Abstract
Calpains are intracellular cysteine proteases, which include widely expressed μ- and m-calpains (1). Both μ-calpains and m-calpains are heterodimers consisting of a large catalytic subunit and a small regulatory subunit. The calpain small subunit encoded by the gene Capn4 directly binds to the intracellular C-terminal tail (C-tail) of the receptor for parathyroid hormone and parathyroid hormone-related peptide and modulates its cellular functions in osteoblasts in vitro (2). To investigate a potential role of the calpain small subunit in osteoblasts in vivo, we generated osteoblast-specific Capn4 knock-out mice using the Cre-LoxP system (3). Mutant mice had smaller bodies with shorter limbs, reduced trabecular bone with thinner cortices, and decreased osteoblast number. In vitro analysis confirmed that deletion of Capn4 in osteoblasts severely affected multiple osteoblast functions including proliferation, differentiation, and matrix mineralization. Collectively, our findings provide the first in vivo demonstration that the calpain small subunit is essential for proper osteoblast activity and bone remodeling.
Calpains are a family of Ca2+-dependent intracellular cysteine proteases that include ubiquitously expressed μ- and m-calpains (1, 4). Both μ-calpains and m-calpains form heterodimers consisting of a large catalytic subunit (80 kDa) encoded by the genes Capn1 and Capn2, respectively, and a small regulatory subunit (28 kDa) encoded by the gene Capn4 (1). Disruption of Capn4 eliminates both μ-calpains and m-calpain activities in embryonic fibroblasts (5), suggesting that the calpain small subunit is essential for maintenance of calpain stability and activity. Notably, genetic ablation of Capn4 results in early embryonic lethality, which demonstrates an essential role of the calpain small subunit during development (5, 6).
Several lines of evidence suggest that calpains are necessary for proper osteoblast function in vitro (7–11). However, a physiological role of calpains in osteoblasts in vivo remains to be established. Chemical inhibition of calpain activity reduces osteoblast proliferation and differentiation in the MC3T3-E1 osteoblastic cell line (11). Moreover, we have previously reported that the calpain small subunit directly binds to the intracellular C-tail of the receptor for PTH2 and PTH-related peptide and modulates its ligand-mediated cellular functions (2). Both ligands are known regulators of bone homeostasis in vivo through their direct actions on cells of the osteoblast lineage (12–14). Taken together, these findings suggest that the calpain small subunit could play a critical but yet unknown role in osteoblast biology in vivo.
To test this hypothesis, we have conditionally ablated Capn4 in cells of the osteoblast lineage in vivo by using the Cre-LoxP system. Lack of the calpain small subunit in osteoblasts caused a significant decrease of both trabecular and cortical bone, which was associated with a severe impairment of osteoblast proliferation and differentiation. These findings are the first in vivo evidence that the calpain small subunit plays a crucial role in osteoblast function and bone homeostasis.
EXPERIMENTAL PROCEDURES
Generation of Osx-Cre+/- Capn4flox/flox Mice—Mice homozygous for floxed Capn4 alleles (Capn4flox/flox) were crossed with those expressing Cre under the control of osterix promoter (Osx-Cre+/-) to generate Osx-Cre+/-Capn4flox/+ mice (15–17). These mice were then crossed with either Capn4flox/flox or Capn4flox/+ mice to generate Osx-Cre+/-Capn4flox/flox mutant mice. All experiments were performed in compliance with the guiding principles of the Guide for the Care and Use of Laboratory Animals and approved by the subcommittee on Research Animal Care of the Massachusetts General Hospital (MGH).
Genotype Analysis—Genomic DNA was isolated from tail biopsies as described previously (18). PCR-based strategies were used to genotype these mice. The Cre transgene was detected by PCR using the primers P1 (5′-CGCGGTCTGGCAGTAAAAACTATC-3′) and P2 (5′-CCCACCGTCAGTACGTGAGAT ATC-3′) to generate an internal sequence of Cre. Taking an advantage of a floxed construct, in which an intron 9 was shortened by 1.6 kilobase pairs (kbp), the Capn4 floxed and wild-type alleles were determined using two sets of PCR reactions (15, 19). The floxed and wild-type alleles were determined as 0-bp (no amplified product) and 1.0-kbp PCR products, respectively, using primers P3 (5′-GTCAGGCTAGATGCCATGTTCC-3′) in exon 9 and P4 (5′-CGACTATCCGAGCGCTGCC-3′) within a sequence deleted in floxed allele in intron 9 and as 0.4-kbp and 2.0-kbp PCR products, respectively, using primers P3 (5′-GTCAGGCTAGATGCCATGTTCC-3′) and P5 (5′-GTTCACTTGGATCTGTCCGGTGCC-3′) corresponding to sequences in exons E9 and E10.
Serology—Ionized calcium was measured with the Ciba-Corning 634 Ca2+/pH analyzer (Ciba-Corning Diagnostics Corp., Medfield, MA). Serum levels of osteocalcin (Biomedical Technologies Inc., Stoughton, MA) and tartrate-resistant acid phosphatase (TRACP) 5b (Immunodiagnostic Systems, Tyne and Wear, UK) were determined using enzyme-linked immunosorbent assays as described by the manufacturers' instructions.
Whole Mount Skeletal Staining—The whole mount skeletal staining was performed as described previously (20). Briefly, newborn mice were fixed in ethanol for 5 days and then in acetone for 2 days. Staining with Alizarin red S and Alcian blue was performed for 3 days at 37 °C. After washing with distilled water, the skeleton was cleared in 1% KOH and taken through graded steps into 100% glycerol.
Sample Preparation and Histological Analysis—For histological analysis, Osx-Cre+/-Capn4flox/flox and sex-matched control littermates (Osx-Cre+/-Capn4flox/+ and Capn4flox/flox) were sacrificed at birth and at 2, 4, 9, and 12 weeks of age. Tissues from Osx-Cre+/-Capn4flox/flox, and control littermates were fixed and stored as described previously (21). In selected cases, hind limbs were decalcified, and paraffin blocks were prepared by standard histological procedures. To detect osteoclast-like cells, tartrate-resistant acid phosphatase (TRAP) staining was performed using an acid phosphatase detection kit (Sigma-Aldrich) for some selected samples.
In Situ Hybridization—In situ hybridization analysis was performed as described previously (22). Complementary 35S-labeled riboprobes were transcribed from the plasmids encoding mouse type I collagen (Col.1), mouse matrix metalloproteinase (MMP)13, mouse osteocalcin (OC), mouse osteopontin (OP), and rat TRAP using Riboprobe systems from Promega (Madison, WI).
