Abstract
The phenomenon of heterosis describes the increased agronomic performance of heterozygous F1 plants compared to their homozygous parental inbred plants. Heterosis is manifested during the early stages of root development in maize. The goal of this study was to identify nonadditive gene expression in primary roots of maize hybrids compared to the average expression levels of their parental inbred lines. To achieve this goal a two-step strategy was used. First, a microarray preselection of nonadditively expressed candidate genes was performed. Subsequently, gene expression levels in a subset of genes were determined via high-throughput quantitative real-time (qRT)–PCR experiments. Initial microarray experiments identified 1941 distinct microarray features that displayed nonadditive gene expression in at least 1 of the 12 analyzed hybrids compared to the midparent value of their parental inbred lines. Most nonadditively expressed genes were expressed between the parental values (>89%). Comparison of these 1941 genes with nonadditively expressed genes identified in maize shoot apical meristems via the same experimental procedure in the same genotypes revealed significantly less overlap than expected by pure chance. This finding suggests organ-specific patterns of nonadditively expressed genes. qRT–PCR analyses of 64 of the 1941 genes in four different hybrids revealed conserved patterns of nonadditively expressed genes in different hybrids. Subsequently, 22 of the 64 genes that displayed nonadditive expression in all four hybrids were analyzed in 12 hybrids that were generated from four inbred lines. Among those genes a superoxide dismutase 2 was expressed significantly above the midparent value in all 12 hybrids and might thus play a protective role in heterosis-related antioxidative defense in the primary root of maize hybrids. The findings of this study are consistent with the hypothesis that both global expression trends and the consistent differential expression of specific genes contribute to the organ-specific manifestation of heterosis.
HETEROSIS describes the superior performance of heterozygous F1-hybrid plants compared to the average of their homozygous parental inbred lines (Shull 1952; Falconer and Mackay 1996) and is of paramount importance in maize breeding. Heterosis was first described by Charles Darwin (Darwin 1876) and independently rediscovered by Shull (1908) and East (1908). Heterosis is most evident for adult traits like plant biomass or yield but is also apparent during embryo (Meyer et al. 2007) and early seedling development (Hoecker et al. 2006).
Several models to explain the genetic basis of heterosis have been suggested, including the dominance, overdominance, and epistasis hypotheses (Birchler et al. 2003, 2006; Hochholdinger and Hoecker 2007). All these hypotheses suggest that the contribution of many genes is responsible for the more vigorous phenotypes of hybrids over inbred lines. The dominance hypothesis explains heterosis by the complementing action of superior dominant alleles from both parental inbred lines at multiple loci over the corresponding unfavorable alleles, leading to improved vigor of hybrid plants (Davenport 1908; Bruce 1910; Keeble and Pellow 1910; Jones 1917). The overdominance hypothesis attributes heterosis to allelic interactions at one or multiple loci in hybrids that result in superior traits compared to the homozygous parental inbred lines (Shull 1908). Finally, the epistasis hypothesis considers epistatic interactions between nonallelic genes at two or more loci as the main factor for the superior phenotypic expression of a trait in hybrids (Powers 1945). It is important to keep in mind that these quantitative genetics hypotheses cannot be directly associated with the quantitative behavior of phenotypic traits or with molecular principles and that no strong consensus has emerged concerning the question of which of these hypotheses can explain heterosis best (Birchler et al. 2003, 2006).
Recently, the molecular analysis of heterosis in maize has been initiated. On the level of genome organization it has been demonstrated that the genetic colinearity is frequently violated between different inbred lines of maize (Fu and Dooner 2002; Song and Messing 2003; Brunner et al. 2005). This implies that genes are frequently present in one inbred line but are missing in another. Hemizygous complementation of many such genes with minor quantitative effects in hybrids might thus lead to a significantly increased performance of hybrid plants and would be consistent with the dominance hypothesis (Fu and Dooner 2002). However, in some instances the apparent loss of gene colinearity might be due to the movement of genes or gene fragments by helitron transposons to other genomic regions and thus the contribution of noncolinear regions of the genome to heterosis is unclear (Lai et al. 2005). Moreover, a number of studies have compared gene expression patterns of selected genes in inbred lines and hybrids (Song and Messing 2003; Auger et al. 2005; Meyer et al. 2007) or examined global gene expression analyses (Guo et al. 2003, 2006; Stupar and Springer 2006; Swanson-Wagner et al. 2006; Uzarowska et al. 2007) to test the hypothesis that nonadditive gene expression in hybrids is associated with heterosis (Song and Messing 2003). These studies differed significantly in their experimental design, analyzed plant tissues and developmental stages, genotypes, and statistical procedures, making it difficult to compare them. In general, global trends of gene expression were not uniform in these surveys. The terms additive and nonadditive are used to describe gene expression levels in hybrids with respect to the average (midparent) expression value of the two parental inbred lines. Nonadditive gene expression patterns in hybrids are significantly different from the midparent value, while additive expression patterns are not. While in some studies additive gene expression was prevalent (Stupar and Springer 2006; Swanson-Wagner et al. 2006; Meyer et al. 2007), in other studies nonadditive gene expression (Uzarowska et al. 2007) or a similar number of genes that provided additive and nonadditive expression (Guo et al. 2006) prevailed. Although this discrepancy in global gene expression patterns might be related to the different experimental approaches this could also be an indication of different global expression patterns in different tissues and developmental stages that might nevertheless be related to heterosis (Hochholdinger and Hoecker 2007). This notion is supported by the observations that different organs of a hybrid plant display significant differences in their degree of heterosis (Melchinger 1999) and that hybrid yield and heterosis for immature maize ears were positively associated with the proportion of allelic additivity in gene expression (Guo et al. 2006). Remarkably, in studies that analyzed the expression of more than one hybrid in a particular tissue (Guo et al. 2006; Uzarowska et al. 2007) it was not yet possible to identify key genes of heterosis that were nonadditively expressed between all studied inbred–hybrid combinations.
