Abstract
Why eukaryotes encode multiple Hsp70 isoforms is unclear. Saccharomyces cerevisiae Ssa1p and Ssa2p are constitutive 98% identical Hsp70's. Stress-inducible Ssa3p and Ssa4p are 80% identical to Ssa1/2p. We show Ssa1p-4p have distinct functions affecting [PSI+] and [URE3] prions. When expressed as the only Ssa, Ssa1p antagonized [URE3] and Ssa2p antagonized [PSI+]. Ssa3p and Ssa4p influenced [URE3] and [PSI+] somewhat differently but overall their effects paralleled those of Ssa1p and Ssa2p, respectively. Additionally, Ssa3p suppressed a prion-inhibitory effect of elevated temperature. Our previously described Ssa1-21p mutant weakens [PSI+] in SSA1-21 SSA2 cells and abolishes it in SSA1-21 ssa2Δ cells. To test if the same mutation affected other prions or altered Ssa2p similarly, we compared effects of a constructed Ssa2-21p mutant and Ssa1-21p on both prions. Surprisingly, [URE3] was unaffected in SSA1-21 SSA2 cells and could propagate in SSA1-21 ssa2Δ cells. Ssa2-21p impaired [URE3] considerably and weakened [PSI+] strongly but in a manner distinct from Ssa1-21p, highlighting functional differences between these nearly identical Hsp70's. Our data uncover exquisite functional differences among isoforms of a highly homologous cytosolic Hsp70 subfamily and point to a possibility that variations in Hsp70 function that might improve fitness under optimal conditions are also important during stress.
HSP70 is a highly conserved and ubiquitous protein chaperone that helps proteins adopt and maintain native conformations. By binding exposed hydrophobic surfaces on partially unfolded proteins Hsp70 plays a vital role in many cellular processes such as translation and translocation, and it is a major factor in protecting cells from stress by preventing protein aggregation. Eukaryotic genomes encode multiple highly conserved Hsp70 isoforms, some of which are expressed constitutively and others that are restricted to conditions of stress or specific tissues. Although these isoforms are structurally similar and have the same reaction cycle of binding and releasing partially unfolded proteins, the extent they possess specific functional differences rather than simply having their expression regulated to change Hsp70 abundance in response to need is only recently beginning to be appreciated.
In plants, fungi, and bacteria, Hsp70 also contributes to stress protection by assisting nonessential Hsp100 family chaperones (Hsp104 in yeast) in resolubilizing aggregated proteins (Parsell et al. 1994; Glover and Lindquist 1998; Gurley 2000; Weibezahn et al. 2005). In vitro this disaggregation reaction requires only Hsp104, Hsp70, and Hsp40 (Glover and Lindquist 1998). Hsp40's also bind hydrophobic peptides and act as Hsp70 cochaperone partners that recruit, regulate, and enhance Hsp70 function. This Hsp104 machinery is required for propagation of yeast prions [URE3], [PSI+], and [PIN+] (Chernoff et al. 1995; Derkatch et al. 1997; Moriyama et al. 2000; Wickner et al. 2007), which are aggregated, self-replicating amyloid forms of proteins Ure2p, Sup35p, and Rnq1p, respectively (Cox 1965; Wickner 1994; Derkatch et al. 2001). The disaggregation function of the Hsp104 machine acts in prion propagation by disrupting prion aggregates to generate more numerous prion particles, or seeds (Paushkin et al. 1996; Kryndushkin et al. 2003).
Altered abundance or function of Hsp70/40 chaperones affects yeast prions differently. Increased expression of the cytosolic Hsp70 Ssa1p does not weaken [PSI+] but cures [URE3] (Schwimmer and Masison 2002). Depleting Ssa1p weakens [PSI+] but not [URE3] while mutating or depleting the nearly identical Ssa2p weakens [URE3] but not [PSI+] (Roberts et al. 2004; Hung and Masison 2006). Altered Ssa1p function can reduce the numbers of [PSI+] seeds per cell without affecting Hsp104 function, suggesting that Hsp70 might have an Hsp104-independent role in prion seeding processes (Jung et al. 2000). Among Hsp40's, overproducing Ydj1p eliminates [URE3] (Moriyama et al. 2000), elevated Ydj1p or Apj1p weakens some strains of [PSI+] (Kushnirov et al. 2000; Kryndushkin et al. 2002), and a specific activity of Sis1p is required for propagation of [PIN+] (Sondheimer and Lindquist 2000; Lopez et al. 2003; Aron et al. 2007). These Hsp40 effects could occur through direct interaction with prion proteins or through disruption of coordinated activity of the chaperone machinery. For example, in vitro Ydj1p binds Ure2p and impairs its ability to form amyloid, and in vivo Hsp40's and other Hsp70 cochaperones, such as the tetratricopeptide repeat (TPR)-containing Sti1p, Cpr7p, and Cns1p, can affect prion propagation through their ability to regulate Hsp70 (Kushnirov et al. 2000; Jones et al. 2004; Lian et al. 2007). Taken together these observations point to Hsp70 as having a key role in yeast prion propagation.
The Saccharomyces cerevisiae SSA Hsp70 subfamily consists of four functionally redundant genes (SSA1-4), of which abundant expression of at least one is necessary for growth (Werner-Washburne et al. 1987). Constitutively expressed Ssa1p and Ssa2p are 98% identical. This near identity becomes even more significant when considering that over half of the nonidentical residues are conservative substitutions. Ssa3p and Ssa4p, which are 88% identical to each other and 80% identical to Ssa1/2p, are expressed only under nonoptimal growth conditions and help protect cells from the adverse effects of stress. Under optimal conditions Ssa2p is roughly fourfold more abundant than Ssa1p because the SSA1 promoter is partially repressed in the absence of stress. When Ssa2p is depleted, expression of Ssa1p is induced, maintaining overall Hsp70 abundance.
Here we present a comprehensive study of effects of an entire Hsp70 subfamily on prions by monitoring [PSI+] and [URE3] propagation in cells expressing the individual Ssa proteins. We also investigated in more detail the differences between the nearly identical Ssa1p and Ssa2p by assessing how a previously identified Ssa1p mutant known to impair [PSI+] affected [URE3], and how Ssa2p carrying the same mutation affected [PSI+] and [URE3]. We find that small amino acid differences among Hsp70's are enough to impart functional distinctions that have considerable yet specific effects on propagation of amyloid in vivo. Differences and similarities we find between stress-inducible Ssa3/4p and constitutively expressed Ssa1/2p suggest that differences in Hsp70 function are not only important under different conditions, but also that similar functional variations between Hsp70's are important under either optimal or stressful conditions.