Histomorphometry—For dynamic histomorphometry, animals were injected intraperitoneally with fluorochromes, calcein, and demeclocycline (20 μg/g of body weight, Sigma-Aldrich), 3 and 10 days before sacrifice, respectively. Bones were fixed and embedded in methyl methacrylate resin as described previously (21). Five-micrometer sections were stained with Masson Trichrome or coverslipped unstained, and histomorphometric analysis was performed with the Osteomeasure system (Osteometrics Inc., Atlanta, GA) using standard procedures. Tibial sections were measured in the proximal metaphysis beginning 340 μm below the chondro-osseous junction.
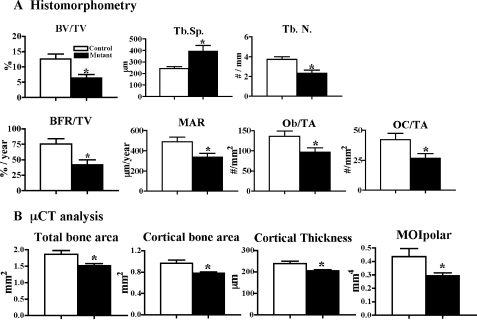
Microcomputed Tomography (μCT)—High resolution images of the femur and lumber vertebra were acquired by using a desktop microtomographic imaging system (MicroCT40: Scanco Medical, Basserdorf, Switzerland) as described previously (23–25). We assessed trabecular bone morphology in the distal femoral metaphysis and the fifth lumbar vertebral body and cortical bone morphology at the femoral mid-shaft (voxel size = 12 μm). For trabecular bone, we measured bone volume fraction (BV/TV, %), trabecular number (Tb.N, 1/mm), and trabecular separation (Tb.Sp, μm) (see Fig. 3). For cortical bone, we assess the total cross-sectional area (TA, mm2), cortical bone area (BA, mm2), cortical thickness (μm), and the polar moment of inertia (MOIpolar, mm4) (see Fig. 3).
FIGURE 3.
Lack of Capn4 in osteoblasts causes reduced trabecular bone with thinner cortices in adult mutant mice. A, histomorphometric analysis was performed using tibias of Capn4flox/flox control (n = 6, open column) and Osx-Cre+/-Capn4flox/flox mutant (n = 6, closed column) male mice. Bone volume per total volume (BV/TV); trabecular spacing (Tb.Sp.); trabecular number (Tb.N.); bone formation rate per total volume (BFR/TV); mineral apposition rate (MAR); osteoblast number per total area (Ob/TA); and osteoclast number per total area (OC/TA), as determined by static and dynamic histomorphometry. B, lack of Capn4 in osteoblasts reduces total and cortical bone areas, cortical thickness, and indices of mechanical strength as determined by μCT analysis of the femoral mid-shaft diaphysis. The polar moments of inertia (MOIpolar). Capn4flox/flox control (n = 5, open column) and Osx-Cre+/-Capn4flox/flox mutant (n = 6, closed column) male mice. *, p < 0.05.
Analysis of Bromodeoxyuridine Incorporation—Two-week-old mice were injected intraperitoneally with 100 μg of bromodeoxyuridine (BrdUrd) and 12 μg of fluorodeoxyuridine per g of body weight 2 h before sacrifice (Sigma-Aldrich). After sacrifice, hind limbs were dissected, fixed, decalcified, and embedded in paraffin, and longitudinal sections across the tibia and femur were obtained. To identify actively proliferating cells, nuclei that had incorporated BrdUrd were detected using a Zymed BrdUrd immunostaining kit (Zymed Laboratories Inc., South San Francisco, CA). For each section, high power field images of the primary spongiosa and the proliferating chondrocyte layer were photographed. All BrdUrd-positive (black) and BrdUrd-negative (light blue) nuclei in these zones were counted separately. Four or five nonconsecutive sections were counted for each of two or three Capn4flox/flox, Osx-Cre+/- Capn4flox/+, and Osx-Cre+/-Capn4flox/flox littermate specimens at each time point.
Determination of Apoptotic Osteoblasts in Vivo—Apoptotic osteoblasts were detected in sections of decalcified tibias of 2-week-old Capn4flox/flox, Osx-Cre+/-Capn4flox/+ mice, and Osx-Cre+/-Capn4flox/flox littermates by a terminal deoxynucleotidyl transferase-mediated nick-end labeling reaction using reagents from the in situ cell death detection kit, POD (Roche Diagnostics). The sections were counterstained with 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA).
Osteoblast Isolation and Culture—Osteoblasts were isolated from calvariae of newborn mice by serial digestion in α-minimum essential medium (αMEM) (Invitrogen) containing 0.25 mg/ml type I and 0.75 mg/ml type II collagenases (Worthington, Lakewood, NJ). Calvariae were digested for 15 min at 37 °C with constant agitation. The digestion solution was collected, washed with fresh medium, and digested five additional times. Digestions 3–6 were collected, washed with αMEM, and cultured in αMEM supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 1% penicillin/streptomycin (Invitrogen Corp.) for 48 h. Medium was changed every other day.
Adenovirus Infection—Monolayer Capn4flox/flox osteoblasts were infected with either control (adeno-lacZ) or Cre-recombinase adenovirus (adeno-Cre) at a multiplicity of infection of 100 for most experiments (26). Osteoblasts were harvested after 48 h. Genomic DNA and total RNA were extracted from osteoblasts for determination of efficiency of Cre-recombinase.
Alkaline Phosphatase (ALP) Activity—ALP activity was measured in cell layers using a p-nitrophenyl phosphate substrate (Sigma-Aldrich) as described previously (27). Cells were cultured in αMEM containing 10% fetal bovine serum, 1% penicillin/streptomycin for 7, 14, and 21 days, respectively.
Bone Nodule Assay—Cells were cultured in αMEM with 10% fetal bovine serum and 1% penicillin/streptomycin supplemented with 50 μg/ml l-ascorbic acid and 10 mm β-glycerophosphate (Sigma-Aldrich) (differentiation medium) for 21 and 28 days, respectively. Cells were then fixed with 70% ethanol, and mineralized calcium-phosphate deposits were stained with either Alizarin red S or von Kossa. Bone nodules were counted using a dissecting microscope. In some experiments, Alizarin red S dye was eluted and quantified as described elsewhere (27).