The goal of this study was to test three hypotheses: first, that nonadditive gene expression is observed during the early stages of the phenotypic manifestation of heterosis; second, that there is a consensus data set of nonadditively expressed genes in hybrids for different plant organs, e.g., primary roots and shoot apical meristems; and third, that there are genes that are consistently expressed in a nonadditive manner when different hybrid genotypes are analyzed for a particular plant organ.
MATERIALS AND METHODS
Plant material:
The maize inbred lines UH002 [National Listing of Plant Varieties (NLPV), accession no. (AC) M7830, European flint] and UH005 (NLPV AC M9379, European flint) from the flint pool, the inbred lines UH250 (NLPV AC M9005, Iowa Stiff Stalk) and UH301 (NLPV AC M8652, IoDent) from the dent pool, and the 12 possible reciprocal hybrid combinations derived from these inbred lines were generated in the nursery of the University of Hohenheim near Eckartsweier (Germany) in the summer season of 2003. Genetic distances of the different hybrids used in this study indicating the relation of the parental inbred lines are given in Hoecker et al. (2006) and range from 0.643 to 0.819. Although the genetic distances of the four inbred lines used in this study are relatively similar, intragroup crosses displayed a smaller genetic distance than intergroup crosses. However, the phenotypical differences of root traits between inbred lines and hybrids did not correlate with genetic distances since we surveyed the very early stages of heterosis manifestation a few days after germination (Hoecker et al. 2006).
Microarray hybridization, scanning, and spot quantification:
Seeds were surface sterilized with 6% hypochlorite for 6 min, thoroughly rinsed in distilled water, and germinated on moistened filter paper (20 × 70-cm grade 603 N; Sartorius, Göttingen, Germany) that was rolled up with 20 seeds per filter paper in a phytochamber at 26°, with a 16-hr light, 8-hr dark cycle and 60% humidity in twice-distilled water according to Hoecker et al. (2006). For subsequent molecular analyses 3.5-day-old primary roots were manually dissected with a razor blade, immediately frozen in liquid nitrogen, and stored at −80°. For each biological replicate ∼20 roots per germination roll and genotype were pooled and homogenized using the Micro-Dismembrator U (Sartorius). Isolation of total RNA with Trizol (Invitrogen, Carlsbad, CA), followed by mRNA purification using Oligotex mRNA columns (QIAGEN, Hilden, Germany), was performed according to the manufacturers' instructions. Reverse transcription of the mRNA to cDNA with concurrent incorporation of aminoallyl dUTPs as well as Cy3 and Cy5 labeling was performed according to Nakazono et al. (2003). Microarray probe hybridization of spotted 12k maize cDNA microarray chips (GenII vB, GPL 1996; http://www.plantgenomics.iastate.edu/maizechip) representing 10,649 nonredundant gene fragments was conducted as described in Woll et al. (2005). Samples from all 16 genotypes were paired on 84 arrays (supplemental material 1), following a hybridization scheme including a dye swap according to Keller et al. (2005). A tailor-made design was developed for this purpose, using simulated annealing that was optimized for estimating nonadditive effects while at the same time allowing estimation of additive effects with comparable precision. Optimizing precision for estimation of nonadditivity required the design to be unbalanced with respect to the number of genotype pairings and the number of replicates per genotype. Hence, two RNA samples were hybridized to each of the 84 microarray chips. Each RNA sample represented an independent biological replicate (supplemental material 1). Dried slides were scanned six times with an Array Scanner (Genetic MicroSystems) for each channel (Cy3 and Cy5), with laser power settings fixed at 90%. A series of up to six scans for each channel, in ascending order of PMT gain, was performed at 10-μm resolution individually adjusted to overall spot intensities per array according to Piepho et al. (2006). ImaGene software (Biodiscovery, Marina Del Rey, CA) was used to quantify the spot intensities on the slides by the use of the default settings.