MATERIALS AND METHODS
Yeast strains and plasmids:
Strains used are listed in Table 1. In a manner similar to SSA1-21, SSA2-21 was incorporated by integrative transformation using genes with hisG-URA3-hisG cassettes 200 bp 3′ to the termination codon (Alani et al. 1987; Jung et al. 2000). Strains with combinations of alleles were obtained through crosses. To monitor effects of individual Ssa proteins, strains lacking all four chromosomal SSA genes were constructed. In the progenitor strain SSA4 was replaced by URA3 and the ura3-2f mutant was isolated on 5-fluoroorotic acid (5-FOA) (Boeke et al. 1984; Jones and Masison 2003). [PSI+] propagated normally in the parental ade2-1 strain (1135), which expresses Ssa1p from a URA3-based plasmid (data not shown). This strain was transformed by LEU2-based plasmids carrying SSA1-4 under control of the SSA2 promoter (see below), and transformants having lost the resident SSA1 plasmid were isolated on 5-FOA. An isogenic [URE3] strain (1161) was made by replacing ade2-1 with the wild-type ADE2 allele controlled by the DAL5 promoter. Strain 1161 expressing Ssa2p from a URA3-based plasmid, which propagates [URE3] normally (data not shown), was transformed by the SSA1-4 plasmids and cells having lost the resident SSA2 plasmid were selected. In these 1135 and 1161 strains the only SSA gene expressed is encoded on the LEU2-based plasmid. Ssa protein is essential so these strains retain the SSA plasmids without selection.
TABLE 1.
Yeast strains
| Straina | Genotype |
|---|---|
| 779-6A | MATα, kar1-1, SUQ5, ade2-1, his3Δ202, leu2Δ1, trp1Δ63, ura3-52 (Jung and Masison 2001) |
| 792 | 779-6A with ura2∷KanMX |
| 793 | 792 with SSA1-21 |
| 794 | 792 with ssa1∷KanMX |
| 812 | 792 with ssa2∷HIS3 |
| 813 | 792 with SSA1-21, ssa2∷HIS3 |
| 814 | 792 with ssa1∷KanMX, ssa2∷HIS3 |
| 912 | 792 with SSA2-21 |
| 913 | 792 with SSA1-21, SSA2-21 |
| 914 | 792 with ssa1∷KanMX, SSA2-21 |
| 1075 | 779-6A with PDAL5∷ADE2 in place of ade2-1 |
| 1079 | 1075 with SSA1-21 |
| 1076 | 1075 with ssa1∷KanMX |
| 1080 | 1075 with ssa2∷HIS3 |
| 1085 | 1075 with SSA1-21, ssa2∷HIS3 |
| 1081 | 1075 with ssa1∷KanMX, ssa2∷HIS3 |
| 1082 | 1075 with SSA2-21 |
| 1083 | 1075 with SSA1-21, SSA2-21 |
| 1084 | 1075 with ssa1∷KanMX, SSA2-21 |
| 1135 | 779-6A with ssa1∷KanMX, ssa2∷HIS3, ssa3∷TRP1, ssa4∷ura3-2f (Tutar et al. 2006) |
| 1161 | 1075 with ssa1∷KanMX, ssa2∷HIS3, ssa3∷TRP1, ssa4∷ura3-2f |
All are isogenic and except where cited all originated in this study. Strains 1135 and 1161 carry URA3-based pRDW10 (SSA1) and pJ401 (SSA2), respectively, or a LEU2-based SSA plasmid as indicated in the text.
Single-copy SSA1 plasmids pRDW10 (URA3) and pC210 (LEU2) have been described (Jung et al. 2000; Schwimmer and Masison 2002). Plasmid pJ401 is single-copy URA3-based pRS316 carrying SSA2 and 500 bp of 5′ and 3′ flanking DNA on a BamHI fragment. Plasmid pDCM62 is single-copy LEU2-based pRS315 with the same SSA2-containing DNA fragment. Plasmids pA3 and pA4 are pC210 with the coding region of SSA3 or SSA4, on NdeI-SphI fragments, in place of SSA1. Plasmid pYSGal104, used for galactose-inducible expression of HSP104, has been described (Lindquist and Kim 1996). Plasmids pH 218 and pH 220, obtained from Herman Edskes (National Institutes of Health), are HIS3-based single-copy pRS313 and multicopy pRS423, respectively, containing Hsp104 with its own promoter.
Media and growth conditions:
Rich medium for plates (1/2YPD) contains 0.5% yeast extract, 2% peptone, 2% dextrose, and 2% agar. Liquid-rich medium (YPAD) is similar but lacks agar and contains 1% yeast extract and 400 mg/liter adenine. Synthetic-defined (SD) media contain 7 g/liter yeast nitrogen base and 2% glucose supplemented with amino acids and uracil as needed, with or without excess (400 mg/liter) or limiting (9 mg/liter) concentrations of adenine to monitor the prions. Growth conditions were as described (Jung et al. 2000) or as indicated in results.
Monitoring and verification of prions:
[PSI+] propagates as an insoluble amyloid form of the translation termination factor Sup35p. The reduced amount of soluble Sup35 in [PSI+] cells reduces efficiency of translation termination, which causes suppression of the ade2-1 nonsense allele in our strains (Cox 1965; Jung et al. 2000). Nonsuppressed ade2-1 strains do not grow without adenine and, when adenine is limiting, accumulate an intermediate of adenine biosynthesis that causes red coloration. Thus, [psi−] cells require adenine and are red when grown on limiting adenine (e.g., 1/2YPD), while [PSI+] cells grow without adenine and are white. Under conditions where [PSI+] propagation is weaker than normal, cells can have an intermediate pink color due to an increased relative amount of soluble Sup35p or an increased rate of loss of [PSI+] as cells divide due to a decrease in number of prion particles. The presence of [PSI+] was confirmed by its dominant phenotype and ability to be eliminated by growth on medium containing 3 mm guanidine–hydrochloride (Tuite et al. 1981; Jung and Masison 2001).
[URE3] was monitored using strains with the wild-type ADE2 gene regulated by the DAL5 promoter in place of ade2-1 (Schlumpberger et al. 2001; Brachmann et al. 2005). Since Ure2p represses expression of nitrogen catabolic genes, on standard ammonium-containing media the DAL5 promoter is inactive so ADE2 is not expressed and cells are ade− and red. When [URE3] (the amyloid prion form of Ure2p) is present the depletion of functional Ure2p into insoluble prion aggregates activates the DAL5 promoter and ADE2 expression, making cells ade+ and white. As with [PSI+], weakened [URE3] propagation can be seen as intermediate colony color or increased frequency of appearance of [ure-o] cells in a population. The presence of [URE3] was confirmed as for [PSI+].
Prion curing by guanidine:
Quantitative assays were performed as described previously (Jung et al. 2000). Briefly, cells maintained in log phase in YPAD containing 3 mm guanidine were removed periodically and spread onto YPD plates to determine the percentage of prion-containing cells as a function of cell doubling. Colony color reflects prion status of the cell that gave rise to the colony. Because prions rarely appear spontaneously, colonies with any detectable white ade+ cells were scored as arising from a prion-containing cell. Routine prion curing was done by growing cells on 1/2YPD plates containing 3 mm guanidine, transferring them to 1/2YPD without guanidine and isolating red colonies.