Real-time Quantitative PCR (qPCR)—The efficiency of Cre-recombinase driven by the osterix promoter or adeno-Cre was assessed using real-time qPCR on genomic DNA isolated from tibiae of Osx-Cre+/-Capn4flox/flox and Capn4flox/flox mice and from primary osteoblasts harvested from Capn4flox/flox mice infected with either adeno-lacZ or adeno-Cre. In brief, tibial diaphyses of Osx-Cre+/-Capn4flox/flox and Capn4flox/flox mice were isolated, extensively flushed in phosphate-buffered saline, and incubated in αMEM containing collagenases I and II at 37 °C for 45 min to remove hematopoietic cells and soft tissues. The diaphyses were then crushed and further digested with collagenase in αMEM medium. Genomic DNA was isolated using standard protocols. Real-time qPCR was then performed at 60 °C for 40 cycles in the Opticon continuous fluorescence detector by using a SYBR Green PCR kit (Qiagen) and primers P3 and P5. To determine RNA expression of various genes, total RNA was extracted from cells by using the RNeasy Mini kit (Qiagen). First-strand cDNA was synthesized using the First Strand cDNA synthesis kit for reverse transcription-PCR (Roche Diagnostics) followed by real-time qPCR. Samples were run in duplicate, and the results were normalized to GAPDH expression (26). Primer sequences were as follows: ALP, P6 (5′-CACGCGATGCAACACCACTCAGG-3′) and P7 (5′-GCATGTCCCCGGGCTCAAAGA-3′); Capn4, P8 (5′-CCAGCTGGCTGGAGACGAC-3′) and P9 (5′-GCGGCCTCAAAGGCGCCTG-3′); Col.1, P10 (5′-CACCCTCAAGAGCCTG AGTC-3′) and P11 (5′-GTTCGGGCTGATGTACCAGT-3′); GAPDH, P12 (5′-AACTACATGGTCTACATGTTCCA-3′) and P13 (5′-CCATTCTCGGCCTTGACTGT-3′); NaPi3, P14 (5′-CACCCATATGGCTTCTGCTT-3′) and P15 (5′-CAGGAATTCATAGCCCAGGA-3); OC, P16 (5′-ACCCTGGCTGCGCTCTGTCTCT-3′) and P17 (5′-GATGCGTTTGTAGGCGGTCTTCA-3′); OP, P18 (5′-CTCCTTGCGCCACAGAATG-3′) and P19 (5′-TGGGCAACAGGGATGACA-3′); PGK, P20 (5′-GGAACGGGTCGTGATGA-3′) and P21 (5′-GC CTTGATCCTTTGGTTGTTTT-3′); Runx2, P22 (5′-AGCTACGAAATGCCTCTGCTG-3′) and P23 (5′-GATCGTTGAACCTGGCCACT-3′); and c-fos P24 (5′-CCACGGTGACAGCCATCTCCACCA-3′) and P25 (5′-GGCTGCAGCCATCTTATTCCTTTCCC-3′) (19, 28–30).
Osteoblast Apoptosis Assay in Vitro—Forty-eight h after adenoviral infection, osteoblasts were replated in 24-well plates at a density of 2.5 × 104 cells/cm2. Cells were stained with annexin V-PE and 7-aminoactinomycin D using Guava PCA Nexin kit and analyzed by Guava personal cytometer (Guava Technology Inc., Hayward, CA).
Gene Silencing by siRNA—UMR106-01 (UMR) rat osteosarcoma cells were plated at a density of 5 × 104 cells/cm2 in 24-well plates. Capn4 and control nonfunctional siRNA were transfected into UMR cells using Oligofectamine as recommended by the manufacture (Invitrogen). Cells were harvested 48 h after transfection.
Commercially available Capn4 siRNA sequences are as follows: siRNA/Capn4-1, P26/P27 (5′-ACUGACCGACAGACCAUUGGUA-3′ and 5′-UACCAAUGGUCGAUCGGUCAGU-3′); siRNA/Capn4-2, P28/P29 (5′-GGAUCCGACUCUGCUGCAAACAUGU-3′ and 5′-ACAUGUUUGCAGCAGAGUCGGUC-3′) (Invitrogen). Efficiency of Capn4 siRNA was assessed by calpain activity as described previously (2).
Statistics—Data were calculated from 3 to 5 independent experiments and expressed as the mean ± S.E. of either duplicate or triplicate determinations. Statistical analysis was performed using the analysis of variance or the unpaired Student's t test. Statistical significance was determined using Fisher's projected least significant difference, and p values less than 0.05 were accepted as significant.
RESULTS
Generation of Mice Lacking Capn4 in Cells of the Osteoblast Lineage and Characterization of Their Gross Phenotype—Mice expressing Cre under the control of the osterix promoter (Osx-Cre+/-) were crossed with floxed Capn4 mice (Capn4flox/flox) to produce Osx-Cre+/-Capn4flox/+ mice, which were then mated with either Capn4flox/flox or Capn4flox/+ to generate osteoblast-specific Capn4 knock-out mice, Osx-Cre+/-Capn4flox/flox. Osx-Cre+/-Capn4flox/+, Osx-Cre+/-Capn4+/+ and Capn4flox/flox were also used in the study as control littermates. Efficiency of Cre activity assessed by real-time qPCR of either genomic DNA or RNA isolated from 4-week-old mutant tibiae was ∼85% (genomic DNA, 86.5 ± 2.5%; RNA, 88.7 ± 0.8%).
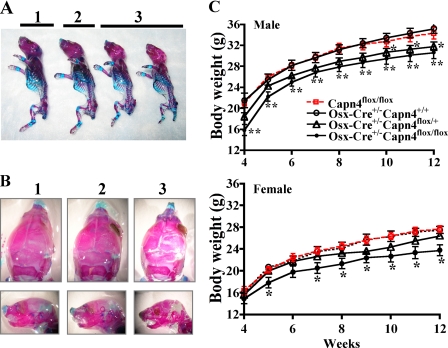
Osx-Cre+/-Capn4flox/flox (mutant) mice were born at the expected Mendelian frequency and were viable into adulthood and fertile. Their mean body weight in both sexes was reduced starting from 4–5 weeks of age in comparison with Osx-Cre+/-Capn4flox/+, Capn4flox/flox, and Osx-Cre+/-Capn4+/+ control littermates. Surprisingly, at 10 weeks of age, body weight of Osx-Cre+/-Capn4flox/+ male mice was also significantly lower than that of the other controls, Capn4flox/flox and Osx-Cre+/-Capn4+/+ (Fig. 1C). Ionized calcium levels of 8-week-old mice were similar between mutant and control littermates.
FIGURE 1.
