Abstract
The Drosophila dRYBP gene has been described to function as a Polycomb-dependent transcriptional repressor. To determine the in vivo function of the dRYBP gene, we have generated mutations and analyzed the associated phenotypes. Homozygous null mutants die progressively throughout development and present phenotypes variable both in their penetrance and in their expressivity, including disrupted oogenesis, a disorganized pattern of the syncytial nuclear divisions, defects in pattern formation, and decreased wing size. Although dRYBP mutations do not show the homeotic-like phenotypes typical of mutations in the PcG and trxG genes, they enhance the phenotypes of mutations of either the Sex comb extra gene (PcG) or the trithorax gene (trxG). Finally, the dRYBP protein interacts physically with the Sex comb extra and the Pleiohomeotic proteins, and the homeotic-like phenotypes produced by the high levels of the dRYBP protein are mediated through its C-terminal domain. Our results indicate that the dRYBP gene functions in the control of cell identity together with the PcG/trxG proteins. Furthermore, they also indicate that dRYBP participates in the control of cell proliferation and cell differentiation and we propose that its functional requirement may well depend on the robustness of the animal.
PATTERN formation during animal development requires the controlled spatial and temporal regulation of gene expression. Once gene transcriptional states have been established, their maintenance during cellular proliferation is crucial for the normal development of the organism. The Polycomb (PcG) and the trithorax (trxG) groups of genes play a pivotal role in this process (for a recent review see Schuettengruber et al. 2007). The PcG genes are required for the maintenance of the repressed state while the trxG are needed for the maintenance of the active state. The PcG and trxG genes were first identified in the fly Drosophila melanogaster, due to their role in morphogenesis as regulators of homeotic gene expression (Lewis 1978; Jürgens 1985; Breen and Harte 1991; for a review see Ringrose and Paro 2004). However, it is now clear that the PcG and trxG genes also have relevant roles in other biological processes, such as hematopoiesis, stem cell renewal, control of cell proliferation, and tumorigenesis (van der Lugt et al. 1994; Valk-Lingbeek et al. 2004; Brock and Fisher 2005; Ferres-Marco et al. 2006; Martinez and Cavalli 2006; Sparmann and Van Lohuizen 2006).
Central to PcG/trxG epigenetic-mediated mechanisms is the recruitment and formation of multimeric protein complexes. In Drosophila, three major protein complexes containing PcG proteins have been isolated. The first identified were the complexes Polycomb repressive complex 1 (PRC1) (Shao et al. 1999) and PRC2 (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). The core of PRC1 includes Polycomb (PC), Posterior sex combs (PSC), the E3 ubiquitin ligase Sex comb extra (SCE), and Polyhomeotic (PH). The core of PRC2 is composed of the histone methyl transferase Enhancer of zeste [E(Z)], Suppressor of zeste 12 [SU(Z)12], Extra sex combs (ESC), and Nurf-55. The third repressive complex, pleiohomeotic repressor complex (PHORC), containing the pleiohomeotic (PHO) protein, has recently been isolated from Drosophila embryos (Klymenko et al. 2006). Three trxG complexes have been identified: trithorax acetylation complex (TAC), NURF, and the SWI/SNF (for reviews see Grimaud et al. 2006b; Schwartz and Pirrotta 2007). There are other PcG/trxG proteins that do not form part of the core of these complexes, but still are associated with them and, therefore, have been classified as PcG/trxG-associated proteins (Otte and Kwaks 2003).
While much is known about the roles of the PcG/trxG proteins in the Drosophila morphogenesis, less is known about their role in biological processes such as the control of cellular proliferation and differentiation during development. Mutations in some of the Drosophila PcG/trxG genes cause phenotypes associated with misregulation of cell proliferation. For example, E(z) was identified in a screen for essential cell-cycle genes (Gatti and Baker 1989), and mutations in the E(z) gene produce proliferation defects and the appearance of small imaginal discs (Phillips and Shearn 1990). Furthermore, the corto protein (a centrosomal and chromosomal factor) colocalizes with PSC binding sites at polytene chromosomes, interacts genetically with PcG mutations, and affects progression through mitosis (Kodjabachian et al. 1998). Moreover, mutations in the PcG genes ph, Pc, and Psc show segregation defects during syncytial embryonic mitosis (O'Dor et al. 2006). Finally, it has been shown recently that the expression of cyclin A is directly regulated by the PcG proteins, showing a clear link between these proteins and the control of the cell cycle (Martinez et al. 2006).
To define the mechanisms by which PcG/trxG function, the isolation and functional characterization of each component of the complexes is necessary. The murine RYBP gene was identified in a two-hybrid screen designed to isolate Ring1A/Ring1B (SCE in Drosophila) interacting proteins (Garcia et al. 1999). Because of its interaction with Ring1A, M33, and YY1 (SCE, PC, and PHO, respectively, in Drosophila)—all of them key components of the PcG complexes—RYBP was proposed to belong to the PcG of proteins (Garcia et al. 1999). Studies of the murine RYBP gene and protein indicate that the gene has several distinct biological roles and, because the protein can bind to several transcription factors, it has been proposed to function as an adaptor protein (Garcia et al. 1999; Trimarchi et al. 2001; Sawa et al. 2002; Schlisio et al. 2002). It has been shown that the murine RYBP protein is a novel ubiquitin-binding protein that is itself ubiquitinated. Furthermore, one of its targets appears to be the ubiquitinated histone H2A, which is also a substrate of Ring1 B E3 ubiquitin ligase (Arrigoni et al. 2006). Recently, a BCOR protein complex has been isolated that, together with the RYBP protein, includes a Posterior sex combs homolog, NSPC1, and RNF2, an E3 ligase with H2A mono-ubiquitylation activity (Gearhart et al. 2006; Sanchez et al. 2007). Together these results suggest that the interaction of RYBP and RNF2/Ring1A/SCE may be necessary for the mono-ubiquitylation of H2A (Arrigoni et al. 2006), an essential mechanism for the maintenance of gene expression (Wang et al. 2003; de Napoles et al. 2004; Fang et al. 2004; Cao et al. 2005). Finally, RYBP knockout mice exhibited lethality at the early postimplantation stage, suggesting an essential role in survival. No homeotic phenotypes were reported in these mutants, but the lack of RYBP function in the central nervous system (CNS) produced brain overgrowth and disrupted neural tube closure (Pirity et al. 2005).
Previously, we described the identification of the dRYBP gene in Drosophila (Bejarano et al. 2005). We showed that the dRYBP protein behaves as a Polycomb-dependent transcriptional repressor throughout development. Furthermore, we showed that high levels of dRYBP protein produce homeotic-like phenotypes that can be modulated by mutations in PcG/trxG genes. These results suggested that the dRYBP protein could function by recruiting the PcG proteins and, therefore, linked dRYBP to the mechanisms of maintenance of gene expression in Drosophila.
We have studied the biological role of dRYBP by characterizing the phenotypes associated with dRYBP mutations. The phenotypes associated with dRYBP mutations are variable both in their expressivity and in their penetrance. dRYBP mutations are pleiotropic, producing progressive lethality during development, arrest in the syncytial nuclear divisions, defects in morphogenesis, reduced size of the wings, and cell differentiation defects. Although dRYBP mutations alone do not result in homeotic-like phenotypes, they do when in combination with mutations in some PcG and trxG genes. We have found that the dRYBP protein is localized in a nuclear pattern and colocalizes with some of the Polycomb nuclear bodies (Buchenau et al. 1998; Saurin et al. 1998; Netter et al. 2001; Ficz et al. 2005; Grimaud et al. 2006a). Moreover, the protein is dynamically distributed during the mitotic cycle in the syncytial embryo. Furthermore, the dRYBP protein physically interacts with SCE and PHO proteins. Finally, we also show that the C-terminal domain of dRYBP protein is required to produce the homeotic-like phenotypes. Our results suggest that dRYBP participates in the control of cell identity and in the control of cell proliferation and cell differentiation through a direct or an indirect interaction with PcG and trxG proteins.