RESULTS
Equivalent mutations in Ssa1p and Ssa2p affect [PSI+] differently:
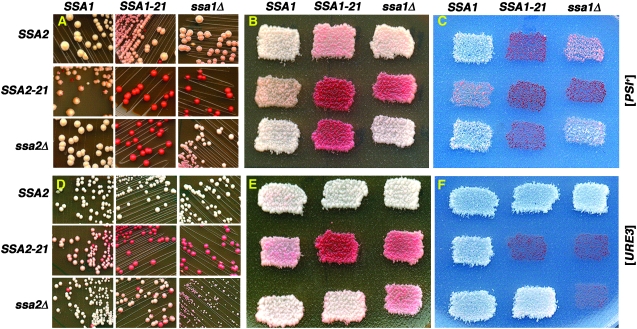
Earlier we showed that cells expressing Ssa1-21p, an Ssa1p mutant with a mutation of a highly conserved residue in the substrate-binding domain (L483W), have a considerably weakened [PSI+] phenotype, and that SSA1-21 cells lacking Ssa2p cannot propagate [PSI+] (Jung et al. 2000). This mutant appears to impair prions by allowing individual prion polymers to more readily self-associate into higher-order aggregates or by inhibiting dismantling of the larger aggregates (Song et al. 2005). A resulting decrease in number of prion seeds per cell increases the relative amount of soluble Sup35p and makes the prion mitotically unstable. Unlike the uniformly white color of wild-type [PSI+] colonies, the weakened [PSI+] propagation in cells expressing Ssa1-21p is seen as pink colony color, reflecting reduced nonsense suppression caused by [PSI+], and frequent appearance of red [psi−] colonies, reflecting spontaneous loss of [PSI+] during mitosis (Figure 1, A and B, top row center; see materials and methods). Thus, both “strength” and stability of the prion are affected.
Figure 1.—
Prion phenotypes of SSA1 and SSA2 mutants. A–C show strains used to monitor [PSI+]; D–F show strains used to monitor [URE3]. (A–C) From left to right, top rows are strains 792, 793, and 794; middle rows are strains 912, 913, and 914; bottom rows are strains 812, 813, and 814 (see Table 1). SSA1 and SSA2 alleles present in each strain are indicated at the top and to the left of the panels, respectively. The wild-type [PSI+] strain is on the top left of each panel. SSA1-21 SSA2-21, SSA1-21 ssa2Δ, and ssa1Δ SSA2-21 strains cannot propagate [PSI+] and show typical [psi−] phenotypes. (A) Strains were streaked onto 1/2YPD and incubated for 3 days at 30° followed by 3 days at 23°. (B) 1/2YPD master plate grown for 1 day at 30° followed by 1 day at 23° contains patches of cells from same strains and in the same pattern as those shown in A. (C) Replica of the plate shown in B onto medium lacking adenine and incubated for 3 days at 30°. (D–F) From left to right, top rows are strains 1075, 1079, and 1076; middle rows are strains 1082, 1083, and 1084; bottom rows are strains 1080, 1085, and 1081 (see Table 1). SSA1/SSA2 allele combinations are in the same pattern as in A–C. Wild-type [URE3] strain is on the top left of each panel. SSA1-21 SSA2-21, ssa1Δ SSA2-21, and ssa1Δ ssa2Δ strains cannot propagate [URE3] and show typical [ure-o] phenotypes. Plates in D and E were incubated for 2 days at 30° followed by 2 days at 23°; the plate in F was incubated for 2 days at 30°. Since pigmentation on 1/2YPD is influenced by adenine abundance, cells in the center (i.e., SSA1-21 SSA2-21) can appear redder than prion-lacking cells on the periphery because less adenine is available due to depletion by surrounding strains.
To assess effects of the homologous mutation in Ssa2p (Ssa2-21p), we constructed an isogenic SSA2-21 strain. SSA2-21 [PSI+] cells had a slight brownish color (Figure 1, A and B, middle row left) that was distinct from the pink of SSA1-21 cells (top row center), yet notably different from the white color of wild-type cells (top row left). As with SSA1-21 cells, red [psi−] colonies appeared on streaks of SSA2-21 cells. Unlike SSA1-21 colonies, SSA2-21 [PSI+] colonies often had obvious red sectors containing progeny of cells that lost [PSI+] during growth of the colony (Figure 1A, middle row left). When restreaked, these sectored colonies gave rise to up to 30% [psi−] cells while SSA1-21 [PSI+] colonies never had more than ∼12% [psi−] cells. Consistent with this difference in frequency of [PSI+] loss we saw a small but reproducible increase in proportion of [psi−] cells in SSA2-21 liquid cultures (Table 2, compare strains 793 and 912). Together these results show that overt effects on [PSI+] phenotype caused by the same mutation in Ssa1p and Ssa2p were clearly distinguishable.
TABLE 2.
Stability of prions in cells expressing different SSA alleles
| Straina: | Prion loss (%)
|
||
|---|---|---|---|
| ([PSI+]/[URE3]) | SSA genotypeb | [PSI+] | [URE3] |
| 792/1075 | SSA1 SSA2 | <0.1 | <0.1 |
| 793/1079 | SSA1-21 SSA2 | 4.5 ± 1.8c | <0.1 |
| 912/1082 | SSA1 SSA2-21 | 8.6 ± 2.5c | 11 ± 4 |
| 794/1076 | ssa1Δ SSA2 | <0.1 | <0.1 |
| 812/1080 | SSA1 ssa2Δ | <0.1 | 7.8 ± 1.6 |
| 814/ | ssa1Δ ssa2Δ | <0.1 | NA |
| /1085 | SSA1-21 ssa2Δ | NA | 17 ± 3 |
| 1135/1161 | SSA1 only | <0.1 | 15 ± 3 |
| 1135/1161 | SSA2 only | <0.1 | <1 |
| 1135/1161 | SSA3 only | <0.1 | 29 ± 4 |
| 1135/1161 | SSA4 only | <1 | <1 |
| /1161 | SSA1-21 only | NA | 13 ± 3 |
Cells from white colonies on 1/2YPD were streaked or diluted in water and spread onto 1/2YPD plates and grown for 3 days at 30° followed by 5 days at 23°. Among a total of 250–1000 resulting colonies for each strain, entirely red colonies were scored as having lost the prion. Data are averages of 3–6 trials ± SE. SSA gene combinations that abolish propagation of both prions (i.e., SSA1-21 SSA2-21 and ssa1Δ SSA2-21) are not included on the table, and NA (not applicable) indicates where SSA gene combination does not allow propagation of one or the other prion.
The slash between strain numbers distinguishes strains used to monitor [PSI+] (left of slash) and [URE3] (right of slash).
Strains except 1135 and 1161 have indicated genes on chromosomes and have genomic SSA3 and SSA4. Strains 1135 and 1161 lack all four chromosomal SSA alleles and express the indicated SSA gene, driven by the SSA2 promoter, from a single-copy plasmid.
Statistically, the values for these numbers overlap.
To assess effects on [PSI+]-mediated nonsense suppression we compared growth of [PSI+] cells on medium lacking adenine. Our wild-type [PSI+] strains are faintly pigmented when grown without adenine at 30° (Figure 1C, top row left). On the same plates SSA1-21 [PSI+] cells were unable to grow, having the appearance of [psi−] cells (Figure 1C, top row center). SSA2-21 cells were pinker and grew more slowly than wild type, but they grew better than SSA1-21 cells (Figure 1C, middle row left). Thus, Ssa2-21p reduced [PSI+]-mediated translational readthrough of the ade2-1 nonsense codon, but this effect was not as great as that caused by Ssa1-21p.