Lack of Capn4 in cells of the osteoblast lineage causes delay in skeletal development. A and B, whole mount skeletal staining with Alizarin red S and Alcian blue. The newborn skeleton of Osx-Cre+/- Capn4flox/flox mice was smaller with no obvious patterning defects (A) but exhibited impaired mineralization of the skull (B) when compared with the controls. 1, 2, and 3 indicate Capn4flox/flox, Osx-Cre+/-Capn4flox/+, and Osx-Cre+/-Capn4flox/flox, respectively. C, the mean body weight of Osx-Cre+/-Capn4flox/flox mice was significantly reduced in both males and females when compared with Capn4flox/flox, Osx-Cre+/-Capn4flox/+, and Osx-Cre+/-Capn4+/+ littermates. Capn4flox/flox (10 males; 10 females), Osx-Cre+/-Capn4+/+ (10 males; 6 females), Osx-Cre+/-Capn4flox/+ (15 males; 11 females), and Osx-Cre+/-Capn4flox/flox (10 males; 6 females) were weighed weekly from 4 to 12 weeks. *, p < 0.05 and **, p < 0.001.
The newborn skeleton of Osx-Cre+/-Capn4flox/flox mice was definitively smaller than that of Capn4flox/flox and Osx-Cre+/- Capn4flox/+ littermates but did not exhibit any obvious patterning defect (Fig. 1A). Notably, mineralization of the skull was strikingly impaired in Osx-Cre+/-Capn4flox/flox mice (Fig. 1B).
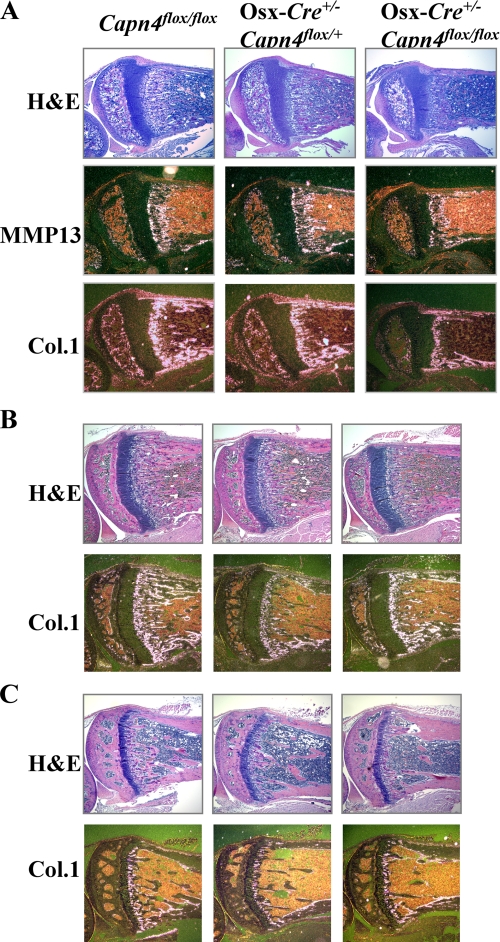
Osx-Cre+/-Capn4flox/flox Mice Have Decreased Trabecular Bone and Reduced Expression of Osteoblast-specific Markers— Detailed histological and in situ hybridization analyses were then performed on hind limbs isolated from 2-, 4-, and 9-week-old male mice, respectively, to investigate a putative role of Capn4 in bone modeling and remodeling. Tibiae of 2-week-old Osx-Cre+/- Capn4flox/flox mice had markedly reduced trabecular bone when compared with both Osx-Cre+/-Capn4flox/+ and Capn4flox/flox control littermates (Fig. 2A). Moreover, expression of osteoblast markers such as MMP13, Col.1, and OC was severely impaired in mutant bones (Fig. 2A and data not shown). A similar phenotype was also detectable in 4- and 9-week-old mutant mice of either sex (Fig. 2, B and C, and data not shown). Surprisingly, at these ages, Osx-Cre+/- Capn4flox/flox/+ mice displayed a loss of trabecular bone, which was virtually indistinguishable from that of Osx-Cre+/- Capn4flox/flox mutants, as shown by routine histology and detection of Col.1 mRNA by in situ hybridization analysis (Fig. 2, B and C). Conversely, Osx-Cre+/-Capn4+/+ mice did not present any detectable bone abnormality at any prenatal or postnatal age. μCT analysis showed no significant differences in 12-week-old male and female Osx-Cre+/-Capn4+/+ versus Capn4flox/flox when compared with parameters such as bone volume per total volume (16.4 ± 2.0 versus 15.3 ± 4.0%), trabecular number (5.1 ± 0.2 versus 4.6 ± 0.8/mm), and trabecular spacing (187.3 ± 10.2 versus 216.85 ± 44.5 μm) (data for female not shown). These results, thus, exclude any adverse effect of the Cre transgene per se on bone homeostasis. It is, therefore, likely that lack of a single Capn4 allele in cells of the osteoblast lineage is sufficient to cause bone loss, which, however, is detectable exclusively in adult mice.
FIGURE 2.
Lack of Capn4 in cells of the osteoblast lineage causes osteoporotic bone phenotype. A, lack of Capn4 in osteoblasts reduces expression of osteoblast-specific markers in tibias of 2-week-old Osx-Cre+Capn4flox/flox male mice. H&E, hematoxylin and eosin. B and C, lack of Capn4 in osteoblasts reduces trabecular bone in Osx-Cre+/-Capn4flox/+ and Osx-Cre+/--Capn4flox/flox mice in 4-week-old (B) and 9-week-old males (C). In situ hybridization was performed using a ribo-probes detecting Col.1 and MMP13 mRNA.
Histomorphometric and μCT analyses were performed on bone specimens of Capn4flox/flox and Osx-Cre+/-Capn4flox/flox mice and confirmed a significant decrease of bone volume and trabecular number with concomitant increase of trabecular spacing in Osx-Cre+/-Capn4flox/flox when compared with Capn4flox/flox control male mice at 12 weeks of age (Fig. 3A and data not shown). Moreover, bone formation rate, osteoblast number, and mineral apposition rate were all significantly reduced in Osx-Cre+/- Capn4flox/flox (Fig. 3A). Osteoclast number was also significantly decreased in mutants versus controls as indicated by both histomorphometry and TRAP staining (Fig. 3A and data not shown). Taken together, these data suggest that bone loss observed in Osx-Cre+/- Capn4flox/flox mice is likely the consequence of decreased osteoblast number and activity. Consistent with the findings described above, serum levels of both osteocalcin and TRACP 5b, markers of osteoblast and osteoclast activities, respectively, were markedly reduced in mutant mice (osteocalcin; Capn4flox/flox (n = 7), 60.0 ± 5.3 ng/ml; Osx-Cre+/-Capn4flox/flox (n = 9), 39.7 ± 2.6* ng/ml. TRACP5b; Capn4flox/flox (n = 6), 3.9 ± 0.6 units/liter; Osx-Cre+/– Capn4flox/flox (n = 10), 2.7 ± 0.3* units/liter. *, p < 0.05).