MATERIALS AND METHODS
Drosophila strains and general procedures:
The following mutations were employed: Pc3, Sce1, pho1, phocv, trxE2, TrlR85, Trl13c, Rpd31, and Df (1) w67c23 flies (y−, w−) (all described in FlyBase, http://flybase.bio.indiana.edu/stocks). The Df(2R) 58B3-59 (kindly provided by T. L. Orr-Weaver) deletes dRYBP and additional genes, and the PC-EGFP transgenic line (kindly provided by R. Paro) reproduces the expression of the Polycomb protein (Dietzel et al. 1999). The recombinant stock dRYBP1, P[Histone-3:GFP]/CyO was made using flies containing P[Histone-3:GFP] on the second chromosome (provided by S. Aldaz). Genetic interactions between dRYBP and PcG/trxG mutations were studied, using stocks containing P[SUPor-P]CG12190 [KG08683] (Bellen et al. 2004) (herein called dRYBP1) in the second chromosome and one of the PcG/trxG mutations on the third or the fourth chromosome. The number of flies examined ranged from 60 to 104. Homeotic transformations observed in these interaction studies appear to be very sensitive to crowding conditions, and therefore care was taken to avoid such conditions. Somatic heat-shocked induced clones were generated by crossing female yw1118 P[hsp70-FLP122]; FRTG13, P[Ubi-GFPnls]2R2 with male FRTG13, dRYBP1/CyO and by crossing female yw1118 P[hsp70-FLP122] f36a ; FRT42D sha1 [forked+]/CyO with male w1118; FRT42D dRYBP1/CyO. The larval progenies of the different crosses were subjected to a 1-hr, 37° heat pulse at 24–48 hr or 48–72 hr or 72–96 hr after egg deposition. For overexpression experiments the GAL4/UAS system was used (Brand et al. 1994) at 25° and 29° with the lines engrailed-GAL4 (en-GAL4) (Brand and Perrimon 1993), cubitus-GAL4 (ci-GAL4), armadillo-GAL4 (arm-GAl4) (Sanson et al. 1996), Ultrabithorax-GAL4 (Ubx-GAL4) (Calleja et al. 1996), scalloped-GAL4 (sd-GAL4) (Calleja et al. 1996), daughterless-GAL4 (da-GAL4) (Wodarz et al. 1995), and tubulin-GAL4 (tub-GAL4). To analyze the ovaries of the females, the virgins were crossed with males and, after 3 days, the ovaries were dissected and stained with DAPI.
The lethal phase of the homozygous dRYBP1 was determined using a dRYBP1/CyOGFP stock. Male and female dRYBP1/CyOGFP were crossed and eggs (0–6 hr old) were collected, counted (∼400 each experiment), and allowed to develop on plates [controls, using the Df (1)w67c23 flies, were done in parallel]. The “non-GFP” embryos were counted and their survival was monitored throughout development.
The fertility of the homozygous dRYBP1 females was determined by making crosses of a single homozygous dRYBP1 female with two Df (1)w67c23 males [controls were performed by crossing a single Df (1)w67c23 female with two Df (1)w67c23 males]. Similarly, the fertility of the homozygous dRYBP1 males was determined by making crosses of a single homozygous dRYBP1 male with two Df (1)w67c23 virgins [controls were made with a single Df (1)w67c23 male crossed to two Df (1)w67c23 females].
To study the effect of the dRYBP mutations on the syncytial nuclear divisions, we made a recombinant stock dRYBP1, P[Histone-3:GFP]/CyO. We have previously confirmed that P[Histone-3:GFP]/P[Histone-3:GFP] embryos do not show an aberrant nuclear division pattern. Female and male dRYBP1, P[Histone-3:GFP]/CyO were crossed, embryos (0–3 hr old) were collected and fixed, and the pattern of nuclear divisions was observed. The syncytial mitotic phenotypes were variable in expressivity and penetrance. Also, the percentage of embryos showing mitotic phenotype was highly variable between experiments, making quantification very difficult. In this cross, the dRYBP1, P[Histone-3:GFP] homozygous embryos were indistinguishable from the heterozygous embryos. Therefore to be sure that the mitotic phenotype was strictly maternal, we studied the pattern of nuclear divisions in embryos from female dRYBP1, P[Histone-3:GFP]/CyO crossed with Df (1)w67c23 males. The syncytial mitotic phenotypes observed in the embryos from this cross were also variable in expressivity and penetrance. Finally, the potential influence of the balancer chromosome on the syncytial mitotic phenotype was excluded by studying the pattern of DAPI staining in the 3-hr-old embryos from females and males P[Histone-3:GFP]/CyO. For this, embryos were collected, fixed, and incubated with DAPI (5 μm in PBS) for 20 min; washed three times in PBS; and mounted for microscopic inspection. The pattern of nuclear divisions in these embryos was normal.
Studies of the lethality and of the cuticle embryonic phenotypes resulting from inactivation of dRYBP were performed using RNA interference. Males and females from the stocks en-GAL4<UAS-dRYBPRNAi or ci-GAL4<UAS dRYBPRNAi were allowed to lay eggs for 6 hr, and eggs were counted (∼400 in each experiment) and allowed to develop on plates. Embryos were collected and mounted for microscopy analysis of the cuticle phenotypes, using standard methods.
Transgenic flies were obtained by standard procedures using Df(1)w67c23 (y−, w−) as host flies. The mounting of larvae and adult flies was performed using standard protocols. Wing size was measured by mounting the wings and next analyzing mounted wings, using the ImageJ image analysis software. With this software, the length of the margin of each mounted wing is determined and used to calculate the wing area. The mean area of multiple mounted wings was used for comparisons.
P-element mutagenesis and screening:
The stock y1w1118; P{SUPor-P}CG12190 [KG08683]/CyO (Bellen et al. 2004) that contains the markers white+ and yellow+ was used. Before starting the mutagenesis experiments, the stock was systematically outcrossed to clean possible second-site mutations. y1w1118; P [y+w+KG08683]/CyO were crossed with female w1118; Sp/CyO; Δ2-3 Dr/TM6Ubx. Male progeny w1118; P [y+w+KG08683]/CyO; Δ2-3 Dr/+ was individually crossed with female y1w1118; Sco/CyOwglacZ and the white− progeny were scored to establish a stock. Genomic DNA was isolated and PCR was performed using the primers 5′-AACACTGGCTGCGCGTACTATCG-3′, 5′-GCGGGAGAGAAGACAACGACTCC-3′, and 5′-GTTCCACTAGCAGCGCCCATCCC-3′. PCR fragments were sequenced after analysis of fragment length. The precise excisions were checked for the lethality phenotype.
Immunohistochemistry:
The antibody staining of embryos and imaginal discs was performed using standard protocols. The primary antibodies used were rabbit anti-DRYBP (1:100) (Bejarano et al. 2005), mouse anti-Ubx (1:10) (White and Wilcox 1984), mouse anti-Abd-B (1:20) (Celniker et al. 1990), rabbit anti-Abd-A (1:20) (Macias et al. 1990), rat anti-Antp (1:1500) (Reuter and Scott 1990), rabbit anti-Scr (1:100) (Glicksman and Brower 1988), mouse anti-en (1:200) (Patel et al. 1989), rat anti-ci (1:50) (Motzny and Holmgren 1995), rabbit anti-Pc (Wang et al. 2004), rabbit anti-Pho (1:10) (Brown et al. 1998), rabbit anti-Sce (1:200) (Gorfinkiel et al. 2004), mouse anti-cyclin A (Hibridoma Bank) (Knoblich and Lehner 1993), rabbit anti-GFP (1:300) (Invitrogen, San Diego), rabbit anti-activated caspase 3 (Cell Signaling Technologies), anti-BrdU (1:10) (Roche, Indianapolis), rabbit anti-β-gal (Cappel), mouse anti-β-gal (Promega, Madison, WI), rat anti-α-tubulin (1:500) (Seralab), mouse anti-Histone-H3 trimethyl Lys 27 (1:500) (Active Motif), and mouse anti-Psc (1:50) (Hybridoma Bank). To-Pro (Kiernan J 2001) and DAPI (Kiernan J 2001) were used to stain the DNA. Daunomycine (Chaires 1983) was used to label the nucleoli. Apoptosis was analyzed by Tunnel (In Situ Cell Death Detection kit TMR, Roche), acridine orange (Abrams et al. 1993), and anti-activated caspase 3 antibody stainings.