The increased spontaneous loss of [PSI+] from SSA1-21 cells appears to be due to a 10-fold reduction in number of prion particles, or seeds, per cell, which increases the chance that a daughter cell fails to inherit prion particles when cells divide (Jung et al. 2000). The number of seeds per cell in a population of [PSI+] cells can be estimated by growing cells in millimolar amounts of guanidine, which inhibits Hsp104 function and thus arrests replication of prion particle, and then monitoring the appearance of [psi−] cells as a function of cell divisions (Eaglestone et al. 2000; Jung and Masison 2001; Ness et al. 2002). Because wild-type cells typically have ∼100 prion seeds per cell, there is a lag of 4–6 cell divisions required to dilute the nonreplicating seeds among progeny before [psi−] cells begin to appear. At this point the proportion of [psi−] cells in the culture increases roughly linearly as cells divide.
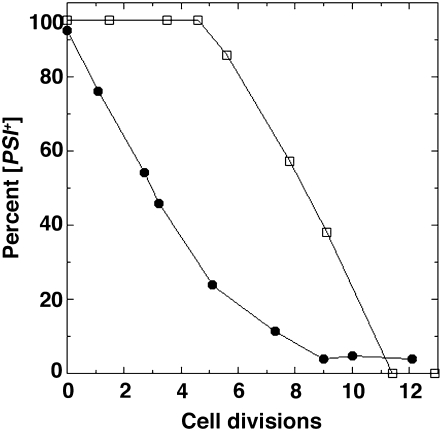
When grown in liquid YPAD without guanidine SSA2-21 cells have ∼2–5% [psi−] cells and SSA1-21 cells have 1–2% [psi−] cells, regardless of how long the cultures are maintained. For both mutants, when guanidine is added to log-phase cultures at a final concentration of 3 mm, [psi−] cells appear immediately and then increase at a rate similar to wild-type cells (Figure 2 and Jung et al. 2000). The absence of the lag indicates that the number of prion seeds before guanidine addition is already at the point where continued dilution by cell division gives rise to daughter cells that fail to inherit a seed. These curing kinetics indicate that Ssa2-21p also causes at least a 10-fold reduction in number of prion seeds per cell, which is at the limit of sensitivity of this assay for our strains. These results are consistent with Ssa2-21p having a similar overall effect as Ssa1-21p on [PSI+] seed replication that results in a considerable reduction in number of prion seeds per cell.
Figure 2.—
SSA2-21 mutant cells have fewer [PSI+] seeds. Guanidine hydrochloride, which arrests prion-seed replication, was added to [PSI+] cells growing in YPAD and the proportion of [PSI+] cells remaining in the cultures was monitored as a function of cell divisions. Data from wild-type (open symbols) and SSA2-21 (solid symbols) cultures are indicated. The absence of a lag before appearance of [psi−] cells in the SSA2-21 culture reflects at least a 10-fold decrease in average number of prion seeds per cell in the starting population (see text).
The effects of Ssa2-21p are dominant in that they occur in the presence of Ssa1p (Figure 1, A–C, middle row left) or Ssa2p (data not shown). Like SSA1-21 cells that lack Ssa2p (Figure 1, A–C, bottom row center; Jung et al. 2000), SSA2-21 cells that lack Ssa1p (Figure 1, A–C, middle row right) did not allow [PSI+] to propagate. Not surprisingly, [PSI+] could not propagate in cells expressing both Ssa1-21p and Ssa2-21p (Figure 1, middle row center in each panel).
Ssa1-21p and Ssa2-21p affect [URE3] differently:
To test the effects of Ssa1-21p and Ssa2-21p on [URE3] propagation we constructed strains to monitor [URE3] by a similar white/red adenine plus/minus phenotype (see materials and methods). As with [PSI+], increases in pigmentation of [URE3] cells or frequency of appearance of [ure-o] cells reflect weakened strength and stability of [URE3], respectively. We showed that the presence of [URE3] causes our strains to grow slowly (Schwimmer and Masison 2002) and note that the [URE3] DAL5/ADE2 system confers adenine prototrophy by a different mechanism than [PSI+]/ade2-1 (see materials and methods). The growth defect is similar to growth of ure2Δ cells (data not shown) suggesting that growth is not being inhibited by the [URE3] prion per se, but more likely by depletion of Ure2p function. In our hands, compared with the [PSI+]/ade2-1 system, the DAL5/ADE2 system for monitoring different strengths of [URE3] is less straightforward. Cells propagating [URE3] weakly can be pink on 1/2YPD but white and grow relatively well on plates lacking adenine (Figure 1, D–F). Because activity of the DAL5 promoter is sensitive to the nitrogen status of the growth medium, this variation might be a consequence of differential regulation of this promoter on the two types of media.
In contrast to its strong dominant inhibitory effects on [PSI+], Ssa1-21p had no obvious effect on [URE3] when Ssa2p was present (Figure 1, D–F, top row center). Moreover, for SSA1-21 ssa2Δ cells, in which [PSI+] cannot propagate (Figure 1A, bottom row center), [URE3] propagated well enough to be maintained through continued subculturing on media where growth does not require presence of the prion, although it was somewhat unstable under these conditions (Figure 1D, bottom row center). Despite this mitotic instability, this strain grew relatively well without adenine (Figure 1F, bottom row center), the condition requiring [URE3] for growth. Thus effects of Ssa1-21p on [URE3] were partial and recessive to Ssa2p.
Unlike Ssa1-21p, and in a manner very similar to its effects on [PSI+], Ssa2-21p dominantly impaired [URE3] when Ssa1p was present (Figure 1, D–F, middle row left) and [URE3] was unable to propagate in ssa1Δ SSA2-21 cells (Figure 1, D–F, middle row right). As for [PSI+], [URE3] was unable to propagate in cells expressing both Ssa1-21p and Ssa2-21p. Thus, Ssa1-21p and Ssa2-21p differed considerably in how they affected [URE3] propagation.
Depleting Ssa1p or Ssa2p affects prions differently:
We earlier showed the [PSI+] phenotype is slightly weakened in ssa1Δ cells but is normal in ssa2Δ cells (Hung and Masison 2006). In contrast, [URE3] propagation appears normal in ssa1Δ cells but unstable in ssa2Δ cells (Roberts et al. 2004). We confirm these results (Figure 1) and present a summary of how the different combinations of Ssa1p and Ssa2p affect prion propagation in Table 3. Together with our data for the Ssa1-21p and Ssa2-21p mutants, results with ssa1Δ and ssa2Δ strains indicate that [URE3] is more sensitive to changes in Ssa2p while [PSI+] is more sensitive to changes in Ssa1p.
TABLE 3.