Lack of Capn4 in Cells of the Osteoblast Lineage Reduces Bone Cortical Thickness and Bone Architecture—Cortical bone morphology was examined by μCT analysis at the mid-femoral diaphysis. Total and cortical bone areas and cortical thickness were significantly reduced in Osx-Cre+/-Capn4flox/flox mice versus controls, suggesting that the bone is smaller and that periosteal apposition is inhibited in Osx-Cre+/-Capn4flox/flox mice (Fig. 3B). In addition, the polar moment of inertia, an index of torsional strength, was significantly lower in mutants than controls (Fig. 3B). Thus, deletion of Capn4 in cells of the osteoblast lineage causes bone loss in both trabecular and cortical compartments and severely affects parameters that predict bone strength.
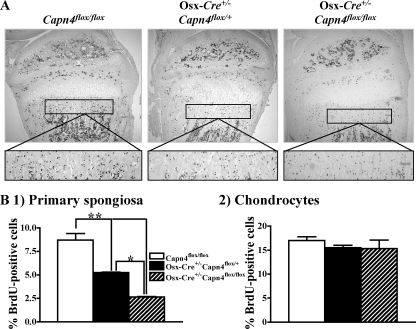
Lack of Capn4 Impairs Proliferation of Cells of the Osteoblast Lineage—The reduced osteoblast number observed in Osx-Cre+/-Capn4flox/flox mice could be a consequence of either decreased proliferation or increased apoptosis. To distinguish between these two possibilities, proliferation of cells of the osteoblast lineage was examined by BrdUrd incorporation in 1- and 2-week-old mice. The number of BrdUrd-positive cells detected in the primary spongiosa was significantly reduced in Osx-Cre+/-Capn4flox/flox when compared with Osx-Cre+/- Capn4flox/+ and Capn4flox/flox littermates (Fig. 4B, panel 1), whereas chondrocyte proliferation, which served as an internal control for proper BrdUrd uptake and distribution, was not affected (Fig. 4B, panel 2). No significant difference in the number of apoptotic cells determined by terminal deoxynucleotidyl transferase-mediated nick-end labeling staining was detected in 2-week-old mutant bones versus controls (data not shown). Collectively, these data indicate that the reduced osteoblast number observed in Osx-Cre+/-Capn4flox/flox is likely due to reduced proliferation of cells of the osteoblast lineage.
FIGURE 4.
Lack of Capn4 impairs proliferation of cell of the osteoblast lineage in vivo. A, incorporated BrdUrd was detected by immunostaining using anti-BrdUrd antibody in proximal tibias of 2-week-old male mice. B, quantitative analysis of BrdUrd (BrdU)-positive osteoblasts in primary spongiosa (panel 1) and of chondrocytes in the proliferative layer of the growth plate (panel 2). *, p < 0.05, **, p < 0.01.
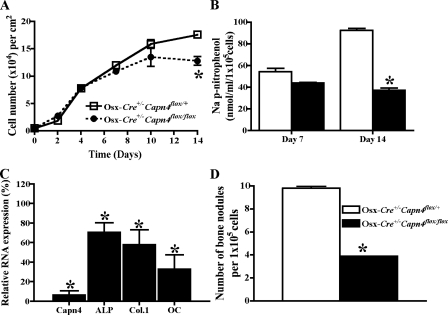
Lack of Capn4 Impairs Osteoblast Differentiation and Mineralization in Vitro—Osteoblast functions were next investigated in vitro. For this purpose, primary calvarial osteoblasts were harvested from Osx-Cre+/-Capn4flox/+ and Osx-Cre+/- Capn4flox/flox newborn mice and seeded at a density of 5.0 × 103 cells/cm2. Levels of Capn4 mRNA expression were reduced by ∼90% in Osx-Cre+/-Capn4flox/flox osteoblasts in comparison with Osx-Cre+/-Capn4flox/+. Cell growth was indistinguishable between Osx-Cre+/-Capn4flox/flox and Osx-Cre+/- Capn4flox/+ calvarial cells until day 10 and became significantly lower in Osx-Cre+/-Capn4flox/flox cells than Osx-Cre+/-Capn4flox/+ cells on day 14. Osteoblast differentiation and mineralization, assessed by ALP activity and mRNA expression of various osteoblast markers, and number of bone nodules, normalized by cell number at the time of assay, respectively, were all significantly lower in Osx-Cre+/-Capn4flox/flox than in Osx-Cre+/-Capn4flox/+ calvarial cells (Fig. 5).
FIGURE 5.
Characterization of primary osteoblasts harvested from Osx-Cre+/-Capn4flox/+ and Osx-Cre+/-Capn4flox/flox mice. A, cell number was reduced in Osx-Cre+/-Capn4flox/flox calvarial cells. Cell number was counted on days 0, 2, 5, 7,10, and 14. B, ALP activity was lower in Osx-Cre+/-Capn4flox/flox calvarial cells. ALP activity was measured on days 7 and 14 and adjusted by cell number at the time of assay. C, expression of markers for osteoblast differentiation was reduced in Osx-Cre+/-Capn4flox/flox calvarial cells. Relative RNA expression of Capn4, ALP, Col.1, and OC was determined using real-time qPCR after 7 days in culture. D, osteoblast mineralization was reduced in Osx-Cre+/-Capn4flox/flox calvarial cells. Cells were seeded at the density of 5 × 103 cells/cm2 and cultured in differentiation medium for 21 days. Formation of bone nodules was assessed by Alizarin red S staining. *, p < 0.05.