Expression constructs:
The dRYBP-ΔCt and dRYBP-ΔZF fragments were generated by PCR amplification from the dRYBP cDNA (LD18758, Drosophila FlyBase). For dRYBP-ΔCt, the primers 5′-CATGTGCGACGTGCGGAAAGGAGGGATACAAGGCCTC-3′ and 5′-GAGGCCTTGTATCCCTCCTTTCCGCACGTCGCACATG-3′ were used; for dRYBP-ΔZF the 5′-CCTCCTCTCCTCCTGTATTATGCCCAACGGGAAGTCC-3′ and 5′-GGACTTCCCGTTGGGCATAATACAGGAGGAGAGGAGG-3′ were used. The PCR fragments were cloned in pGEM and sequenced. The expression construct fragments were cloned in the pUAST vector and transgenic flies P[UAS-dRYBP-ΔCt] and P[UAS-dRYBP-ΔZF] were generated. The pUAST-dRYBPRNAi construct was made with a PCR-amplified 480-bp fragment obtained using 5′-CCCGGTACCGGGCTTTAAACGTGG-3′ and 5′-CCCGGATCCAGGAACCTCCACGC-3′. The PCR product was cloned in pGEM-easy (Promega), generating pGEM-dRYBP-480RNAi, which, after several cloning steps (details upon request) using the vector pHIBS (Nagel et al. 2002) and the vector pUAST, yielded pUAST-dRYBPRNAi. This construct was injected to generate the P[UAS-dRYBPRNAi] transgenic flies.
Quantitative PCR:
RNA from first instar larvae and adult flies of the Df(1)w67c23, dRYBP1/dRYBP1, dRYBP Δ16/dRYBP Δ16, and Df(2R) 58B3-59/CyOGFP genotypes was isolated by lysis and homogenization in TriZOL (Invitrogen), using a Pellet Motor (short pulses during 30 sec), followed by centrifugation (3 min, 13,000 rpm), chloroform/isopropanol extraction, ethanol precipitation, and resuspension in DEPC-water. Both the RT and the PCR reactions were done following the instructions accompanying the TaqMan predesigned gene-expression assay kit (Applied Biosystems, Foster City, CA) for Drosophila CG12190 (reference no.137861.2). RP49RNA (Foley et al. 1993) and 18srRNA primers and probes were used as controls.
Co-immunoprecipitaion, pull-down assays:
arm-GAL4>UAS-dRYBP embryos were pelleted and lysed in 250 μl of lysis buffer containing 1% Triton X-100, 10 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1mm EGTA pH 8, and protease inhibitors (Complete Mini, Roche) at 4° for 15 min. After removing the cellular debris by centrifugation, 400 μg total protein lysate were incubated overnight with 25 μl 10 mg/ml BSA and 4 μg anti-SCE antibody or 6 μg of anti-β-gal antibody as a negative control. This mixture was then incubated with 30 μl of Protein-A Sepharose beads (Sigma-Aldrich) in PBS for 2 hr while rotating at 4°. The beads were collected and washed with lysis buffer four times at 4°. Proteins were eluted from the beads by heating with 20 μl of 2× PAGE loading buffer [200 mm DTT, 4% SDS, 100 mm Tris (pH 6.8), 20% glycerol, 0.1% bromophenol blue] at 100° for 5 min. The eluted fractions were resolved on a 15% PAGE gel system and transferred to a nitrocellulose membrane. After blocking, the membrane was incubated with anti-DRYBP antibody (1:500) followed by the appropriate secondary antibodies conjugated with horseradish peroxidase (HRP). Signals were detected with ECL reagents (Amersham, Arlington Heights, IL). We also attempted immunoprecipitation (IP) experiments with anti-PHO antibody. However, these experiments were not successful and, as a result, we performed pull-down assays to study the interaction with PHO.
Pull-down experiments used crude Drosophila extracts prepared by homogenizing 0.2 ml of third instar larvae in 0.4 ml of lysis buffer [1% NP-40, 1 mm PMSF, and protease inhibitor cocktail (Roche) in PBS]. The homogenates were centrifuged, and the aqueous supernatant was mixed with the full-length His-DRYBP protein [extracted under native conditions and purified by following the QIA express (QIAGEN, Valencia, CA) protocols]. The complexes were purified using Ni-NTA Agarose (QIAGEN) and washed five times in 0.5 m NaCl in PBS and one time in 50 mm Tris (pH 6.8). Proteins were eluted by boiling in loading buffer and resolved by 15% SDS–PAGE. After blotting to nitrocellulose, the membrane was incubated with anti-PHO antibody (1:500) and the signal was detected using the Amersham ECL Western blotting analysis system.
5′-RACE analysis:
Total RNA was extracted from Df(1)w67c23 (control) and from dRYBP Δ55/dRYBP Δ55 embryos as described above for quantitative PCR. cDNAs containing the 5′ transcript ends were identified using the 5′-RLM–RACE reagents (FirstChoice RLM–RACE kit; Ambion, Austin, TX), using the 5′ primer adaptor provided and the primer 5′-TTCCCGTTGGGCATGTTGACACTGGC-3′ based on the dRYBP sequence. The products were analyzed by agarose gel, isolated from the gel, cloned in pBluescript, and sequenced.
RESULTS
The dRYBP gene and dRYBP protein expression:
The dRYBP gene (CG12190, FlyBase) is cytogenetically located at 58F7 and extends over 2.4 kb of genomic DNA (Figure 1A). 5′-RACE analysis shows that the transcription unit produces a single mRNA (Figure 1C). This transcript encodes a 150-aa protein (18 kDa) with a conserved amino terminus that includes an Npl4 zinc finger (NZF) type zinc finger (Meyer et al. 2000, 2002) (Figure 1B). The carboxy terminus is conserved within the dRYBP proteins, but shows no similarity with any domains in the databases (Bejarano et al. 2005). Recently, a subgroup of the NZF domains (Wang et al. 2003; Alam et al. 2004) that include RYBP (Arrigoni et al. 2006) has been shown to possess ubiquitin-binding activity.
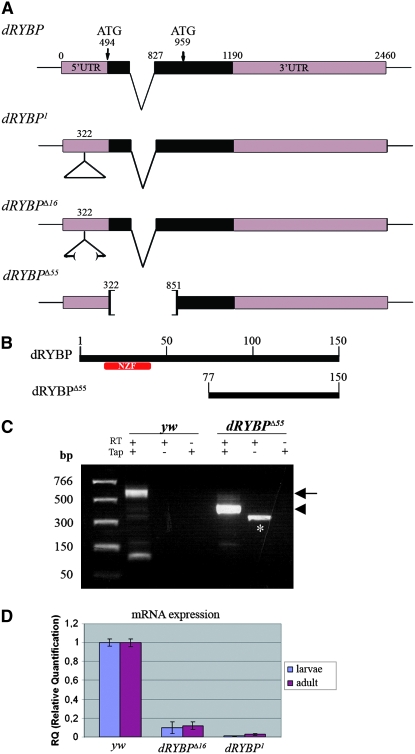
Figure 1.—
Mutations in the dRYBP gene, 5′-RACE and quantitative PCR analysis. (A) The dRYBP gene structure, showing the positions of the two in-frame ATGs codons. dRYBP1 contains the P[KG08683]-element insertion (indicated as a triangle) at nucleotide position 322. dRYBPΔ16 is an incomplete deletion of the P[KG08683] element. dRYBPΔ55 deletes nucleotides 322–853. (B) The dRYBP protein contains an NZF domain (red) located in the amino terminus. The putative dRYBPΔ55 protein encoded by dRYBPΔ55 does not contain the NZF domain (red). (C) 5′-RACE analysis was performed using total RNA isolated from homozygous Df(1)w67c23 (yw) and homozygous dRYBPΔ55 embryos. The reverse-transcription (RT) reaction was performed in the presence and in the absence of Tobacco acid pyrophosphatase (Tap). A control containing “minus-Tap”-treated RNA was also performed. A band of 560 bp (arrow) was observed, corresponding to the dRYBP wt 5′-RACE product (lane yw, “plus-RT/plus-Tap”; the smaller band in this lane is an artifact). A band of 380 bp (arrowhead) corresponds to the dRYBPΔ55 5′-RACE product (lane dRYBPΔ55, plus-RT/plus-Tap). The band in the second lane of dRYBPΔ55 (marked with *) corresponds to “plus-RT/minus-Tap” is an artifact of the 5′-RACE protocol used (Instructions manual, FirstChoice-RLM-RACE; Ambion). (D) Quantitative PCR results showing the relative mRNA expression levels from homozygous Df(1)w67c23 (yw), dRYBP1, and dRYBPΔ16 early first-instar larvae and adults.