Relative “strength” of [PSI+] and [URE3]
| Straina: | Prion phenotype
|
||
|---|---|---|---|
| ([PSI+]/[URE3]) | SSA genotype | [PSI+] | [URE3] |
| 792/1075 | SSA1 SSA2 | +++++ | +++++ |
| 793/1079 | SSA1-21 SSA2 | ++ | ++++ |
| 794/1076 | ssa1Δ SSA2 | ++++ | ++++ |
| 812/1080 | SSA1 ssa2Δ | +++++ | ++ |
| 813/1085 | SSA1-21 ssa2Δ | — | ++ |
| 814/1081 | ssa1Δ ssa2Δ | ++++ | — |
| 912/1082 | SSA1 SSA2-21 | ++ | + |
| 913/1083 | SSA1-21 SSA2-21 | — | — |
| 914/1084 | ssa1Δ SSA2-21 | — | — |
Subjective scoring is based on prion phenotypes in Figure 1 from strongest (+++++) to weakest (+). Minus (−) indicates prion cannot propagate.
The slash between strain numbers distinguishes strains used to monitor [PSI+] (left of slash) and [URE3] (right of slash).
We further find that cells lacking both Ssa1p and Ssa2p could propagate [PSI+] but not [URE3] (Figure 1, D–F, bottom row right). The stable [PSI+] phenotype in ssa1Δ ssa2Δ cells (strain 814, Figure 1, A–C, bottom row right) shows that Ssa3/4p, whose expression is induced in cells lacking both Ssa1p and Ssa2p (Jung et al. 2000), do not significantly affect [PSI+] propagation. The inability of [URE3] to propagate in ssa1Δ ssa2Δ cells, however, suggests Ssa3/4p do not allow [URE3] propagation. Alternatively, as strains lacking both Ssa1p and Ssa2p grow poorly at any temperature, combining [URE3] with deletion of both Ssa1p and Ssa2p could make cells grow too slowly to be considered viable.
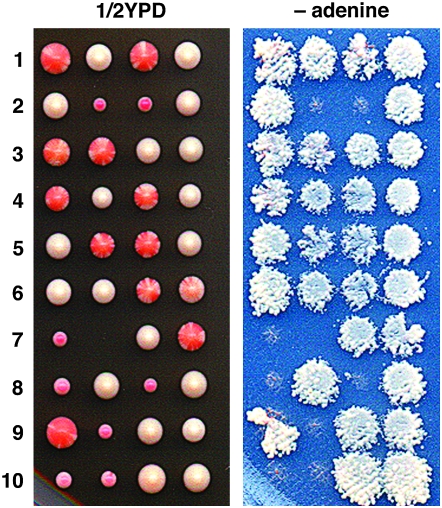
To test if combining [URE3] with ssa1Δ ssa2Δ is lethal, [URE3] diploids heterozygous for ssa1Δ and ssa2Δ were sporulated and 40 tetrads (160 spores) were dissected. The pattern of growth and [URE3] segregation in a sample of these tetrads is shown in Figure 3. Spore viability was normal (∼95%) and [URE3] was present and stable in all wild-type and ssa1Δ spore clones. Moreover, [URE3] is present, although very unstable, in all ssa2Δ colonies. Together these data show that [URE3] is transmitted with very high fidelity among all the spores and is readily detectable in spore colonies that contain predominantly [ure-o] cells. Therefore, all ssa1Δ ssa2Δ spores should have inherited [URE3], and its presence should be detectable. However, all of 36 colonies arising from these spores, and only these colonies, were entirely [ure-o]. These data show that [URE3] is not lethal to ssa1Δ ssa2Δ cells but that Ssa3/4p are incompatible with [URE3] propagation.
Figure 3.—
Cells lacking Ssa1p and Ssa2p cannot propagate [URE3]. Ten tetrads dissected on 1/2YPD (left) were replica plated onto synthetic medium lacking adenine (right). Larger colonies with predominantly red ([ure-o]) cells are ssa2Δ. Although white [URE3] cells within these colonies are sometimes difficult to see, their presence is evident by growth on the −adenine replica plate. All of the slowest growing colonies are ssa1Δ ssa2Δ double mutants. These colonies are entirely red on 1/2YPD and none show detectable growth without adenine, which indicates there are no [URE3] cells in the colonies.
Individual Ssa proteins variously influence prion propagation:
To evaluate how individual Ssa proteins influence prion propagation we used strains 1135 and 1161 (see Table 1) to monitor [PSI+] and [URE3], respectively. These strains lack all four chromosomal SSA genes and express Ssa1p, Ssa2p, Ssa3p, or Ssa4p from plasmid-borne genes regulated by the strong constitutive SSA2 promoter (see materials and methods). Since Ssa protein is essential, the plasmids are retained even when cells are grown on rich medium. All differences in prion phenotype among these strains grown under any conditions are attributed solely to effects of the plasmid-expressed Ssa protein. For strains lacking prions grown at 30°, those expressing Ssa3p or Ssa4p grew somewhat more slowly than the other strains but the remaining strains grew at a wild-type rate (see Table 4) showing that at the levels expressed each Ssa protein functioned well in essential processes that require Hsp70. Cells expressing only Ssa2-21p had a slight cold-sensitive growth defect but otherwise those expressing either Ssa1-21p or Ssa2-21p grew well at the other temperatures. These data are consistent with previously observed lack of growth phenotype for SSA1-21 cells and indicate that although the L483W substitution impairs [URE3] and abolishes [PSI+] prion propagation, it does not significantly affect Hsp70 function in any critical cellular process despite the near universal conservation of leucine at this position.
TABLE 4.
Relative growth of strains expressing individual Ssa proteins at different temperatures
| Growth
|
|||
|---|---|---|---|
| Strain/Ssap | 18° | 30° | 37° |
| 1135/Ssa1p | ++++ | +++++ | +++++ |
| 1135/Ssa2p | ++++ | +++++ | +++++ |
| 1135/Ssa3p | ++ | ++++ | ++++ |
| 1135/Ssa4p | + | +++ | +++ |
| 1135/Ssa1-21p | +++++ | +++++ | +++++ |
| 1135/Ssa2-21p | +++ | +++++ | +++++ |
Prion-lacking cells expressing the indicated Ssap as the only Ssa protein driven by the SSA2 promoter were streaked onto YPAD plates and incubated for 4 days at 18° or 3 days at 30° and 37°. Scoring reflects size of resulting colonies from largest (+++++) to smallest (+).