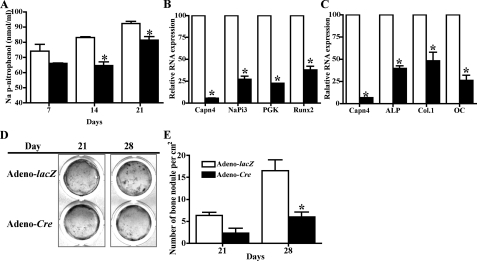
Next, to further investigate a role of Capn4 in osteoblast differentiation, calvarial cells isolated from Capn4flox/flox mice were infected with adenovirus expressing either Cre-recombinase (adeno-Cre) or β-galactosidase (adeno-lacZ). The efficiency of Cre-recombinase was assessed at both genomic DNA and RNA levels using real-time qPCR. Approximately 85% of Capn4flox alleles were successfully excised in adeno-Cre-infected cells, and their levels of Capn4 mRNA expression were reduced to less than 10% of those observed in adeno-lacZ-infected control cells. Forty-eight h after adenoviral infection, cells were replated at the density of 1.3 × 105 cells/cm2 (higher density) to allow them to reach an indistinguishable cell number at the time of assay and, thus, minimize the confounding effect of impaired proliferation on the differentiation process. Notably, cell number of both mutant and control cells similarly increased and reached a plateau around day 10, when cells were seeded at the high density (data not shown). When the number of the apoptotic events was determined by annexin V staining, no significant difference was detected in Capn4-null calvarial cells when compared with the controls (ratio of annexin V-positive early and late apoptotic cells per total cells; control, 14.3 ± 1.8%; Capn4-null, 15.2 ± 1.3%). Consistent with the in vivo data, calvarial cells lacking Capn4 showed significantly reduced ALP activity (Fig. 6A) as well as impaired expression of several genes including NaPi3, PGK, and Runx2 that are early markers of osteoblast differentiation (Fig. 6B) (28). Expression of late markers of osteoblast differentiation such as ALP, Col.1, and OC was also significantly down-regulated in Capn4-null cells versus controls (Fig. 6C). Lastly, Capn4-null cells grown in differentiation medium for 21 and 28 days, respectively, formed only 33% of the bone nodules observed in control cells (Fig. 6, D and E). Taken together, these in vitro results confirm that lack of Capn4 impairs both proliferation and differentiation of cells of the osteoblast lineage as indicated by the in vivo model.
FIGURE 6.
Characterization of primary calvarial cells harvested form Capn4flox/flox mice and infected with adenovirus carrying either lacZ or Cre. Cells were infected with either adeno-LacZ or adeno-Cre and, 48 h later, were replated at the density of 1.3 × 105 cells/cm2. A, lack of Capn4 in osteoblasts reduced ALP activity in 14- and 21-day culture. B and C, relative RNA expression of early (B; 3-day incubation) and late (C; 14-day incubation) markers for osteoblast differentiation was reduced in Capn4-null primary calvarial cells. D and E, lack of Capn4 in calvarial cells impaired osteoblast mineralization. Images of bone nodules detected by von Kossa staining (D) and quantitative analysis of the number of bone nodules (E) are shown. *, p < 0.05.
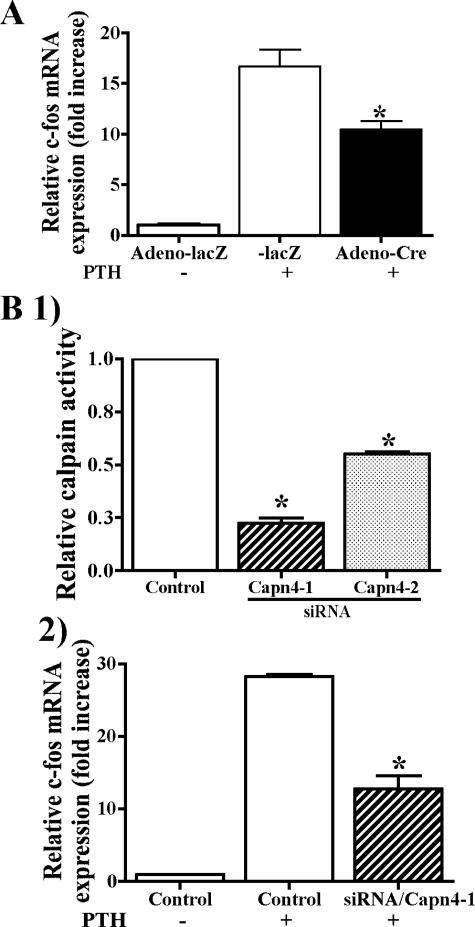
Lack of Capn4 in Cells of the Osteoblast Lineage Reduced PTH-stimulated c-fos mRNA Expression in Vitro—To further examine an effect of Capn4 ablation on gene transcription in cells of the osteoblast lineage, primary calvarial cells were treated with PTH for 0, 1, and 2 h, and levels of c-fos mRNA expression were assessed using real-time qPCR. c-fos is a key downstream target gene of PTH, and its expression increases rapidly and transiently upon PTH stimulation (31–33). In adeno-lacZ-infected control calvarial cells, PTH treatment for 1 and 2 h increased c-fos mRNA levels 17- and 9-fold above basal levels, respectively. This effect was reduced by ∼35% in adeno-Cre-infected Capn4-null calvarial cells (Fig. 7A). We also silenced the calpain small subunit in UMR cells by using siRNA technology. siRNAs that specifically targeted Capn4 (Capn4-1 and Capn4-2) reduced calpain activity to 25 and 55%, respectively, when compared with control siRNA (Fig. 7B, panel 1). Capn4-knock-out in UMR cells resulted in a significant impairment of PTH-mediated increase of c-fos expression (Fig. 7B, panel 2). Cell surface expression of the receptor for PTH and PTH-related peptide determined by ligand binding assay was indistinguishable between control and Capn4-null cells (data not shown). Collectively, these results suggest that Capn4 could regulate osteoblast function, at least in part, by modulating PTH activity in these cells.
FIGURE 7.
Lack of Capn4 reduced PTH-stimulated c-fos mRNA expression in cells of the osteoblast lineage. A, primary calvarial cells harvested from Capn4flox/flox mice were infected with either adeno-lacZ or -Cre. Forty-eight h later, cells were stimulated with 1 × 10-7 m PTH for 1 h, and c-fos mRNA expression was quantified using real-time qPCR. The mean value of c-fos expression before PTH stimulation in control cells was set as 1. B, panel 1, efficiency of Capn4 siRNA in silencing Capn4 was assessed by calpain activity. UMR cells were transfected with siRNAs that specifically targeted Capn4 (Capn4-1 and Capn4-2) for 24 h and incubated in fresh medium for additional 24 h. Calpain activity was reduced to 25 and 55% in UMR cells transfected with siRNA/Capn4-1 and -2, respectively. Panel 2, control and Capn4-null (siRNA/Capn4-1) calvarial cells were treated with 1 × 10-7 m PTH for 1 h, and c-fos mRNA expression was quantified using real-time qPCR. *, p < 0.05.
DISCUSSION
Here we report the novel finding that Capn4 is critically important in bone development and remodeling in vivo. Given the early lethality of the universal Capn4 knock-out embryos, Osx-Cre+/-Capn4flox/flox was an appropriate animal model to investigate the role of the calpain small subunit in cells of the osteoblast lineage (5, 6).