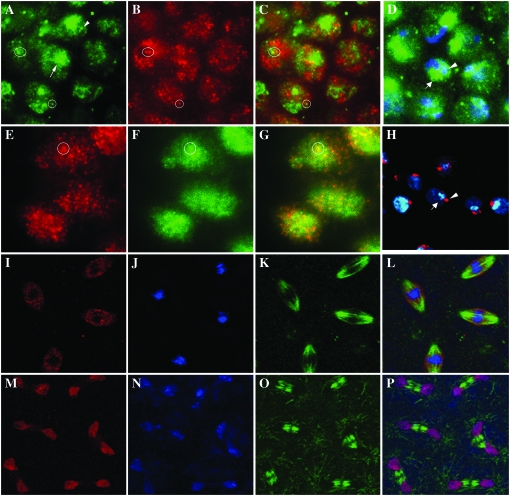
We have shown previously that the dRYBP protein is found in the oocyte nucleus, suggesting a maternal component of the protein (Bejarano et al. 2005). Moreover, its nuclear expression is observed ubiquitously and throughout development. A closer inspection of dRYBP distribution within the nucleus (Figure 2, A–H) reveals that it does not colocalize to either the heterochromatin (labeled with DAPI, Figure 2D) or the nucleoli (labeled with Daunomycine, Figure 2H). However, like the PcG proteins (Buchenau et al. 1998; Saurin et al. 1998; Netter et al. 2001; Ficz et al. 2005; Grimaud et al. 2006a), dRYBP is distributed in the nucleus in a discrete punctate pattern (Figure 2, A and E). We refer to these sites of protein localization as dRYBP nuclear bodies. Typically, there are of the order of ∼30 small discrete spots plus one more prominent spot per nucleus (Figure 2A). The dRYBP nuclear bodies are similar to both PC nuclear bodies (Buchenau et al. 1998; Ficz et al. 2005; Grimaud et al. 2006a), which appear as small discrete spots, and Posterior sex combs nuclear bodies (Buchenau et al. 1998), which appear as a combination of small spots plus one prominent spot. We have studied whether dRYBP nuclear distribution colocalizes with PC [using the transgenic flies PC-EFGP (Dietzel et al. 1999)]. Additionally, we have studied whether dRYBP nuclear distribution colocalizes with the nuclear distribution of Histone H3 trimethyl Lys27 (H3K27me3) because it has been shown that H3K27me3 marks the PC nuclear bodies (T. Cheutin, personal communication). The results shown in Figure 2 indicate that some, but not all, of the dRYBP nuclear bodies colocalize with the PC bodies (Figure 2, A–C) and with the H3K27me3 nuclear bodies (Figure 2, E–G). The pattern of dRYBP distribution during the mitotic progression in the syncytial cell cycles of Drosophila embryos has been also analyzed. The first 13 mitotic divisions are synchronous and very rapid due to very abbreviated G1 and G2 phases and the absence of cytokinesis (Orr-Weaver 1994; Tram et al. 2001). Figure 2 shows the dynamic dRYBP distribution pattern throughout mitosis. dRYBP is dissociated from chromatin in prophase, remains visibly dissociated in metaphase (Figure 2, I–L), becomes associated with chromatin in anaphase (Figure 2, M–P), and remains bound in telophase. A similar dynamic pattern of distribution has also been observed for the Drosophila PC, PH, and PSC proteins (Buchenau et al. 1998).
Figure 2.—
Nuclear localization pattern of dRYBP protein. (A) dRYBP protein localization (green) in the embryonic nuclei. dRYBP nuclear bodies appear as both large (arrows) and small (arrowheads) structures. (B) H3K27me3 (red) colocalizes (circle) with dRYBP in some of the nuclear bodies (circle). (C) Merged image of A and B. The circles indicate colocalization in some of the bodies. (D) Staining of DAPI (blue) and dRYBP (green). (E) Embryonic nuclear distribution of dRYBP (red). (F) GFP nuclear localization of PC-EGFP transgenic embryos (circle indicates colocalization). (G) Merged image of E and F. (H) Daunomycine staining [used to label the nucleoli (red, arrowhead)] and dRYBP protein expression (blue, arrow) do not colocalize in the nuclei of the wing imaginal disc cells. (I–L) Localization of dRYBP protein (I, red), To-PRO (J, blue), and α-tubulin (K, green) during metaphase of syncytial embryonic nuclear divisions. dRYBP does not localize with To-PRO (J). (L) Merged image of I–K. (M–P) Localization of dRYBP protein (M, red), To-PRO (N, blue), and α-tubulin (O, green) during anaphase of syncytial embryonic nuclear divisions. dRYBP (red) colocalizes with To-PRO (blue). (P) Merged image of M–O.
Genetic and molecular characterization of dRYBP mutations:
The P-element P[KG08683] or P{SUPor-P}CG12190KG08683 (Bellen et al. 2004) is inserted in the 5′-UTR of the dRYBP gene (Figure 1A). This insertion, as described below, creates a null mutation in the dRYBP gene and we have therefore named it dRYBP1. We have obtained two imprecise excisions from dRYBP1: dRYBPΔ16 and dRYBPΔ55 (Figure 1A).
dRYBP1 is classified as a null mutation because homozygous dRYBP1 embryos and homozygous dRYBP1 adult flies do not express dRYBP mRNA (Figure 1D). However, dRYBP protein is seen in dRYBP1/dRYBP1 embryos. We interpret this to be a result of the maternal contribution (see materials and methods), as no protein is seen in the imaginal discs of dRYBP1/dRYBP1 larvae.
All the phenotypes observed in embryos, larvae, and adults of the dRYBP1/dRYBP1 genotypes are highly variable both in their penetrance and in their expressivity, and similar results were obtained when the phenotypes were studied in dRYBP1/Df(2R) 58B3–59. Flies dRYBP1/dRYBP1are sublethal and show progressive lethality throughout development. Only 13% of the dRYBP1 homozygous embryos reached the adult stage, with 43% dying during embryogenesis and 44% during larval/pupal development. Moreover, dRYBP1/dRYBP1 larvae show a significant developmental delay: it takes 10 days at 25° for dRYBP1/dRYBP1 first-instar larvae to reach the pupal stage, instead of the normal 4–5 days (Ashburner 1989). Nearly all (90%) homozygous dRYBP1 females are sterile and, in most cases, oogenesis is arrested at stage 8 (Ashburner 1989) (Figure 3J). Finally, dRYBP1/dRYBP1 adult flies from the few dRYBP1/dRYBP1 fertile mothers crossed with dRYBP1/dRYBP1 fathers present the same phenotypes described below for the homozygous dRYBP1 offspring from dRYBP1/+ parents.
Figure 3.—
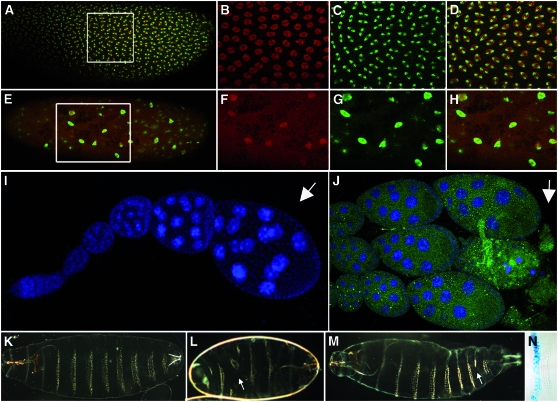
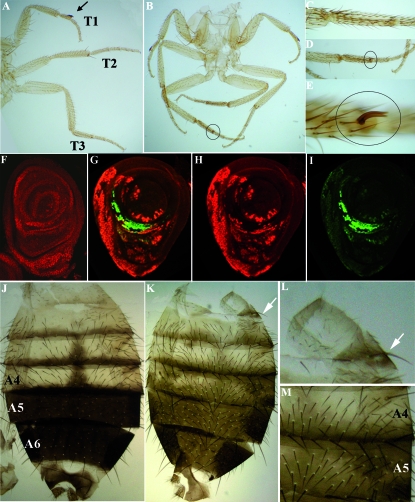
dRYBP mutant phenotypes in the embryo. (A) Wild-type syncytial embryo stained with anti-dRYBP (red) and anti-α-tubulin (green) antibodies. There is a uniform distribution of nuclei along the embryo. (B–D) High magnification of the indicated area: dRYBP (B, red), α-tubulin (C, green), and merged (D). (E) Syncytial embryo from dRYBP1/+ parents showing a mitotic collapse and the distribution of dRYBP protein (red) and α-tubulin (green). There are few nuclei and these are evenly distributed along the embryo. (F–H) High magnification of the indicated area: dRYBP (F, red), α-tubulin (G, green), and merged (H). (I) Wild-type ovariole stained with DAPI (blue) showing the stages of oogenesis (the arrow indicates stage 8 of oogenesis). (J) Ovariole from a homozygous dRYBP1 female (the arrow indicates the degenerated stage 8 of oogenesis). (K) Wild-type first-instar larval cuticle showing the pattern of denticle belts (anterior to the left). (L) Example of an en-Gal4>UAS-dRYBP-RNAi embryo showing a severely disrupted pattern of the denticle belts along the entire embryonic cuticle (arrow indicates region lacking denticles). (M) Example of an en-Gal4>UAS-dRYBP-RNAi embryo showing a weak effect on the pattern of the denticle belts along the embryonic cuticle (arrow indicates the effect on the seventh abdominal segment). (N) β-Gal expression (blue) in an en-Gal4>UAS-lacZ larva showing the domain of expression of the en-Gal4 line in the posterior compartment.