Microarray data analysis:
The experimental design for the microarray experiments was described in Keller et al. (2005). To combine data from different scanning intensities, a nonlinear latent regression model was applied (Piepho et al. 2006). After combining the signals of different scans, a loess regression was performed. To account for differences between arrays the median absolute deviations (MAD) of the slides were normalized. The normalized data were analyzed by a linear mixed model for every gene that also accounted for the unbalanced design of the experiment. Effects that occur during hybridization (genotype, dye, slide) were considered as well as temporal and spatial effects that might affect the genetic material during cultivation in the phytochamber (Keller et al. 2005). The normalized data and raw data of all microarray chips have been deposited in the gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with the accession no. GSE10539. Pairwise contrasts between genotypes were estimated. Furthermore, linear contrasts between hybrid and parental mean were determined. The P-values of these contrasts based on Wald tests were adjusted for multiple testing by controlling the false discovery rate (FDR) at 5%, using the procedure of Benjamini and Hochberg (1995). The analyses for this article were generated using SAS software, Version 9. Linear model analyses were done by PROC MIXED, while the FDR adjustment was performed by PROC MULTTEST.
Quantitative real-time PCR and data analysis:
Total RNA for quantitative real-time (qRT)–PCR experiments was isolated with Trizol reagent from frozen 3.5-day-old maize primary roots germinated in paper rolls as described above. Subsequent mRNA isolation and cDNA synthesis were performed with the Chemagic mRNA Direct kit (Chemagen, Baesweiler, Germany) and the iScipt cDNA Synthesis kit (Bio-Rad Laboratories, Hercules, CA), according to the manufacturers' instructions. Gene expression for each gene in each genotype was determined in three independent biological replicates, i.e., in RNA isolated from three independent pools of plants that were different from the pools that were used for the microarray analyses. Oligonucleotides (supplemental material 4) were designed with Primer3 software (Rozen and Skaletzky 2000). Ten-microliter PCR reactions contained diluted cDNA, 2× Power SYBR Green Master mix (Applied Biosystems, Foster City, CA), and 300 nm of forward and reverse primers. PCRs were performed on a SDS 7900 HT instrument (Applied Biosystems) with the following temperature scheme: 50° for 2 min, 95° for 10 min, followed by 40 cycles of 95° for 15 sec and 60° for 1 min. Each reaction was performed in three replicates on 384-well plates. Raw Ct values obtained with SDS 2.2.2 (Applied Biosystems) were imported in Excel software. Normalization and fold-change calculations were performed using the GeNorm method (Vandesompele et al. 2002). Since we had no prior knowledge of which maize genes display similar expression levels in primary roots of hybrids and inbred lines, we tested a number of different genes. A homolog of a myosin heavy-chain gene (GenBank accession AI941656) and a hobbit homolog (GenBank accession AI621476) displayed the most consistent expression levels between the different genotypes and have therefore been selected for normalization. After log2 transformation of the PCR data a linear model was fitted for each gene with a fixed effect for genotype. The same contrasts as with the microarray data were estimated and multiplicity adjustment of P-values was likewise performed by controlling the FDR at 5% (Benjamini and Hochberg 1995).
RESULTS
Maize seedling roots display heterosis for various traits early after germination:
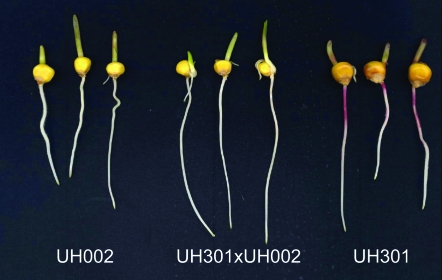
Previously, we demonstrated that heterosis is manifested during the very early stages of maize (Zea mays L.) root development in hybrids derived from the inbred lines UH002 and UH005 of the flint and UH250 and UH301 of the dent pool (Hoecker et al. 2006; Figure 1). Among other traits, enhanced primary root elongation and lateral root density in young hybrids demonstrated that the simple morphological structure of the maize primary root is a suitable model to study the early stages of heterosis manifestation. We chose a developmental stage very early during the phenotypic manifestation of heterosis in primary roots to study gene expression that might be associated with the manifestation of heterosis.
Figure 1.—
Seedling phenotype 3.5 days after germination (DAG) of the exemplary inbred lines UH002 (flint) and UH301 (dent) with their hybrid UH301 × UH002. The primary roots of four different inbred lines and 12 hybrids at this developmental stage were used for microarray and qRT–PCR analyses.