Calpains belong to a family of intracellular cysteine proteases that have been shown to cleave numerous and diverse substrates (1). Our study provided direct evidence that lack of Capn4 severely affects proliferation of cells of the osteoblast lineage both in vivo and in vitro. Consistent with these findings, it was previously reported that a cell-permeable calpain inhibitor attenuates proliferation in the MC3T3-E1 cell line (10, 11). The role of calpain in the cell cycle has been controversial. Capn4-null embryonic fibroblasts with no calpain activity proliferate normally (5). However, several studies have suggested that calpain plays a role in the cell cycle, mainly at G1 to S transition, by regulating proteolysis of various proteins such as cyclin D and A, cyclin-dependent kinase 2, and p53 (31–33). Our novel animal model demonstrates that regulation of cell proliferation is indeed a crucial biological function of the calpain small subunit, at least in osteoblasts.
Lack of Capn4 in osteoblasts significantly reduces ALP activity and expression of differentiation markers such as ALP, Col.1, OC, and Runx2 mRNAs. Consistent with our findings, a cell-permeable calpain inhibitor was reported to attenuate osteoblast differentiation in the MC3T3-E1 cell line (11). The finding constitutes the first in vivo evidence that Capn4 is an essential modulator of cell terminal differentiation in osteoblasts. Notably, immunostaining for μ- and m-calpains performed in the MC3T3-E1 cell line revealed that m-calpain is a cytoplasmic protein, whereas μ-calpain is both nuclear and cytoplasmic, which implies that calpains may control gene expression directly at the transcriptional level (11).
It has been reported that PTH stimulates expression of the immediate early gene, c-fos, which has a critical role in bone biology, by regulating phosphorylation of the cAMP response element binding protein at serine 133 by protein kinase A (34–38). Moreover, we have previously reported that MC3T3-E1 osteoblastic cells stably expressing calpastatin show markedly reduced PTH-mediated cAMP accumulation (2). Lastly, our present study indicates that lack of Capn4 reduces PTH-stimulated expression of c-fos in both primary calvarial and UMR cells. Collectively, these data thus suggest that Capn4 may regulate osteoblast functions, at least in part, by modulating PTH activity in these cells.
A role of calpains in bone homeostasis has been previously proposed by the analysis of universal μ-calpain knock-out mice, which develop osteopenia secondary to an increased osteoclast activity. No detectable osteoblast phenotype was reported in these mutant mice (39). Thus, their bone phenotype clearly differs from that of the osteoblast-specific Capn4 knock-out mice, which showed both impaired osteoblast number and function and reduced osteoclast number and activity. Altogether, the phenotype of the Capn4 knock-out mice may reflect either loss of m-calpain function or loss of both m-calpains and μ-calpains function in osteoblasts.
Several lines of evidence indicated that lack of a single Capn4 allele in cells of the osteoblast lineage causes a detectable bone phenotype. Osx-Cre+/-Capn4flox/+ mice started to lose body weight around 6 weeks of age, and their mean body weight became indistinguishable from that of Osx-Cre+/- Capn4flox/flox mice at 10 weeks of age. Although body weight only serves as an indirect indicator of skeletal growth, the data suggest impaired skeletal growth in Osx-Cre+/-Capn4flox/+ mice. Moreover, histological analysis showed that Osx-Cre+/-Capn4flox/+ mice rapidly develop an osteoporotic phenotype between 2 and 4 weeks of age. Lastly, proliferation of cells of the osteoblast lineage as assessed by BrdUrd incorporation at 2 weeks was significantly reduced in Osx-Cre+/-Capn4flox/+ mice when compared with Capn4flox/flox mice, although it was still ∼2-fold higher than in Osx-Cre+/-Capn4flox/flox specimens. Importantly, Osx-Cre+/- Capn4+/+ were, virtually and as confirmed by μCT analysis, indistinguishable from wild-type controls, which indicates that the bone phenotype of Osx-Cre+/- Capn4flox/+ was not the mere consequence of a nonspecific effect secondary to the presence of the Cre transgene in cells of the osteoblast lineage. These results are suggestive of a possible haploinsufficiency phenotype in mice lacking one allele of Capn4 in cells of the osteoblast lineage.
In summary, lack of Capn4 in cell of the osteoblast lineage negatively affects both trabecular and cortical bone by impairing proliferation and differentiation of osteoblastic cells. Our data thus indicate that calpains could be important molecules in pathological conditions such as osteoporosis.
Acknowledgments
We thank Dr. Henry M. Kronenberg for critical review of the manuscript and helpful discussion. We also thank Clare Thomas, Kimberly Atkin, Cornelia Withington, Dilani Rossa, and Yuko Sumiyama at MGH, and David Panus at Beth Israel Deaconess Medical Center for technical supports.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK072102 (to M. S.). This work was also supported by Canadian Institutes of Health Research Grant MOP-81189 (to P. A. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PTH, parathyroid hormone; μCT, microcomputed tomography; BrdUrd, bromodeoxyuridine; αMEM, α-minimum essential medium; ALP, alkaline phosphatase; qPCR, quantitative PCR; siRNA, small interfering RNA; UMR, UMR106-01; kbp, kilobase pairs.