The homozygous embryos from dRYBP1/+ parents that reach the stage of larval cuticle formation (∼40%) show no detectable morphological cuticle defects. However, some embryos from dRYBP1/+ parents (see materials and methods) that die before cuticle formation (∼60%) show severe defects in their pattern of nuclear division (Figure 3, compare A–D with E–H) and during mitotic progression. The nuclear divisions are asynchronic and large irregularly formed nuclei are often observed, most likely representing nuclei that did not divide (Figure 3, E–H).
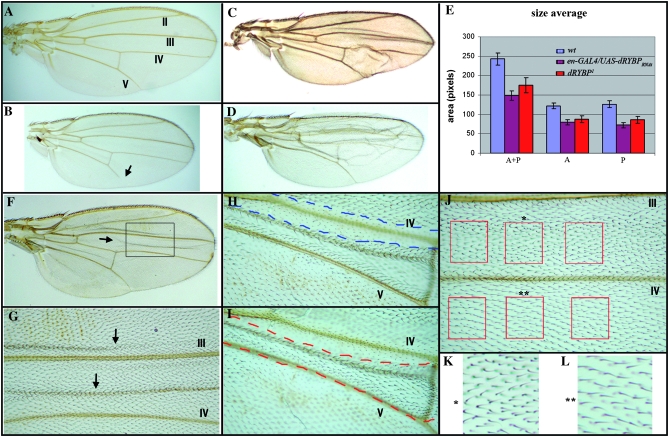
Neither dRYBP1/+ nor dRYBP1/dRYBP1 flies show phenotypic similarities to flies with mutations in the PcG/trxG genes (Sato and Denell 1985, 1987; Busturia and Morata 1988; Breen and Harte 1991). However, dRYBP1/dRYBP1 flies do show other very weak and low-penetrance morphological defects, including the presence of a distal gap in vein L5 (21% of the flies, Figure 4B), malformed legs (11% of the flies, most frequently the mesothoracic leg), umbrella-shaped wings (54% of the flies), and two to three bristles on the sixth sternite in the males (5% of the flies). Finally, the size of the wings of the dRYBP1/dRYBP1 flies is reduced 27% when compared with wild type (Figure 4E).
Figure 4.—
dRYBP mutant phenotypes in the adult wings. (A) Wing from a wild-type male fly. The numbers of the veins are indicated. (B) Wing from a homozygous dRYBP1 male showing a reduction of the wing size and a distal gap in vein V (arrow). (C) Wing from an en-Gal4>UAS-dRYBP-RNAi male showing the blister in the posterior compartment and a reduction in wing size. (D) Wing from a ci-Gal4>UAS-dRYBP-RNAi male showing the blister in the anterior compartment and a reduction in wing size. (E) Area of wild-type male wings (blue), en-Gal4>UAS-dRYBP-RNAi male wings (purple), and homozygous dRYBP1 (red). “A+P” indicates entire wing area; “P” indicates area of the posterior compartment; “A” indicates area of the anterior compartment. (F) Wing containing somatic homozygous dRYBP1 mutant clones marked with forked and twin clones marked with shavenoid. The presence of mutant clones results in the formation of blisters (one of them is indicated with an arrow). (G) High magnification of the area indicated in F showing two blisters (arrows). (H) Ventral view of the wing containing dRYBP1 mutant clones marked with forked (dashed red line labels the forked clone) between veins IV and V, where the presence of the clone induces the appearance of a blister on the dorsal surface of the wing. (I) Dorsal view of the same wing (dashed blue line labels the shavenoid clone). (J) dRYBP1 mutant clone marked with forked between veins III and IV. The red squares identify an example of the areas chosen to calculate the trichome density in the mutant clones (forked) vs. the density of trichomes in the corresponding wild-type areas (red squares at bottom). (K) High magnification of the mutant forked area (marked with *). (L) High magnification of the wild-type area (marked with **). The density of trichomes is higher in K than in L.
Even though we see no homeotic-like phenotypes in dRYBP mutant flies, given that the gene has been proposed to belong to the PcG, we have studied the expression of the homeotic proteins Ultrabithorax (UBX) (White and Wilcox 1984), Abdominal-A (ABD-A) (Macias et al. 1990), and Abdominal-B (ABD-B) (Celniker et al. 1990; De Lorenzi and Bienz 1990) in homozygous dRYBP1 embryos and larval imaginal discs. Expression of all these proteins was indistinguishable from wild type (data not shown).
The dRYBPΔ16 mutation is an imprecise excision of the original dRYBP1 that deletes 9 kb of the P[KG08683] element, but leaves the coding sequence of the dRYBP gene intact. dRYBP mRNA levels in dRYBPΔ16/dRYBPΔ16 embryos and adults are severely reduced (Figure 1D), indicating that dRYBPΔ16 is a strong hypomorphic. The phenotypes of homozygous dRYBPΔ16 adult flies are highly variable in their penetrance and expressivity and extremely similar to the phenotype described above for dRYBP1.
dRYBPΔ55 is a complete deletion of the P[KG08683] element that also removes 508 bp of the dRYBP gene (nucleotides 323–831; Figure 1A), including sequences of the 5′-UTR as well as the amino-terminal sequences encoding the NZF of the dRYBP protein. dRYBPΔ55/dRYBPΔ55 flies die progressively throughout development, are developmentally delayed, and, very infrequently, show the distal gap in the L5 vein, the umbrella-shaped wing, or the malformed mesothoracic leg phenotypes observed in the dRYBP1/dRYBP1 flies. However, dRYBPΔ55/dRYBPΔ55 females are not sterile and the dRYBPΔ55 homozygotes can be maintained as a stock. 5′-RACE analysis indicates that a shortened dRYBP mRNA is produced in dRYBPΔ55/dRYBPΔ55 embryos (Figure 1C). This suggests that a truncated version of the dRYBP protein, most probably using the second in-frame ATG (Figure 1A), is produced in these flies. Finally, the expression levels and the cellular localization pattern of the truncated dRYBP protein in dRYBPΔ55/dRYBPΔ55 embryos and imaginal discs are very similar to those of the full-length proteins in wild-type (wt) tissues (Figure 2).
The analysis of dRYBP mutant phenotypes indicates that the gene is required for progression through mitosis during the nuclear divisions of the syncytial embryos, perhaps accounting in part for the developmental delay that is also observed in dRYBP mutant larvae. Moreover, dRYBPΔ55/dRYBPΔ55 females are fertile, suggesting that the amino-terminal domain of the dRYBP protein is not required for its function in oogenesis.
Inactivation of dRYBP function by RNA interference:
Phenotypes associated with the inactivation of the dRYBP function have been also studied by RNA interference (Montgomery 2004), using UAS-dRYBP-RNAi constructs. The specificity of the inactivation was tested by examining the expression of dRYBP in sd-GAL4>UAS-dRYBP>UAS-dRYBP-RNAi imaginal discs. A strong reduction of dRYBP protein expression was observed (not shown). Moreover, the sd-GAL4>UAS-dRYBP>UAS-dRYBP-RNAi flies (not shown) showed rescue of the overexpression phenotypes seen in sd-GAL4>UAS-dRYBP (Figure 7D). The rescue might have resulted from a dilution of the sd-GAL4 driver cause by the presence of two UAS-containing constructs (UAS-dRYBP and UAS-dRYBPRNAi) instead of one (UAS-dRYBP). To control for this dilution possibility, we studied the dRYBP protein expression in imaginal discs and adult phenotypes of sd-GAL4>UAS-dRYBP>UAS-GFP (not shown).