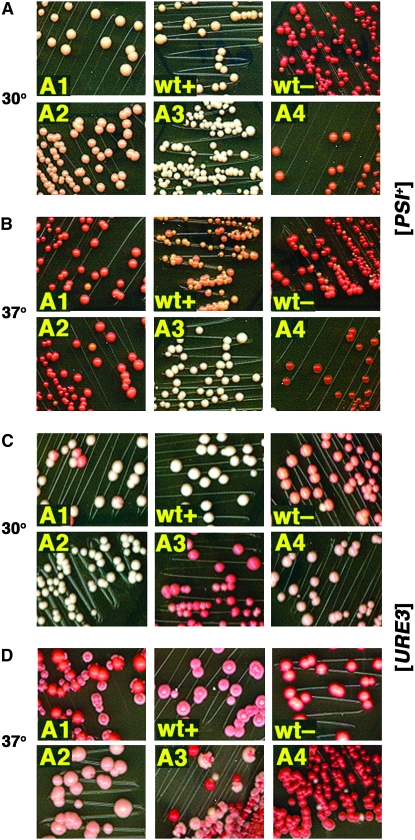
Strain 1135 cells expressing only Ssa1p had a normal [PSI+] phenotype (Figure 4A, top row left) while those expressing only Ssa2p (from plasmid pDCM62) had a slightly weakened [PSI+] phenotype (Figure 4A, bottom row left). These phenotypes are very similar to those we saw for ssa2Δ cells (Figure 1A, bottom row left) or ssa1Δ cells (Figure 1A, top row right), respectively. Thus, differences in the [PSI+] phenotype of ssa1Δ or ssa2Δ strains are not due simply to changes in overall expression levels of Ssa1p and Ssa2p, which normally are expressed from promoters that are regulated differently, or to any level of expression of Ssa3/4p. We showed earlier that deleting SSA1 induces expression of Hsp104 more so than deleting SSA2 (Jung et al. 2000). Modest increases in Hsp104 abundance can weaken the [PSI+] phenotype, so this difference might explain why [PSI+] is weaker in cells lacking Ssa1p.
Figure 4.—
Prion phenotypes of cells expressing individual Ssaps. Representative sections of streaks of cells from the same 1/2YPD plate, magnified to show detail, are shown. (A and B) Wild-type cells (strain 792) carrying (+) or lacking (−) [PSI+], and [PSI+] cells of strain 1135 expressing Ssa1p (1), Ssa2p (2), Ssa3p (3), or Ssa4p (4) as the only Ssap, as indicated, were incubated at 30° (A) or 37° (B) for 3 days. (C and D) As in A and B, respectively, except [URE3], strains are 1075 (wild type) and 1161 (expressing only Ssa1p-Ssa4p, as indicated). After incubation at the indicated temperature, plates were incubated 1 ([PSI+]) or 2 ([URE3]) more days at 23°.
When present as the only Ssa protein Ssa3p seemed to enhance [PSI+] propagation. Strain 1135 cells expressing only Ssa3p were whiter than wild-type cells when grown on 1/2YPD at 30° (Figure 4A, bottom row center). We earlier showed that elevated temperature considerably weakens [PSI+]-mediated nonsense suppression without affecting [PSI+] stability (Jung et al. 2000). The basis for this effect is unknown. Unlike cells expressing any of the other Ssa proteins, those expressing only Ssa3p did not show this characteristic weakening of [PSI+] when grown at 37° (Figure 4B). Moreover, wild-type cells do not get as red at 37° as those expressing the other individual Ssaps, which could be explained by increased expression of Ssa3p in these cells at this temperature. Thus, Ssa3p improved the [PSI+] phenotype at optimal temperature and suppressed prion-weakening effects of elevated temperature on [PSI+].
In contrast, the other stress-inducible Hsp70, Ssa4p, had an opposite effect on [PSI+]. When 1135 cells expressing only Ssa4p were grown on 1/2YPD at 30° they were brownish pink (Figure 4A), although loss of [PSI+] did not occur frequently enough to be detected upon continued restreaking. At 37° these cells were redder than any of the others. Thus, in a manner similar to the constitutively expressed Hsp70's, the conditionally expressed and Ssa3p and Ssa4p also functioned differently. Here, effects of Ssa3p and Ssa4p on [PSI+] resembled those of Ssa1p and Ssa2p, respectively.
In line with the contrasting effects that depleting Ssa1p or Ssa2p has on [PSI+] and [URE3], [URE3] was unstable when only Ssa1p was present but was normal in cells expressing only Ssa2p (Figure 4C). The effects Ssa3p and Ssa4p had on [URE3] also were reversed compared with their effects on [PSI+]. Here [URE3] propagation was weaker in cells expressing only Ssa3 than in cells expressing only Ssa4. Thus, with regard to prion propagation, opposing activities of the constitutively expressed Hsp70's were also seen for the conditionally expressed isoforms. Here again, Ssa3p resembled Ssa1p while Ssa4p resembled Ssa2p. Although Ssa3/4p functioned differently than Ssa1/2p, our data uncover functional similarities between Ssa1p and Ssa3p and between Ssa2p and Ssa4p.
When [URE3] cells expressing the individual Ssaps were grown at 37° they also showed a weakened phenotype that was very similar to the effect on [PSI+] at this temperature. In general, [URE3] cells grown at 37° were dark pink to red in color (Figure 4D). As with [PSI+], when cells grown at 37° were regrown at 30° their typical [URE3] phenotype returned, indicating that like [PSI+], [URE3] was weakened but not cured by growth at 37°. Thus, although elevated temperature causes solubilization of prion aggregates that causes pigment accumulation, it did not result in destruction of prion seeds leading to mitotic instability.
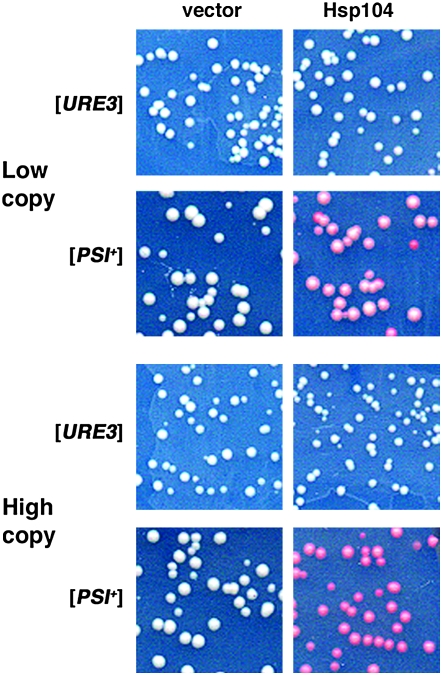
A possible explanation for the prion-weakening effect is that prions are partially solubilized by the increased abundance of Hsp104 induced by elevated temperature. To test this possibility we increased Hsp104 abundance under noninducing conditions either modestly or highly in [PSI+] and [URE3] strains by transformation with single-copy or multicopy plasmids carrying HSP104 controlled by its own promoter. As shown in Figure 5, [PSI+] was weak and unstable in cells carrying the low-copy plasmid and further weakened and destabilized in cells carrying the high-copy plasmid. In contrast, neither plasmid had any effect on the [URE3] phenotype. These results show that increasing abundance of Hsp104 alone does not produce the same general prion-weakening effect without prion elimination that is caused by elevated temperature and thus does not adequately explain the effect of elevated temperature on [PSI+] or [URE3].
Figure 5.—
Effects of elevated expression of Hsp104 on [URE3] and [PSI+]. Representative sections of primary transformation plates with limiting adenine of strains 1075 ([URE3]) and 792 ([PSI+]) transformed by low- and high-copy plasmids, as indicated, are shown. Strains transformed by plasmids carrying HSP104 (Hsp104) or lacking it (vector) are indicated.