Analysis of nonadditive gene expression in young primary roots—the strategy:
The goal of this study was to identify genes that are expressed in a nonadditive manner between hybrids and the average of their parental inbred lines. To achieve this goal we utilized a two-step strategy. First, we screened for candidate genes that are expressed in a nonadditive manner in hybrids compared to their parental inbred lines with spotted cDNA microarray chips. Spotted cDNA microarray chips have some limitations and can therefore provide only initial clues on nonadditive gene expression that need to be substantiated with independent gene expression experiments, e.g., via qRT–PCR analyses. The limited statistical power of expression data obtained via the analysis of spotted microarray chips in this particular study is due to several reasons. First, maize is a segmental allotetraploid species. Therefore, the transcript fragments spotted on the microarray chips used in this analysis could potentially cross-hybridize with closely related members of gene families. Second, while the sequences spotted on the microarray chips used in this study were deduced from the inbred line B73, the RNA samples that were hybridized with these microarray chips were of different genetic origin (see materials and methods). Third, while spotted microarray chips can be hybridized to two different samples, gene expression comparisons of reciprocal hybrids and the two parental inbred lines require indirect comparisons of four genotypes. For these reasons we used the microarray experiments only as a primary screen for candidate genes that might be expressed different from additivity in hybrids compared to their parental inbred lines. Subsequently, determination of gene expression levels of a subset of nonadditive candidate genes was performed using qRT–PCR experiments.
Microarray screening of genes that are expressed nonadditively between hybrids and the midparent value of the related inbred lines:
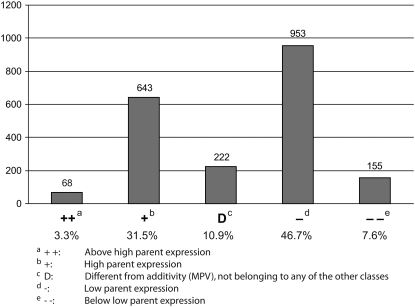
The initial microarray screening for nonadditively expressed candidate genes was performed according to the experimental design devised in Keller et al. (2005), including the four inbred lines UH250, UH005, UH002, and UH301 and the hybrids generated from these inbred lines. Hence, six inbred hybrid quadruplets each containing two inbred lines and the two corresponding reciprocal hybrids were analyzed. Gene expression of each quadruplet was analyzed using 12–16 microarray chips (supplemental material 1). Each hybridization probe used for the microarray experiments represented an independent biological replicate. Microarray hybridization was performed with a spotted maize cDNA microarray chip containing 10,649 informative distinct features (GenII vB, GPL 1996; http://www.plantgenomics.iastate.edu/maizechip). After hybridization each chip was scanned at six different laser intensities as previously described (Piepho et al. 2006). After false discovery rate adjustment (FDR < 5%) in total 1941 of the 10,649 distinct microarray features (18%) displayed an expression level that was significantly different from additivity in at least one inbred–hybrid combination. Several features were nonadditively expressed in more than one hybrid, resulting in a total of 2041 nonadditive expression patterns. These 2041 nonadditive expression patterns were grouped into different expression classes (Figure 2) as suggested by Stupar and Springer (2006). Expression that was significantly higher in the hybrid than in both parental inbred lines was designated as “above high parent” (Figure 2: ++). Similarly, if expression in the hybrid was significantly lower than that in both parents, expression was classified as “below low parent” Figure 2: −−). Hybrid expression patterns that were significantly different from that in the parent with the lower expression but not different from that in the parent with the higher expression were classified as “high parent” (Figure 2: +). Similarly, if the expression in the hybrid was not significantly different from the expression in the parent with the lower expression but significantly different from that in the parent with the higher expression, the expression pattern was designated in the category “low parent” (Figure 2: −). Furthermore, if expression in the hybrid was significantly higher than that in one parent but significantly lower than that in the other parent, the pattern displayed partial dominance (+/−). However, none of the analyzed expression profiles fulfilled this criterion in the microarray experiments. Finally, genes that displayed an expression pattern that was significantly different from the midparent value but not from the expression levels of the two parental inbred lines were designated as “different from midparent value” (Figure 2: D). Most of the analyzed genes (1818/2041: 89%) displayed nonadditive expression levels between the parental expression levels (classes +, D, −), while only a minority of genes displayed expression levels that fell into the extreme classes of above high parent (++) or below low parent (−−) expression (223/2041: 11%). A detailed account of the statistically significant microarray features including their GenBank accessions, the genotypes in which they were nonadditively expressed, their linear contrasts and standard errors, P-values, δ/α-ratios (for definition see supplemental material 3), mode of action, and blastx hits are summarized in supplemental material 2. Functional annotation of the microarray features summarized in supplemental material 2 was performed according to the MIPS system (http://mips.gsf.de/projects/annotation), which revealed that these genes are involved in a wide variety of cellular processes. A general overview of the expression trends of nonadditively expressed genes in the 12 different hybrids is depicted in the scatter plots in supplemental material 3, although the values summarized in supplemental material 3 are not necessarily statistically significant.
Figure 2.—
Relative distribution of 2041 nonadditively expressed genes identified via spotted cDNA microarray analyses in different expression classes. The numbers above the columns indicate the absolute numbers of genes in each category.