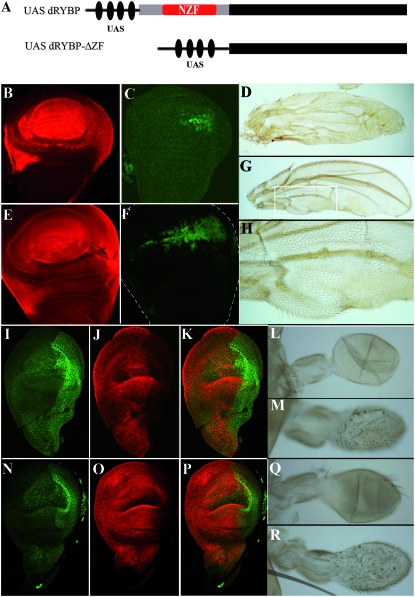
Figure 7.—
Overexpression of dRYBP in the wing and haltere imaginal discs: function of the carboxy-terminal domain. (A) Scheme of the UAS-dRYBP and UAS-dRYBP-ΔZF (lacking the NFZ domain) constructs. (B) Expression of dRYBP (red) in a sd-Gal4>UAS-dRYBP wing imaginal disc. (C) Expression of UBX (green) in a sd-Gal4>UAS-dRYBP wing imaginal disc. (D) Wing of a sd-Gal4>UAS-dRYBP fly. (E) Expression of dRYBP (red) in a sd-Gal4>UAS-dRYBP-ΔZF wing imaginal disc. (F) Expression of UBX (green) in a sd-Gal4>UAS-dRYBP-ΔZF wing imaginal disc. (G) Wing of a sd-Gal4>UAS-dRYBP-ΔZF fly showing the trichomes typical of the haltere. (H) Higher magnification of the indicated area in G to show the haltere-like trichomes. (I–K) Expression of UBX (I, green) and DRYBP (J, red) in an Ubx-Gal4>UAS-dRYBP haltere disc (K, merged). (L) Wild-type haltere. (M) Ubx-Gal4>UAS-dRYBP haltere, showing the trichomes similar to the wing trichomes. (Q) Ubx-Gal4 haltere. The Ubx-Gal4 line is inserted in the Ubx gene and shows the haplo-insufficient Ubx phenotype consisting of the appearance of some hairs in the capitelum (compare with L). (N–P) Expression of UBX (N, green) and dRYBP (O, red) in an Ubx-Gal4>UAS-dRYBP-ΔZF haltere disc (P, merged). (R) Haltere of an Ubx-Gal4>UAS-dRYBP-ΔZF fly.
The ci-GAL4 and the en-GAL4 lines drive expression throughout development in the anterior and the posterior compartments, respectively, and have been used to inactivate dRYBP function. en-Gal4>UAS-dRYBP-RNAi flies exhibit progressive lethality throughout embryonic and larval development (∼50% of the embryos die during embryogenesis). The larval cuticle phenotypes of the en-Gal4>UAS-dRYBP-RNAi embryos that survive to secrete cuticle show an aberrant pattern of segmentation with severe disruption in the pattern of the denticle belts, but no detectable homeotic transformations (Figure 3, L and M). The phenotype is highly variable both in penetrance and in expressivity. Interestingly, the inactivation of dRYBP in the cells of the posterior compartments in the en-Gal4>UAS-dRYBP-RNAi embryos appears to have a nonautonomous effect in the adjacent cells of the anterior compartment: the pattern of both the anterior and the posterior compartments is disrupted (Figure 3L).
The wings of surviving en-Gal4>UAS-dRYBP-RNAi (Figure 4C) and of ci-Gal4>UAS-dRYBP-RNAi (Figure 4D) flies are blistered in the posterior and anterior compartments, respectively. Curiously, the overall size of the wings is reduced (Figure 4, compare A with C and D). Although the blistered-wings phenotype complicates the size quantifications, when wing size in the en-Gal4>UAS-dRYBP-RNAi flies is measured, an overall reduction of 40% is observed compared to the wild type (Figure 4E), with a 43% reduction in the posterior compartment and a 35% reduction in the anterior compartment. This, once again, indicates a nonautonomous effect in the wild-type cells of the anterior compartment. The most likely explanation for this phenomenon is the “accommodation effect” first described by Garcia-Bellido et al. (1994). Interestingly, the inactivation of the dRYBP function, by RNA interference, in the whole wing using the ubiquitous arm-GaL4, da-Gal4, and tub-GaL4 drivers did not produce blistered wings (not shown), suggesting that the generation of the wing phenotypes observed in en-Gal4>UAS-dRYBP-RNAi and ci-Gal4>UAS-dRYBP-RNAi requires the contact of wild-type and mutant cells. Finally, the expression of the UBX protein in the wing and haltere imaginal discs from en-Gal4>UAS-dRYBP-RNAi and ci-Gal4>UAS-dRYBP-RNAi larvae was normal (data not shown).
The disrupted-pattern phenotypes observed in embryos and the reduction of the wing size observed in the adults lacking dRYBP function prompted us to characterize the pattern of apoptosis in embryos and imaginal discs of the genotypes dRYBP1/dRYBP1, en-Gal4>UAS-dRYBP-RNAi, and ci-Gal4>UAS-dRYBP-RNAi. This was done by staining imaginal discs and embryos with acridine orange (Abrams et al. 1993) as well as analyzing the expression of both Tunnel and the activated form of caspase-3 (Lee and Luo 1999), each serving as markers of cell death. In all these assays, no differences in the pattern of apoptosis between dRYBP mutant and wild-type tissues were seen (data not shown).
The proliferation and aberrant mitosis phenotypes found in embryos from dRYBP1/+ parents, as well as the reduced wing size in en-Gal4>UAS-dRYBP-RNAi and dRYBP1/dRYBP1, could be caused by a disruption in cell-cycle progression. The Cdk1/cyclin A and Cdk1/cyclin B complexes control progression through mitosis (for a review see Deshpande et al. 2005). We therefore analyzed cyclin A (Knoblich and Lehner 1993) expression in en-Gal4>UAS-dRYBP-RNAi wing imaginal discs. Cyclin A expression was identical in both the anterior and the posterior compartments of the wing disc (not shown).
Requirement of dRYBP throughout larval development:
Somatic mutant clones of dRYBP were induced at different times during development, using the null allele dRYBP1 and the FLP/FRT system. We first induced homozygous dRYBP1 mutant clones in the wing disc, marked with GFP expression, to study the size of the mutant clones compared to the corresponding twin clones and to analyze the expression of the homeotic UBX and ABD-B proteins. The homozygous dRYBP1 mutant clones and the corresponding wild-type twin clones were of similar size (not shown) and no ectopic expression of either UBX or ABD-B proteins was seen (data not shown). To study the phenotype of the homozygous dRYBP1 mutant clones in the adult fly, we induced mutant clones marked with forked (f) (Lindsley and Zimm 1992) and wild-type twin clones marked with shavenoid (sha) (Lindsley and Zimm 1992). No homeotic transformations were found in the adult mutant clones. However, and independently of the time of clone induction, a number of observations were made: first, forked clones and shavenoid clones were of similar size, suggesting that the proliferation rates were not affected in the mutant clones; second, in ∼30% of the clones studied, the presence of a forked clone on either one of the wing surfaces (ventral or dorsal) caused the appearance of a blister on the opposite surface (Figure 4, F–I). Finally, the forked mutant clones showed an increase in cell density. We calculated a 20% reduction in the number of wild-type trichomes compared to the forked clones (Figure 4, J–L). The phenotypes of these clones indicate that dRYBP is required for cellular differentiation throughout larval development. The developmental requirement of dRYBP was also studied, using the UAS-dRYBP-RNAi constructs to repress dRYBP expression in the imaginal discs. The blistered-wing phenotype (similar to that shown in Figure 4, C and D) was observed regardless of the time of clone induction (at 24–48, 48–72, and 72–96 hr after egg laying).
dRYBP genetic and molecular interactions:
The observed absence of homeotic phenotypes associated with lack of dRYBP function is not consistent with the proposed classification of dRYBP as a member of the PcG genes. Therefore, we studied whether dRYBP mutations genetically interact with mutations in the PcG and trxG genes. To this end, we used the dRYBP1, Pc3, Sce1, pho1, phocv, TrlR85, trxE2, and Rpd31 mutant alleles and scored for either enhancement or suppression of the dRYBP- and the PcG/trxG-associated phenotypes. Adult flies dRYBP1/dRYBP1; Pc3/+, dRYBP1/dRYBP1; TrlR85/+, dRYBP1/dRYBP1; Rpd31/+, dRYBP1/dRYBP1; pho1/+, and dRYBP1/dRYBP1; pho1/phocv showed no enhancement or suppression of homeotic phenotypes. Furthermore, embryos and imaginal discs from dRYBP1/CyO; Pc3/+, dRYBP1/CyO; Rpd31/+, and dRYBP1/CyO; TrlR85/+ showed no altered expression of the UBX or ABD-B proteins (data not shown).