DISCUSSION
Structural homology and functional redundancy among the very highly conserved Hsp70 molecular chaperones is significant enough that results from many studies of single isoforms are generalized to explain functions of others. Nevertheless, it has begun to be appreciated that different isoforms have specialized roles or possess different functions (Tutar et al. 2006; Daugaard et al. 2007; Genevaux et al. 2007; Brocchieri et al. 2008). Here we show that each of the four yeast cytosolic Ssa Hsp70's function differently with regard to propagation of yeast prions. The functional distinctions between the nearly identical Ssa1p and Ssa2p are highlighted by our finding that the same amino acid alteration in these two proteins had different affects on prion strength and stability. The most significant example is that [URE3] could propagate in cells expressing Ssa1-21p as the only Ssap but not in cells expressing only Ssa2-21p. The mutation in these proteins resides in the Hsp70 structural domain that binds partially unfolded protein substrates. However, given that these ∼150 residue domains of Ssa1p and Ssa2p differ by a single conservative substitution outside the substrate-binding pocket, we suspect the likelihood that the mutation could be affecting different substrate-binding activities is low. We favor the explanation that the functional differences between Ssa1p and Ssa2p are due to differences in the way they are regulated by ATP or other factors.
For prion particles (which are presumed to be composed of amyloid fibers) to propagate stably in yeast, they must grow and replicate. Growth occurs by addition of soluble prion protein onto fiber ends, and replication occurs by division of fibers. Aggregation of individual polymers into higher-order aggregates also occurs, and this aggregation might preclude replication resulting in “dead-end” aggregates. Alterations of the yeast prion phenotype reflect alterations in the processes of prion growth and replication. For example, the mitotic instability of [URE3] in cells expressing only Ssa1p could be explained by decreased efficiency of prion replication, which would result in a reduced number of prion particles per cell and a correspondingly higher chance that daughter cells fail to inherit the prion during cell division. Nevertheless these cells are white, indicating that despite a reduced number of fiber ends available to sequester Ure2 protein, fiber growth must occur fast enough to deplete Ure2p to a level that confers a normal [URE3] phenotype. In contrast, [URE3] is much more stable in cells expressing only Ssa4p, which implies they have more seeds, but they are pigmented, indicating that they have more soluble Ure2p and less Ure2p in prion aggregates. Here, it would be expected that replication occurs efficiently but growth is impaired. [URE3] was strong and stable in cells expressing only Ssa2p, but weak and unstable in cells expressing only Ssa3p. Remarkably therefore, each of the highly homologous Ssa proteins affected [URE3] prion propagation differently so that all of the four possible effects on growth and replication are represented (see Table 5).
TABLE 5.
Model for relative differences in protein solubility, processes influencing prion phenotype, and Ssa protein responsible for prion-specific effects
| Protein
|
Propagation
|
Ssapa
|
||||
|---|---|---|---|---|---|---|
| Prion phenotype | Soluble | Insoluble | Growth | Replication | [PSI+] | [URE3] |
| Strong, stable | Low | High | Fast | Fast | Ssa1p, 3p | Ssa2p |
| Strong, unstable | Low | High | Fast | Slow | NO | Ssa1p |
| Weak, stable | High | Low | Slow | Fast | Ssa2p, 4p | Ssa4p |
| Weak, unstable | High | Low | Slow | Slow | NO | Ssa3p |
NO, not observed.
The indicated wild-type Hsp70 is the only Ssap in the cell.
Although the effects of the stress-inducible Ssa3p and Ssa4p differed somewhat from those of Ssa1p and Ssa2p, the overall differences between Ssa3p and Ssa4p in terms of their opposing effects on [PSI+] and [URE3] paralleled those of Ssa1p and Ssa2p, respectively. Thus, although the stress-inducible Hsp70's functioned differently than their constitutively expressed counterparts, there was a similar distinction in how different isoforms of the constitutive and inducible Hsp70 interacted with the two prions. Together, our data uncover functional differences between constitutive and stress-inducible Hsp70's as well as a similarity in functional differences between the isoforms expressed under different conditions. These observations suggest first that stress-inducible Hsp70 isoforms possess functions important for cell survival during stress that are different from those of isoforms expressed under optimal conditions, and second that the subtle variations in Hsp70 function that might exist to improve fitness under optimal conditions are also important during stress.
An unexplained observation here and in earlier studies is that the [PSI+] phenotype, but not stability, is inhibited by elevated temperature (Eaglestone et al. 1999; Jung et al. 2000). Elevated temperature increased solubility of prion protein without loss of prion seeds, suggesting that protein from preexisting prion aggregates is somehow solubilized without completely destroying prion seeds. We now also show that this effect is a general one as [URE3] responded similarly to elevated temperature. When Hsp104 is modestly elevated under conditions it is not normally induced, it weakened [PSI+]. Unlike increased temperature, however, it also eliminated [PSI+] with reasonable efficiency and did not effect [URE3]. Thus, in addition to or independently of Hsp104, other factors are involved in the general stress-induced weakening of the prion phenotype. An interesting exception to the stress effect was that [PSI+] was unaffected by heat in cells expressing only Ssa3p. It is possible that among the Ssa proteins the stress-inducible Ssa3p has specialized or enhanced Hsp70 function that is important not only for health of cells exposed to stressful growth conditions, but also for processes important for optimal [PSI+] replication.
Hsp104 is the major component of the protein disaggregation machinery, which includes Hsp70 and its major cochaperone partner Hsp40, that is required for generating new prion seeds from preexisting material. Although Hsp104 is unique in the cell, in addition to the four Ssa Hsp70's yeast encode over a dozen Hsp40's, three of which are known to influence prion propagation (Kryndushkin et al. 2002; Moriyama et al. 2000; Sondheimer et al. 2001). Thus, there is a large number of combinations of Hsp70/40 whose differences in expression, specificity of interactions with each other or substrates, or cellular localization might explain the different effects on the prions through differences in their contributions to efficiency of protein disaggregation reactions. For example, when Ssa1p is paired with one Hsp40 (e.g., Sis1p), it could act differently than when partnered with another (e.g., Ydj1p), or the different Ssa proteins might have different physical and functional interactions with the various Hsp40's or the machinery as a whole. Such possibilities are currently under investigation. It is also possible that specific Hsp70/40 pairings might alter prions independently of Hsp104, either directly or through effects on as yet unidentified cellular processes important for prion propagation or that the different Hsp70 isoforms differentially alter expression of other factors that influence prions.
Although the fungal prions analyzed to date all have parallel in-register β-sheet structure, different prions, or even strains of the same prions, have different overall conformations and dynamics of growth and replication (Kushnirov and Ter-Avanesyan 1998; Shewmaker et al. 2006; Tanaka et al. 2006; Baxa et al. 2007; Wickner et al. 2008). Such conformational differences are likely a basis for differences in how well the chaperone machinery interacts with the different amyloids in vivo and how efficiently it is able to influence dynamics of growth and replication required for prion propagation. It is also likely that the different prion proteins contain structural elements that affect how they are recognized as substrates by the chaperone machinery. In the bacterial system the homologous Hsp70 and Hsp40 components, DnaK and DnaJ, respectively, act first on protein aggregates and recruit the Hsp104 homolog ClpB (Acebron et al. 2008). If the situation in yeast is analogous, then the Ssa proteins and Hsp40's could determine substrate recognition of the prions and initiate interaction of the machinery. It is known, for example, that Ydj1p interacts with Ure2 protein and diminishes its ability to form amyloid (Lian et al. 2007), and such a direct interaction in vivo could influence or be influenced by Hsp70 in different ways by the different Hsp70 isoforms.
