green fluorescent protein; AF-1, activation function-1; GST, glutathione S-transferase; LBD, ligand-binding domain.
References
- 1.McCaffery, P., Zhang, J., and Crandall, J. E. (2006) J. Neurobiol. 66 780–791 [DOI] [PubMed] [Google Scholar]
- 2.Maden, M. (2007) Nat. Rev. Neurosci. 8 755–765 [DOI] [PubMed] [Google Scholar]
- 3.Chiang, M. Y., Misner, D., Kempermann, G., Schikorski, T., Giguere, V., Sucov, H. M., Gage, F. H., Stevens, C. F., and Evans, R. M. (1998) Neuron 21 1353–1361 [DOI] [PubMed] [Google Scholar]
- 4.Misner, D. L., Jacobs, S., Shimizu, Y., de Urquiza, A. M., Solomin, L., Perlmann, T., De Luca, L. M., Stevens, C. F., and Evans, R. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11714–11719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocco, S., Diaz, G., Stancampiano, R., Diana, A., Carta, M., Curreli, R., Sarais, L., and Fadda, F. (2002) Neurosci. 115 475–482 [DOI] [PubMed] [Google Scholar]
- 6.Etchamendy, N., Enderlin, V., Marighetto, A., Pallet, V., Higueret, P., and Jaffard, R. (2003) Behav. Brain Res. 145 37–49 [DOI] [PubMed] [Google Scholar]
- 7.Etchamendy, N., Enderlin, V., Marighetto, A., Vouimba, R. M., Pallet, V., Jaffard, R., and Higueret, P. (2001) J. Neurosci. 21 6423–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton, M. A., and Schuman, E. M. (2005) J. Neurobiol. 64 116–131 [DOI] [PubMed] [Google Scholar]
- 9.Sutton, M. A., and Schuman, E. M. (2006) Cell 127 49–58 [DOI] [PubMed] [Google Scholar]
- 10.Kosik, K. S., and Krichevsky, A. M. (2002) Sci. STKE 2002, PE16. [DOI] [PubMed]
- 11.Martin, K. C., and Zukin, R. S. (2006) J. Neurosci. 26 7131–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju, W., Morishita, W., Tsui, J., Gaietta, G., Deerinck, T. J., Adams, S. R., Garner, C. C., Tsien, R. Y., Ellisman, M. H., and Malenka, R. C. (2004) Nat. Neurosci. 7 244–253 [DOI] [PubMed] [Google Scholar]
- 13.Chen, N., and Napoli, J. L. (2008) FASEB J. 22 236–245 [DOI] [PubMed] [Google Scholar]
- 14.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999) Nat. Biotechnol. 17 1030–1032 [DOI] [PubMed] [Google Scholar]
- 15.Damm, K., Heyman, R. A., Umesono, K., and Evans, R. M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 2989–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svitkin, Y. V., Ovchinnikov, L. P., Dreyfuss, G., and Sonenberg, N. (1996) EMBO J. 15 7147–7155 [PMC free article] [PubMed] [Google Scholar]
- 17.Borman, A. M., Michel, Y. M., and Kean, K. M. (2000) Nucleic Acids Res. 28 4068–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillai, R. S., Bhattacharyya, S. N., Artus, C. G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., and Filipowicz, W. (2005) Science 309 1573–1576 [DOI] [PubMed] [Google Scholar]
- 19.Kanai, Y., Dohmae, N., and Hirokawa, N. (2004) Neuron 43 513–525 [DOI] [PubMed] [Google Scholar]
- 20.Elvira, G., Wasiak, S., Blandford, V., Tong, X. K., Serrano, A., Fan, X., del Rayo Sánchez-Carbente, M., Servant, F., Bell, A. W., Boismenu, D., Lacaille, J. C., McPherson, P. S., DesGroseillers, L., and Sossin, W. S. (2006) Mol. Cell Proteomics 5 635–651 [DOI] [PubMed] [Google Scholar]
- 21.Bach, M. E., Hawkins, R. D., Osman, M., Kandel, E. R., and Mayford, M. (1995) Cell 81 905–915 [DOI] [PubMed] [Google Scholar]
- 22.Sutton, M. A., Ito, H. T., Cressy, P., Kempf, C., Woo, J. C., and Schuman, E. M. (2006) Cell 125 785–799 [DOI] [PubMed] [Google Scholar]
- 23.Gray, N. K., Coller, J. M., Dickson, K. S., and Wickens, M. (2000) EMBO J. 19 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minshall, N., Thom, G., and Standart, N. (2001) RNA 7 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nott, A., Le Hir, H., and Moore, M. J. (2004) Genes Dev. 18 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbee, S. A., Estes, P. S., Cziko, A. M., Hillebrand, J., Luedeman, R. A., Coller, J. M., Johnson, N., Howlett, I. C., Geng, C., Ueda, R., Brand, A. H., Newbury, S. F., Wilhelm, J. E., Levine, R. B., Nakamura, A., Parker, R., and Ramaswami, M. (2006) Neuron 52 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson, P., and Kedersha, N. (2006) J. Cell Biol. 172 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, Z., Jakymiw, A., Wood, M. R., Eystathioy, T., Rubin, R. L., Fritzler, M. J., and Chan, E. K. (2004) J. Cell Sci. 117 5567–5578 [DOI] [PubMed] [Google Scholar]
- 29.Zalfa, F., Achsel, T., and Bagni, C. (2006) Curr. Opin. Neurobiol. 16 265–269 [DOI] [PubMed] [Google Scholar]
- 30.Forman, B. M., Umesono, K., Chen, J., and Evans, R. M. (1995) Cell 81 541–550 [DOI] [PubMed] [Google Scholar]
- 31.Dilworth, F. J., and Chambon, P. (2001) Oncogene 20 3047–3054 [DOI] [PubMed] [Google Scholar]
- 32.Bour, G., Lalevée, S., and Rochette-Egly, C. (2007) Trends Cell Biol. 17 302–309 [DOI] [PubMed] [Google Scholar]