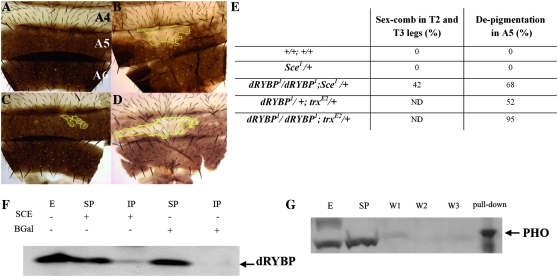
In contrast, dRYBP1/dRYBP1; Sce1/+ males showed an increased number of sex combs in the meso- and metathoracic legs when compared with Sce1/+ males or with siblings dRYBP1/+; Sce1/+. Surprisingly, dRYBP1/dRYBP1; Sce1/+ males showed depigmentation of the fifth abdominal segment (Figure 5, B and E), a phenotype never seen in either Sce1/+ or dRYBP1/+; Sce1/+ males. Additionally, dRYBP1/dRYBP1; trxE2/+ males (Figure 5D) showed an increased percentage of individuals exhibiting depigmentation of the fifth abdominal segment when compared with sibling dRYBP1/+; trxE2/+ males or trxE2/+ males (Figure 5C). The expressivity of the depigmentation phenotype was strongly increased compared to the depigmentation of the abdomen of trxE2/+ males. No differences in ABD-B protein expression pattern were seen between dRYBP1/dRYBP1; Sce1/Sce1 and Sce1/Sce1 embryos. Likewise no differences in ABD-B protein expression pattern were observed between dRYBP1/dRYBP1; trxE2/trxE2 and trxE2/trxE2 embryos (not shown).
Figure 5.—
dRYBP genetic and molecular interactions. (A) Dorsal abdomen of a wild-type adult male showing the A4, A5, and A6 segments. The A5 and A6 segments are pigmented. (B) Dorsal abdomen of a dRYBP1/dRYBP1; Sce1/+ male showing patches of depigmentation of the A5 segment. (C) Dorsal abdomen of a trxE2/+ male showing patches of depigmentation of the A5 segment. (D) Dorsal abdomen of an dRYBP1/dRYBP1; trxE2/+ male showing patches of depigmentation. (E) Percentage of flies showing the extra sex comb phenotype in T2 and T3 legs and the depigmentation phenotype in the A5 segment of the males. ND: not determined. (F) dRYBP protein co-immunoprecipitates with SCE but not with BGAL used as a control. Shown is a Western blot of the IP with SCE antibody and BGAL antibody revealed with anti-dRYBP antibody. E, extract; SP, supernatant; IP, immunoprecipitation. (G) dRYBP protein pulls down the PHO protein. Shown is a Western blot of the dRYBP pull down revealed with anti-PHO antibody. E, extract; SP, supernatant; W, washes.
Possible molecular interactions between dRYBP and SCE and PHO were studied using immunoprecipitation and pull-down experiments. The dRYBP protein was found to interact with SCE and PHO proteins (Figure 5, F and G). The results of both the genetic and the molecular-interaction experiments indicate that the dRYBP protein interacts genetically with both PcG and the trxG proteins and molecularly with the PcG proteins SCE and PHO.
Function of the dRYBP protein domains in the generation of homeotic–like phenotypes:
Overexpression of the dRYBP protein has been shown to generate homeotic-like phenotypes that can be modulated by mutations in the PcG/trxG genes (Bejarano et al. 2005). Interestingly, and depending on the cellular context, high levels of dRYBP caused either the ectopic expression or the repression of the homeotic UBX protein (Bejarano et al. 2005 and Figure 7, C and I). To further investigate the role dRYBP plays in the regulation of homeotic genes, we analyzed the effects of overexpression of dRYBP in different cellular contexts and studied the function of the domains of the dRYBP protein in the generation of the homeotic-like phenotypes. We used the arm-GAL4 line, which drives the expression ubiquitously and throughout development (Sanson et al. 1996), the Ubx-Gal4 line, which drives the expression in the haltere and third leg imaginal disc (de Navas et al. 2006), and the sd-GAL4 line, which drives the expression in the wing and haltere imaginal discs.
Ubx-Gal4>UAS-dRYBP flies exhibit ectopic sex combs on the metathoracic (third) legs (Figure 6, B–E). Sex comb identity is determined by the Sex comb reduced (Scr) gene (Struhl 1982; Pattatucci and Kaufman 1991), and the homeotic Sex comb reduced (SCR) protein is expressed in the prothoracic (first) leg imaginal disc (Figure 6F), but not in the second (mesothoracic) or third leg discs. We looked at the expression of SCR (Glicksman and Brower 1988) in Ubx-Gal4>UAS-dRYBP imaginal leg discs and found that it is ectopically expressed in the third imaginal leg discs (Figure 6, G–I). It is important to note that SCR expression only partially overlaps with the domain where dRYBP is overexpressed (Figure 6, G–I). The arm-Gal4>UAS-dRYBP flies also occasionally showed ectopic sex combs in the second and third legs (not shown). Moreover, arm-Gal4>UAS-dRYBP males showed the Miscadestral pigmentation (Mcp) (Lewis 1978) and the Ultra-abdominal (Uab) (Lewis 1978) phenotypes. The Mcp phenotype consists of ectopic pigmentation in the fourth abdominal segment (Figure 6, K and M) and the Uab phenotype consists of the appearance, in the first abdominal segment, of long bristles morphologically characteristic of posterior abdominal segments (Figure 6, K and L). Although these phenotypes result from the misexpression of the homeotic proteins (Lewis 1978; Busturia et al. 1989; Celniker et al. 1990; Macias et al. 1990; Sanchez-Herrero 1991), we could find no misregulation of ABD-A and ABD-B homeotic proteins in arm-Gal4>UAS-dRYBP embryos.
Figure 6.—
Overexpression of dRYBP in the leg and abdomen. (A) Legs (T1, T2, and T3) of a wild-type male. The T1 leg shows the sex comb in the basitarso (arrow). (B) Legs from an Ubx-Gal4>UAS-dRYBP male showing the appearance of an extra sex comb on the T3 leg (circle). (C) Basitarso of a wild-type male T3 leg. (D–E) Higher magnification of the basitarso shown in B (circles). (F) Expression of the SCR protein (red) in a wt male T1 leg imaginal disc. (G) Merged expression of dRYBP (red) and SCR (green) in the Ubx-Gal4>UAS-dRYBP T3 leg imaginal disc. The expression of SCR does not completely overlap with the expression of dRYBP. (H) Expression of dRYBP (red). (I) Expression of SCR (green). (J) Abdomen of a wild-type male showing the A4, A5, and A6 segments. A5 and A6 are pigmented. (K) Abdomen of an arm-Gal4>UAS-dRYBP male showing the Uab phenotype (long bristles in the A1 segment marked by an arrow) and the Mcp phenotype (ectopic pigmentation in the A4 segment). (L) Higher magnification of Uab phenotype shown in K. (M) Higher magnification of Mcp phenotype shown in K.
A functional analysis of dRYBP protein domains was performed by expressing the UAS-dRYBP-ΔZF and UAS-dRYBP-ΔCt constructs. dRYBP-ΔZF deletes the protein's amino-terminal domain that contains the NZF sequences (amino acids 1–70, materials and methods, Figure 7A); dRYBP-ΔCt deletes the protein's carboxy-terminal domain (amino acids 70–150).
sd-Gal4>UAS-dRYBP-ΔCt flies showed no wing or haltere phenotypes. We looked at the expression of dRYBP protein in the wing imaginal discs and observed a weak and diffused expression (not shown), suggesting either that the dRYBP-ΔCt protein is not stable or that it is not fully recognized by the anti-dRYBP antibody.
sd-Gal4>UAS-dRYBP-ΔZF flies exhibit transformation of wings toward haltere, as indicated by the appearance of haltere-like trichomes in the wing (Figure 7H). This transformation is likely mediated by the ectopic expression of UBX in the wing disc (Figure 7, C and F). The phenotype in the wings and the ectopic expression of UBX in the sd-Gal4>UAS-dRYBP-ΔZF wing imaginal discs were very similar to those of sd-Gal4>UAS-dRYBP (Figure 7, D and G, and Bejarano et al. 2005). Moreover, in both cases the ectopic expression of UBX did not completely overlap with the domain of dRYBP overexpression. Curiously, we observed that the dRYBP protein in the imaginal discs of sd-Gal4>UAS-dRYBP-ΔZF larvae is, primarily, localized outside the nucleus, suggesting that this truncated protein might be functional outside the nucleus.