High-throughput qRT–PCR analysis of nonadditively expressed candidate genes:
The microarray analyses performed in this survey provided a large data set of candidate genes that were expressed in a nonadditive way between inbred lines and hybrids. However, one has to be cautious concerning the interpretation of these data since fold changes obtained via microarray experiments do not always correspond to actual expression levels for the reasons discussed above. We therefore decided to perform qRT–PCR experiments with a considerable number of candidate genes because this technique allows for the detection of subtle gene expression differences between the different genotypes by generating a specific pair of oligonucleotide primers for each transcript (supplemental material 4). A subset of 64 genes that displayed nonadditive expression in hybrid UH250 × UH005 and in hybrid UH002 × UH301 was subjected to detailed qRT–PCR analyses. In a first round of qRT–PCR the 64 genes were analyzed in three independent biological replicates of the inbred lines UH002, UH005, UH250, and UH301 and the hybrids UH250 × UH005 and UH002 × UH301 and their reciprocal hybrids UH005 × UH250 and UH301 × UH002 (supplemental materials 5 and 6). In hybrid UH250 × UH005 for 42 of 64 analyzed genes (66%) an expression that was significantly different from the midparent value was confirmed. In the reciprocal hybrid UH005 × UH250 44 of 64 genes (69%) displayed expression levels different from the midparent value although such differences were not detected in the previous microarray experiments most likely due to the limited statistical power of these microarray analyses. Similarly, for the hybrid UH002 × UH301 43 of 64 genes (67%) were confirmed to be differentially expressed from the midparent value and 44 of 64 genes (69%) in the reciprocal hybrid UH301 × UH002 were differentially expressed from the midparent value although these differences were not detected in the microarray experiments. Hence, in all four genotypes a similar proportion of genes were detected to be nonadditively expressed, suggesting that the primary microarray screen enriched for candidate genes that displayed nonadditive expression in hybrids.
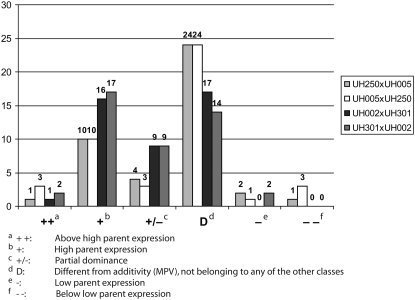
Genes that displayed nonadditive expression in qRT–PCR experiments were categorized into different classes (Figure 3) as outlined for the microarray experiments (Figure 2). In general, the relative frequency of the different classes was similar among the nonadditively expressed genes in the four analyzed hybrids. While most genes fell into the category that displayed only slight but significant deviations from the midparent value (D: 44.9% of all genes), a considerable number of genes displayed high parent expression (+: 30.8%) or partially dominant expression (+/−: 14.5%). In summary, high parent and above high parent expression (++ and +: 34.3%) prevailed among the analyzed genes compared to low parent or below low parent expression (− and − −: 5.2%). Only a minority of genes displayed expression levels that fell into the extreme classes of above high parent or below low parent expression (++ and −−: 5.8%). A reversal of expression, i.e., high parent (+) or above high parent (++) expression of a gene in one hybrid and low parent (−) or below low parent (−−) expression of the same gene in a different hybrid was not observed for any of the 64 genes subjected to qRT–PCR analyses.
Figure 3.—
Relative distribution of nonadditively expressed genes analyzed via qRT–PCR in four hybrids revealed conserved expression trends of selected nonadditive gene expression among the different hybrids. The numbers above the columns indicate the absolute numbers of genes in each category.
qRT–PCR analysis of 22 genes in 12 hybrids:
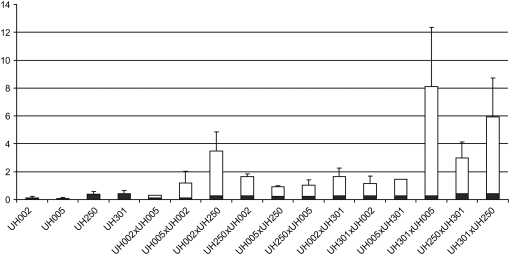
Among the 64 analyzed genes 22 displayed an expression profile that was different from the midparent value in all four hybrids. These 22 genes were subsequently subjected to additional qRT–PCR analyses of three independent biological replicates per genotype in the remaining eight hybrids that can be generated from the four inbred lines investigated in this study (supplemental materials 5 and 6). The eight hybrids displayed for 3–15 of the 22 analyzed genes nonadditive expression compared to their midparent values (supplemental material 5). Remarkably, one of these genes, a superoxide dismutase 2 (Cu-Zn) (GenBank accession P11428), showed an expression pattern that was significantly higher than the midparent values in all 12 hybrid genotypes (Figure 4) and might thus be a candidate gene for further molecular analyses of heterosis-related gene expression in the young seedling root system.
Figure 4.—
Relative expression levels of a superoxide dismutase 2 (GenBank accession P11428) as detected by qRT–PCR in four parental inbred lines and 12 hybrids. Parental expression levels in inbred lines and midparent expression levels in hybrids are indicated by the solid sections of the columns. Expression above the midparent value in hybrids is indicated by the open sections of the columns. Error bars indicate standard error of the mean.