References
- 1.Goll, D. E., Thompson, V. F., Li, H., Wei, W., and Cong, J. (2003) Physiol. Rev. 83 731-801 [DOI] [PubMed] [Google Scholar]
- 2.Shimada, M., Mahon, M. J., Greer, P. A., and Segre, G. V. (2005) Endocrinology 146 2336-2344 [DOI] [PubMed] [Google Scholar]
- 3.Sternberg, N., and Hamilton, D. (1981) J. Mol. Biol. 150 467-486 [DOI] [PubMed] [Google Scholar]
- 4.Cong, J., Goll, D. E., Peterson, A. M., and Kapprell, H. P. (1989) J. Biol. Chem. 264 10096-10103 [PubMed] [Google Scholar]
- 5.Arthur, J. S., Elce, J. S., Hegadorn, C., Williams, K., and Greer, P. A. (2000) Mol. Cell Biol. 20 4474-4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerman, U. J., Boring, L., Pak, J. H., Mukerjee, N., and Wang, K. K. (2000) IUBMB Life 50 63-68 [DOI] [PubMed] [Google Scholar]
- 7.Tram, K. K., Spencer, M. J., Murray, S. S., Lee, D. B., Tidball, J. G., and Murray, E. J. (1993) Biochem. Mol. Biol. Int. 29 981-987 [PubMed] [Google Scholar]
- 8.Tram, K. K., Murray, S. S., Lee, D. B., and Murray, E. J. (1993) Kidney Int. 43 693-699 [DOI] [PubMed] [Google Scholar]
- 9.Murray, E. J., Murray, S. S., Tram, K. K., and Lee, D. B. (1994) Exp. Cell Res. 215 241-248 [DOI] [PubMed] [Google Scholar]
- 10.Murray, E. J., Grisanti, M. S., Bentley, G. V., and Murray, S. S. (1997) Metab. Clin. Exp. 46 1090-1094 [DOI] [PubMed] [Google Scholar]
- 11.Murray, S. S., Grisanti, M. S., Bentley, G. V., Kahn, A. J., Urist, M. R., and Murray, E. J. (1997) Exp. Cell Res. 233 297-309 [DOI] [PubMed] [Google Scholar]
- 12.Reeve, J., Hesp, R., Williams, D., Hulme, P., Klenerman, L., Zanelli, J. M., Darby, A. J., Tregear, G. W., and Parsons, J. A. (1976) Lancet 1 1035-1038 [DOI] [PubMed] [Google Scholar]
- 13.Kronenberg, H. M., Lanske, B., Kovacs, C. S., Chung, U. I., Lee, K., Segre, G. V., Schipani, E., and Juppner, H. (1998) Recent Prog. Horm. Res. 53 283-303 [PubMed] [Google Scholar]
- 14.Schipani, E., and Provot, S. (2003) Birth Defects Res. Part C. Embryo Today Rev. 69 352-362 [DOI] [PubMed] [Google Scholar]
- 15.Tan, Y., Dourdin, N., Wu, C., De Veyra, T., Elce, J. S., and Greer, P. A. (2006) Genes. J. Genet. Dev. 44 297-303 [DOI] [PubMed] [Google Scholar]
- 16.Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R., and de Crombrugghe, B. (2002) Cell 108 17-29 [DOI] [PubMed] [Google Scholar]
- 17.Rodda, S. J., and McMahon, A. P. (2006) Development (Camb.) 133 3231-3244 [DOI] [PubMed] [Google Scholar]
- 18.Shimada, M., Shimano, H., Gotoda, T., Yamamoto, K., Kawamura, M., Inaba, T., Yazaki, Y., and Yamada, N. (1993) J. Biol. Chem. 268 17924-17929 [PubMed] [Google Scholar]
- 19.Arthur, J. S., Greer, P. A., and Elce, J. S. (1998) Biochim. Biophys. Acta 1388 247-252 [DOI] [PubMed] [Google Scholar]
- 20.McLeod, M. J. (1980) Teratology 22 299-301 [DOI] [PubMed] [Google Scholar]
- 21.Calvi, L. M., Sims, N. A., Hunzelman, J. L., Knight, M. C., Giovannetti, A., Saxton, J. M., Kronenberg, H. M., Baron, R., and Schipani, E. (2001) J. Clin. Investig. 107 277-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, K., Lanske, B., Karaplis, A. C., Deeds, J. D., Kohno, H., Nissenson, R. A., Kronenberg, H. M., and Segre, G. V. (1996) Endocrinology 137 5109-5118 [DOI] [PubMed] [Google Scholar]
- 23.Bouxsein, M. L., Pierroz, D. D., Glatt, V., Goddard, D. S., Cavat, F., Rizzoli, R., and Ferrari, S. L. (2005) J. Bone Miner. Res. 20 635-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouxsein, M. L., Myers, K. S., Shultz, K. L., Donahue, L. R., Rosen, C. J., and Beamer, W. G. (2005) J. Bone Miner. Res. 20 1085-1092 [DOI] [PubMed] [Google Scholar]
- 25.Glatt, V., Canalis, E., Stadmeyer, L., and Bouxsein, M. L. (2007) J. Bone Miner. Res. 22 1197-1207 [DOI] [PubMed] [Google Scholar]
- 26.Pfander, D., Kobayashi, T., Knight, M. C., Zelzer, E., Chan, D. A., Olsen, B. R., Giaccia, A. J., Johnson, R. S., Haase, V. H., and Schipani, E. (2004) Development (Camb.) 131 2497-2508 [DOI] [PubMed] [Google Scholar]
- 27.Gori, F., Divieti, P., and Demay, M. B. (2001) J. Biol. Chem. 276 46515-46522 [DOI] [PubMed] [Google Scholar]
- 28.Liu, X., Bruxvoort, K. J., Zylstra, C. R., Liu, J., Cichowski, R., Faugere, M. C., Bouxsein, M. L., Wan, C., Williams, B. O., and Clemens, T. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2259-2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahdjoudj, S., Kaabeche, K., Holy, X., Fromigue, O., Modrowski, D., Zerath, E., and Marie, P. J. (2005) Exp. Cell Res. 303 138-147 [DOI] [PubMed] [Google Scholar]
- 30.Okahashi, N., Inaba, H., Nakagawa, I., Yamamura, T., Kuboniwa, M., Nakayama, K., Hamada, S., and Amano, A. (2004) Infect Immun. 72 1706-1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi, Y. H., Lee, S. J., Nguyen, P., Jang, J. S., Lee, J., Wu, M. L., Takano, E., Maki, M., Henkart, P. A., and Trepel, J. B. (1997) J. Biol. Chem. 272 28479-28484 [DOI] [PubMed] [Google Scholar]
- 32.Carragher, N. O., Westhoff, M. A., Riley, D., Potter, D. A., Dutt, P., Elce, J. S., Greer, P. A., and Frame, M. C. (2002) Mol. Cell Biol. 22 257-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, W., Lu, Q., Xie, Z. J., and Mellgren, R. L. (1997) Oncogene 14 255-263 [DOI] [PubMed] [Google Scholar]
- 34.Caubet, J. F., and Bernaudin, J. F. (1988) Biol. Cell 64 101-104 [DOI] [PubMed] [Google Scholar]
- 35.Lee, K., Deeds, J. D., Chiba, S., Un-No, M., Bond, A. T., and Segre, G. V. (1994) Endocrinology 134 441-450 [DOI] [PubMed] [Google Scholar]
- 36.Demiralp, B., Chen, H. L., Koh, A. J., Keller, E. T., and McCauley, L. K. (2002) Endocrinology 143 4038-4047 [DOI] [PubMed] [Google Scholar]
- 37.Pearman, A. T., Chou, W. Y., Bergman, K. D., Pulumati, M. R., and Partridge, N. C. (1996) J. Biol. Chem. 271 25715-25721 [DOI] [PubMed] [Google Scholar]
- 38.Tyson, D. R., Swarthout, J. T., and Partridge, N. C. (1999) Endocrinology 140 1255-1261 [DOI] [PubMed] [Google Scholar]
- 39.Marzia, M., Chiusaroli, R., Neff, L., Kim, N. Y., Chishti, A. H., Baron, R., and Horne, W. C. (2006) J. Biol. Chem. 281 9745-9754 [DOI] [PMC free article] [PubMed] [Google Scholar]