Because prion propagation depends upon coordinated function of chaperone machinery, antiprion effects could result from alterations of individual components that influence specific functions of the machinery. The dominant impairment of [PSI+] by Ssa1-21p or Ssa2-21p could be due to partial disruption of machinery function in a process required by [PSI+] that is buffered by the presence of the wild-type Hsp70 counterpart. Notably, for the individual wild-type Ssa proteins a negative effect on one prion was opposed by a positive effect on the other, and none of the individual Hsp70's was optimal with regard to propagation of both prions. Thus, subtle variations in how the different Hsp70 isoforms function as a component of the chaperone machinery could cause the differences in prion phenotype that in turn can reveal which prion-specific processes are being affected. The differences in machinery function would affect prions differently if the prions have different requirements of the machinery for specific processes. For example, if [PSI+] prions are more fragile, and therefore less dependent on chaperones for division, but grow more slowly than [URE3], then changes in chaperone function that affect prion growth would affect [PSI+] more than [URE3], while changes that affect replication would affect [URE3] more than [PSI+]. Although some altered Hsp70 functions we observed are generally incompatible with prion propagation, they do not affect normal cellular processes enough to affect cell growth. This combination is an attractive feature of these alterations in terms of considerations for design of therapies to combat amyloid disorders.
Acknowledgments
We thank Christine Schwimmer and Seyung Chung for assistance with preliminary work on SSA2-21 and ssa2Δ strains and plasmid constructions, and Mehdi Kabani for constructing plasmids pA3 and pA4. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). D.S. is a recipient of a NIDDK Nancy Nossal Fellowship award.
References
- Acebron, S. P., V. Fernandez-Saiz, S. G. Taneva, F. Moro and A. Muga, 2008. DnaJ recruits DnaK to protein aggregates. J. Biol. Chem. 283 1381–1390. [DOI] [PubMed] [Google Scholar]
- Alani, E., L. Cao and N. Kleckner, 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron, R., T. Higurashi, C. Sahi and E. A. Craig, 2007. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 26 3794–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa, U., R. B. Wickner, A. C. Steven, D. E. Anderson, L. N. Marekov et al., 2007. Characterization of beta-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 46 13149–13162. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., U. Baxa and R. B. Wickner, 2005. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24 3082–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri, L., E. Conway de Macario and A. J. Macario, 2008. hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 8 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov and S. W. Liebman, 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268 880–884. [DOI] [PubMed] [Google Scholar]
- Cox, B. S., 1965. “Ψ” a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20 505–521. [Google Scholar]
- Daugaard, M., M. Rohde and M. Jaattela, 2007. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581 3702–3710. [DOI] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, J. Y. Hong and S. W. Liebman, 2001. Prions affect the appearance of other prions: the story of [PIN+]. Cell 106 171–182. [DOI] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, P. Zhou, Y. O. Chernoff and S. W. Liebman, 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone, S. S., B. S. Cox and M. F. Tuite, 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 18 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone, S. S., L. W. Ruddock, B. S. Cox and M. F. Tuite, 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux, P., C. Georgopoulos and W. L. Kelley, 2007. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 66 840–857. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., and S. Lindquist, 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94 73–82. [DOI] [PubMed] [Google Scholar]
- Gurley, W. B., 2000. HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12 457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, G. C., and D. C. Masison, 2006. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. W., and D. C. Masison, 2003. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. W., Y. Song, S. Chung and D. C. Masison, 2004. Propagation of yeast [PSI+] is prion impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24 3928–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., and D. C. Masison, 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43 7–10. [DOI] [PubMed] [Google Scholar]
- Jung, G., G. Jones, R. D. Wegrzyn and D. C. Masison, 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin, D. S., V. N. Smirnov, M. D. Ter-Avanesyan and V. V. Kushnirov, 2002. Increased expression of Hsp40 chaperones, transcriptional factors and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277 23702–23708. [DOI] [PubMed] [Google Scholar]
- Kryndushkin, D. S., I. M. Alexandrov, M. D. Ter-Avanesyan and V. V. Kushnirov, 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278 49636–49643. [DOI] [PubMed] [Google Scholar]
- Kushnirov, V. V., and M. D. Ter-Avanesyan, 1998. Structure and replication of yeast prions. Cell 94 13–16. [DOI] [PubMed] [Google Scholar]
- Kushnirov, V. V., D. S. Kryndushkin, M. Boguta, V. N. Smirnov and M. D. Ter-Avanesyan, 2000. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10 1443–1446. [DOI] [PubMed] [Google Scholar]
- Lian, H. Y., H. Zhang, Z. R. Zhang, H. M. Loovers, G. W. Jones et al., 2007. Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J. Biol. Chem. 282 11931–11940. [DOI] [PubMed] [Google Scholar]
- Lindquist, S., and G. Kim, 1996. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl. Acad. Sci. USA 93 5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, N., R. Aron and E. A. Craig, 2003. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol. Biol. Cell 14 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, H., H. K. Edskes and R. B. Wickner, 2000. [URE3] Prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness, F., P. Ferreira, B. S. Cox and M. F. Tuite, 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22 5593–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell, D. A., A. S. Kowal, M. A. Singer and S. Lindquist, 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372 475–479. [DOI] [PubMed] [Google Scholar]
- Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov and M. D. Ter-Avanesyan, 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Roberts, B. T., H. Moriyama and R. B. Wickner, 2004. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast 21 107–117. [DOI] [PubMed] [Google Scholar]
- Schlumpberger, M., S. B. Prusiner and I. Herskowitz, 2001. Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 21 7035–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer, C., and D. C. Masison, 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker, F., R. B. Wickner and R. Tycko, 2006. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc. Natl. Acad. Sci. USA 103 19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer, N., and S. Lindquist, 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5 163–172. [DOI] [PubMed] [Google Scholar]
- Sondheimer, N., N. Lopez, E. A. Craig and S. Lindquist, 2001. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 20 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Y. X. Wu, G. Jung, Y. Tutar, E. Eisenberg et al., 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., S. R. Collins, B. H. Toyama and J. S. Weissman, 2006. The physical basis of how prion conformations determine strain phenotypes. Nature 442 585–589. [DOI] [PubMed] [Google Scholar]
- Tuite, M. F., C. R. Mundy and B. S. Cox, 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar, Y., Y. Song and D. C. Masison, 2006. Primate chaperones Hsc70 (constitutive) and Hsp70 (induced) differ functionally in supporting growth and prion propagation in Saccharomyces cerevisiae. Genetics 172 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn, J., C. Schlieker, P. Tessarz, A. Mogk and B. Bukau, 2005. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol. Chem. 386 739–744. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., D. E. Stone and E. A. Craig, 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7 2568–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R. B., 1994. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science 264 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., H. K. Edskes, F. Shewmaker and T. Nakayashiki, 2007. Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 5 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R. B., F. Dyda and R. Tycko, 2008. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc. Natl. Acad. Sci. USA 105 2403–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]