Only a small fraction of genes display reciprocal expression differences:
Differential expression of a gene in reciprocal hybrids is observed either when a gene displays additive behavior in one hybrid and nonadditive expression in the reciprocal hybrid or if both alleles are expressed in a nonadditive manner but one allele is expressed significantly above and the reciprocal allele is expressed significantly below the midparent value. Among 216 reciprocal expression patterns of nonadditively expressed genes analyzed in the six inbred hybrid quadruplets via qRT–PCR (supplemental material 5), only 13 (6%) displayed significant reciprocal effects (t-test; P < 0.05).
Identification of genes that are nonadditively expressed between hybrids and inbred lines in maize primary roots and shoot apical meristems:
Recently, a number of gene expression studies analyzing different genotypes, developmental stages, and plant organs and applying different experimental designs and techniques and data analyses tools have been published but with no strong consensus view on the mechanism underlying heterosis. Among those studies, the analysis by Uzarowska et al. (2007), which profiled gene expression in maize shoot apical meristems (SAM), was very similar to our study. In that study the same genotypes were analyzed in a similar experimental design with the same maize microarray chips that were analyzed with the same data analysis tools. Hence, these data sets allowed for the comparison of overlapping gene expression that was different from additivity, in different plant organs. Among the 434 genes that were identified in SAMs as nonadditively expressed between inbred lines and hybrids 32 genes (8%) were also expressed nonadditively in primary roots (supplemental material 7). Interestingly, the observed frequency of 32 is significantly less than expected purely by chance (expected frequency ≈ 1941 × 434/10,649 = 79) based on a chi-square test for independence (P < 0.001). This might suggest that nonadditive gene action tends to be organ specific at least when young primary roots and shoot apical meristems are compared. Gene expression analyses in other maize organs may reveal a set of genes that are globally correlated with heterosis.
DISCUSSION
Primary roots provide a model for molecular studies of heterosis manifestation in maize:
Despite its wide exploitation in agriculture the molecular basis of heterosis remains enigmatic (Birchler et al. 2003, 2006; Hochholdinger and Hoecker 2007). A number of studies have compared gene expression profiles of inbred lines vs. hybrids in different maize organs and developmental stages, including embryos (Stupar and Springer 2006; Meyer et al. 2007), endosperm (Guo et al. 2003; Song and Messing 2003), whole seedlings (Stupar and Springer 2006; Swanson-Wagner et al. 2006), immature ears (Guo et al. 2006; Stupar and Springer 2006), adult leaves (Auger et al. 2005), and shoot apical meristem (Uzarowska et al. 2007). In addition to these studies in maize, the transcriptomes of the first Arabidopsis leaf (Vuylsteke et al. 2005) and rice panicles (Huang et al. 2006) have been analyzed. These gene expression surveys revealed global expression trends that ranged from predominant nonadditivity to additivity (summarized in Hochholdinger and Hoecker 2007). These results could be an indication that in different tissues or developmental stages different global expression patterns might prevail that could nevertheless be related to heterosis. For immature maize ears gene expression patterns were correlated with yield and heterosis and it was demonstrated that the proportion of allelic additivity was positively associated with hybrid yield and heterosis and that there is no correlation between the over- and the underexpression of specific genes in hybrids with either yield or heterosis (Guo et al. 2006).
While most of these studies analyzed gene expression in inbred lines and hybrids after the manifestation of heterosis, we studied the very early stages of heterosis manifestation in young primary roots 3.5 DAG (days after germination). In an initial screen 1941 unique microarray features were nonadditively expressed between inbred lines and hybrids. For all other 8708 distinct microarray features, evidence for a nonadditive expression pattern could not be provided. This, however, does not indicate that these transcripts truly behave additively, because nonsignificance may also result from lack of statistical power. The functional classification of the genes with expression patterns different from additivity revealed that their relative abundance in the different functional categories was similar to the relative gene distribution in the completely sequenced Arabidopsis (Arabidopsis Genome Initiative 2000) and rice (Goff et al. 2002) genomes. This might indicate that no specific function is required during heterosis manifestation in maize primary roots but rather the interplay of genes related to diverse functions. For 66% of the genes that displayed nonadditive expression in microarray experiments, this difference from additivity was confirmed in qRT–PCR experiments. This confirmation rate was similar to the values obtained for maize seedlings (Swanson-Wagner et al. 2006) where among 45 genes tested in qRT–PCR 31 were confirmed to be nonadditively expressed (69%), for maize embryos where 8 of 12 (67%) nonadditively expressed genes were confirmed via qRT–PCR (Meyer et al. 2007), and for shoot apical meristems where 2 of 3 genes (67%) were confirmed via qRT–PCR (Uzarowska et al. 2007).
Global expression trends among selected nonadditively expressed genes in maize roots are conserved between different hybrids:
Genes for which expression is significantly different from the midparent value in hybrids can be classified in different categories depending on their relative expression levels compared to the parental inbred lines (Stupar and Springer 2006). In general, nonadditively expressed genes are either expressed between the two parental expression levels or their expression is above the better-expressing inbred line or below the lower-expressing parental inbred line. In this study ∼89% of the nonadditively expressed genes identified in microarray experiments displayed expression levels between the expression levels of the parental inbred lines. Similarly, in subsequent qRT–PCR experiments with a subset of 64 genes >94% of the nonadditively expressed genes displayed transcript levels in hybrids that were between the levels of their parental inbred lines. These results were in line with gene expression studies of young maize seedlings, immature ears, and embryos (Guo et al. 2006; Stupar and Springer 2006; Swanson-Wagner et al. 2006), where the majority of genes that were expressed distinctly from additivity in hybrids displayed expression levels between the levels of the parental inbred lines. Remarkably, all 64 genes analyzed via qRT–PCR displayed a similar tendency for nonadditive gene expression in young roots; i.e., there was no instance in which a gene displayed above high parent or high parent expression in one hybrid and low parent or below low parent in another hybrid. Hence, for the subset of genes analyzed in this study the similar tendency of expression in hybrids that was distinct from additivity might also support a role of these genes that might be associated with heterosis manifestation in the primary root.
Overlapping nonadditive gene expression between maize primary roots and shoot apical meristems:
In contrast to several studies in maize where the majority of nonadditively expressed genes fell between the parental expression values (summarized in Hochholdinger and Hoecker 2007), in a survey of maize shoot apical meristems >50% of all nonadditively accumulated transcripts displayed an expression above the high or below the low parent level (Uzarowska et al. 2007). This might imply that in different tissues or developmental stages different global expression patterns might prevail that might nevertheless be related to heterosis (summarized in Hochholdinger and Hoecker 2007). This notion is supported by the observation that different tissues and organs within a hybrid plant display significant differences in their degree of phenotypic heterosis (Melchinger 1999). In this context we compared the overlap of nonadditive gene expression in maize seedling roots and shoot apical meristems (Uzarowska et al. 2007). Both experiments utilized the same genotypes, applied a similar experimental design, used the same maize microarray chips, and adopted the same data analysis tools. Interestingly, the observed frequency of 32 overlapping nonadditively expressed microarray features was significantly less than expected purely by chance on the basis of a chi-square test for independence (P < 0.001). This finding supports the notion that nonadditive gene action tends to be organ specific (Hochholdinger and Hoecker 2007) at least for the analyzed organs primary root and shoot apical meristem. Despite the little overlap between nonadditively expressed genes in both organs this data set might nevertheless be an interesting starting point for the identification of genes that might play a global role in heterosis manifestation in maize.
Nonadditive expression of a superoxide dismutase 2 in all analyzed hybrids might imply a protective role of this gene during heterosis manifestation in primary roots:
Only a few studies are currently available that analyzed gene expression in a variety of different hybrids and they did not reveal any genes consistently under- or overexpressed (Guo et al. 2006; Uzarowska et al. 2007). qRT–PCR analyses of a subset of genes that was nonadditively expressed in at least two hybrids in microarray experiments identified a superoxide dismutase 2 (GenBank AC P11428) that displayed an expression that exceeded the midparent value in all 12 hybrids. Superoxide dismutases are antioxidative enzymes in plants that catalyze the dismutation of the superoxide anion to molecular oxygen and hydrogen peroxide, which are subsequently degraded by the enzyme catalase to oxygen and water and hence protect cells from the damaging effects of reactive oxygen species (Cannon et al. 1987; Scandalios 2005). It could be hypothesized that the enhanced defense from reactive oxygen species could be related to the superior performance of hybrids compared to their parental inbred lines. This finding is consistent with the hypothesis that not only global expression trends (Guo et al. 2006) but also the consistent expression of selected key genes (Hochholdinger and Hoecker 2007) might be relevant during the organ-specific manifestation of heterosis.
In summary, the analysis of gene expression during the early stages of heterosis manifestation in combination with the future detailed analysis of nonadditively expressed genes identified in such studies might contribute to a better understanding of the molecular networks regulating the phenotypic variations between inbred lines and hybrids.
Acknowledgments
We thank A. Melchinger (University of Hohenheim) and his coworkers for providing seeds of the inbred lines and hybrids analyzed in this study and Dieter Steinmetz and Sandra Kaiser (University of Tuebingen) for their help with data organization. Heterosis research in F.H.'s laboratory and H.-P.P.'s work group is funded by the Deutsche Forschungsgemeinschaft framework program SPP1149, “Heterosis in Plants.”
References
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Auger, D. L., A. D. Gray, T. S. Ream, A. Kato, E. H. Coe, Jr. et al., 2005. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. [Google Scholar]
- Birchler, J. A., D. L. Auger and N. C. Riddle, 2003. In search of the molecular basis of heterosis. Plant Cell 15 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., H. Yao and S. Chundalayandi, 2006. Unraveling the genetic basis of hybrid vigor. Proc. Natl. Acad. Sci. USA 103 12957–12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, A. B., 1910. The Mendelian theory of heredity and the augmentation of vigor. Science 32 627–628. [DOI] [PubMed] [Google Scholar]
- Brunner, S., K. Fengler, M. Morgante, S. Tingey and A. Rafalski, 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, R. E., J. A. White and J. G. Scandalios, 1987. Cloning of cDNA for maize superoxide dismutase 2 (SOD2). Proc. Natl. Acad. Sci. USA 84 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C. R., 1876. The Effects of Cross- and Self-Fertilization in the Vegetable Kingdom. John Murray, London.
- Davenport, C. B., 1908. Degeneration, albinism and inbreeding. Science 28 454–455. [DOI] [PubMed] [Google Scholar]
- Keeble, F., and C. Pellow, 1910. The mode of inheritance of stature and time of flowering in peas (Pisum sativum). J. Genet. 1 47–56. [Google Scholar]
- East, E. M., 1908. Inbreeding in corn. Conn. Agric. Exp. Stn. Rep. 1907 419–428. [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, New York.
- Fu, H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., D. Ricke, T.-H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100. [DOI] [PubMed] [Google Scholar]
- Guo, M., M. A. Rupe, O. N. Danilevskaya, X. Yang and Z. Hu, 2003. Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36 30–44. [DOI] [PubMed] [Google Scholar]
- Guo, M., M. A. Rupe, X. Yang, O. Crasta, C. Zinselmeier et al., 2006. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 113 831–845. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F., and N. Hoecker, 2007. Towards the molecular basis of heterosis. Trends Plant Sci. 12 427–432. [DOI] [PubMed] [Google Scholar]
- Hoecker, N., B. Keller, H.-P. Piepho and F. Hochholdinger, 2006. Manifestation of heterosis during early maize (Zea mays L.) root development. Theor. Appl. Genet. 112 421–429. [DOI] [PubMed] [Google Scholar]
- Huang, Y., L. Zhang, J. Zhang, D. Yuan, C. Xu et al., 2006. Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs. Plant Mol. Biol. 62 579–591. [DOI] [PubMed] [Google Scholar]
- Jones, D. F., 1917. Dominance of linked factors as a means of accounting for heterosis. Genetics 2 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, B., K. Emrich, N. Hoecker, M. Sauer, F. Hochholdinger et al., 2005. Designing a microarray experiment to estimate heterosis. Theor. Appl. Genet. 111 57–64. [DOI] [PubMed] [Google Scholar]
- Lai, J., Y. Li, J. Messing and H. K. Dooner, 2005. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchinger, A. E., 1999. Genetic diversity and heterosis. International Symposium on Genetics and Exploitation of Heterosis in Crop Plants, Mexico City, August 17–22, 1997, pp. 99–118.
- Meyer, S., H. Pospisil and S. Scholten, 2007. Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol. Biol. 63 381–391. [DOI] [PubMed] [Google Scholar]
- Nakazono, M., F. Qiu, L. A. Borsuk and P. S. Schnable, 2003. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15 583–596 (erratum: Plant Cell 15: 1049). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho, H.-P., B. Keller, N. Hoecker and F. Hochholdinger, 2006. Combining signals from spotted cDNA microarrays obtained at different scanning intensities. Bioinformatics 22 802–807. [DOI] [PubMed] [Google Scholar]
- Powers, L., 1945. An expansion of Jones's theory for the explanation of heterosis. Am. Nat. 78 275–280. [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Scandalios, J. G., 2005. Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res. 38 995–1014. [DOI] [PubMed] [Google Scholar]
- Shull, G. F., 1908. The composition of a field of maize. Rep. Am. Breed. Assoc. 5 51–59. [Google Scholar]
- Shull, G. F., 1952. Beginnings of the heterosis concept, pp. 14–48 in Heterosis, edited by J. W. Gowen. Iowa State College Press, Ames, IA.
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on nonlinear haplotypes. Proc. Natl. Acad. Sci. USA 100 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., and N. M. Springer, 2006. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1-hybrid. Genetics 173 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner, R. A., Y. Jia, R. De Cook, L. A. Borsuk, D. Nettleton et al., 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1-hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 103 6805–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzarowska, A., B. Keller, H.-P. Piepho, G. Schwarz, C. Ingvardsen et al., 2007. Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol. Biol. 63 21–34. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy et al., 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 RESEARCH0034. [DOI] [PMC free article] [PubMed]
- Vuylsteke, M., F. van Eeuwijk, P. Van Hummelen, M. Kuiper and M. Zabeau, 2005. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 171 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woll, K., L. A. Borsuk, H. Stransky, D. Nettleton and P. S. Schnable, 2005. Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 139 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]