A haltere phenotype consisting of small-size haltere covered with wing-like trichomes was seen in both sd-Gal4>UAS-dRYBP-ΔZF and Ubx-Gal4>UAS-dRYBP-ΔZF flies (Figure 7, M and R). The haltere discs of both sd-Gal4>UAS-dRYBP-ΔZF and Ubx-Gal4>UAS-dRYBP-ΔZF (Figure 7, N–P) larvae showed repression of the UBX expression, as was seen with the overexpression of UAS-dRYBP (Figure 7, I–K).
These results indicate that the carboxy terminus is required while the amino terminus is dispensable for the repression of UBX in the haltere disc and for the ectopic expression of UBX in the wing discs, both mediated by the overexpression of the dRYBP protein.
DISCUSSION
The dRYBP loss-of-function phenotypes are remarkable in the high variability of both their penetrance and their expressivity. Given the current knowledge of dRYBP function, it is difficult to explain the dramatic phenotypic variability. A striking illustration of this is that some embryos experience complete mitotic collapse (Figure 3, E–H) while others (Figure 4B) develop completely to adulthood. As there are no genes in the Drosophila genome that are clearly homologous to dRYBP, it is unlikely that this is due to the existence of homologous proteins that replace dRYBP function at a given developmental time.
It is possible that dRYBP phenotypic variability reflects on the robustness of the embryos, which is dependent on the fitness of the mother as well as the extrinsic and intrinsic environmental perturbations to which the flies are exposed (Stelling et al. 2004; Friedman and Perrimon 2007). dRYBP function might remain “latent” in the cell until the requirement becomes essential in response, for example, to specific stress conditions. Stress of this type could happen during the rapid nuclear divisions that take place in the syncytial embryo or when, in addition to the removal of the dRYBP protein, an intrinsic perturbation is introduced, i.e., modifying the genetic background by introducing mutations in other genes. In both cases, depending on the robustness of the individual, cells lacking dRYBP function may respond either by terminating mitosis (Figure 3, E–H) or by affecting morphological development (Figure 5, A–E).
Another characteristic of dRYBP inactivation are the differences between the phenotypes caused by null mutations and those caused by RNA interference. For example, dRYBP1/dRYBP1 embryos that reach the stage of cuticle formation do not show larval cuticle morphological defects. In contrast, en-Gal4>UAS-dRYBP-RNAi larvae show severe cuticle defects (Figure 3, L and M). Additionally, the wings of dRYBP1/dRYBP1 flies do not show the blistered phenotype observed in en-Gal4>UAS-dRYBP-RNAi or ci-Gal4>UAS-dRYBP-RNAi flies (Figure 4, C and D). A trivial explanation for these phenotypic differences is that the UAS-dRYBP-RNAi construct, in addition to inactivating the expression of dRYBP, is also inactivating the function of other genes. We have seen reduced dRYBP expression and rescue of the overexpression phenotypes in sd-Gal4>UAS-dRYBP, UAS-dRYBP-RNAi imaginal discs and adults, suggesting that dRYBP inactivation using UAS-dRYBP-RNAi is quite specific. A more attractive and plausible explanation is that the phenotypes caused by inactivation of dRYBP function are strongly revealed when wild-type cells are in physical contact with dRYBP mutant cells. This idea is supported by several observations. First, dRYBP 1/dRYBP 1 mutant clones in the wing, where wild-type and dRYBP mutant cells are in contact, cause a blistered phenotype (Figure 4, F–L) similar to that seen in en-Gal4>UAS-dRYBP-RNAi and ci-Gal4>UAS-dRYBP-RNAi (Figure 4, C and D). Second, dRYBP 1/dRYBP 1 larvae do not show cuticle defects while en-Gal4>UAS-dRYBP-RNAi, in which mutant cells physically contact wild-type cells, show strong and nonautonomous phenotypes (Figure 3, L and M). Third, wings of da-Gal4>UAS-dRYBP-RNAi, tub-Gal4>UAS-dRYBP-RNAi, and arm-Gal4>UAS-dRYBP-RNAi flies, in which mutant cells are not in contact with wild-type cells because da-Gal4, tub-Gal4, and arm-Gal4 inactivate dRYBP in all the wing cells, did not show the blistered phenotype. The study of the factors involved in the generation of these nonautonomous defects will facilitate the understanding of dRYBP function in the process of cellular differentiation.
dRYBP involvement in the mechanisms of PcG/trxG—expression of the protein, genetic analysis, and molecular interactions:
dRYBP expression is restricted to the nonnucleoli portion of the nucleus and forms a pattern of discrete spots very similar to the so-called PcG nuclear bodies reported for the PC, PH, and PSC proteins. Like PSC, the dRYBP nuclear expression pattern consists of both large and small bodies, numbering ∼30 per embryonic nucleus (Buchenau et al. 1998; Saurin et al. 1998; Dietzel et al. 1999; Netter et al. 2001; Ficz et al. 2005; Grimaud et al. 2006a) (Figure 2A). The partial overlap of dRYBP and PC distribution in the nuclear bodies (Figure 2, C and G) suggests that dRYBP functions, together with PcG, in the regulation of a subset of PcG target genes. Moreover, it also indicates that dRYBP, independently of PcG, is involved in the regulation of other processes.
dRYBP (Figure 2, I–P) and PcG proteins show similar dynamic mitotic distribution patterns during the nuclear cycles in the syncytial embryo (Abrams et al. 1993; Buchenau et al. 1998; Netter et al. 2001; Martinez et al. 2006; O'Dor et al. 2006). This distribution pattern may indicate a role for the protein in the regulation of cell-cycle progression because dRYBP mutant syncytial embryos exhibit a severe disruption in the pattern of nuclear divisions (Figure 3, E–H). The involvement of PcG/trxG proteins in the regulation of cell-cycle progression has been proposed. For example, Polycomb response elements (PREs) have been identified in the cell-cycle regulator cyclin A gene (Martinez et al. 2006) while mutations in ph, Pc, and Psc, all members of the PcG, show severe defects in chromosomal segregation (O'Dor et al. 2006). The dRYBP protein might very well participate with PcG/trxG in this process.
A role for dRYBP in mitotic progression and control of cell proliferation may explain the slow larval growth observed in dRYBP1/dRYBP1 and the reduction of the wing size when dRYBP is inactivated (Figure 4, B–D). Furthermore, the described increase in cell density observed in dRYBP mutant clones (Figure 4, F–L) suggests that the gene might also have a role in cell differentiation.
The interactions described between loss-of-function dRYBP mutations and mutations in Sce (a PcG) and the trx (a trxG) genes show that dRYBP participates in the maintenance of both the repressed transcriptional states controlled by the PcG genes and the active transcriptional states controlled by the trxG genes. The interaction with mutations of the Sce gene exhibits enhancement of the Pc-like phenotype (i.e., increased number of sex combs) and enhancement of the trx-like phenotype (i.e., increased depigmentation of the fifth abdominal segment). We have observed a molecular interaction between PHO and dRYBP proteins (Figure 5G). However, we did not find a genetic interaction between mutations in both genes, using the dRYBP1 and the pho1 and phocv mutant alleles. The dRYBP1/dRYBP1; pho1/+ flies do not show homeotic phenotypes and the dRYBP1/dRYBP1; pho1/phocv flies do not show enhancement or suppression of the number of ectopic sex combs observed in pho1/phocv flies. We believe that this is probably due to the strong maternal effect of the PHO protein (Breen and Duncan 1986). We have shown that the dRYBP and SCE proteins physically interact. It has been shown that the murine RYBP protein is a novel ubiquitin-binding protein that is itself ubiquitinated (Arrigoni et al. 2006). Furthermore, it has been shown that the RING1B, the mouse SCE homolog and a known ubiquitin E3 ligase, promotes RYBP ubiquitination (Arrigoni et al. 2006). The observed interaction between dRYBP and Sce suggests that this may be the case in Drosophila as well. The interaction with mutations in the trx gene clearly shows that dRYBP is able to interact not only with PcG complexes, but also with trxG.
dRYBP involvement in the mechanisms of PcG/trxG—analysis of the overexpression phenotypes:
High levels of the dRYBP protein generate homeotic-like phenotypes in the legs, wings, haltere, and abdomen of flies (Figures 6 and 7). The carboxy-terminal domain of the dRYBP protein is sufficient to cause misexpression of the homeotic proteins to generate the homeotic-like phenotypes (Figures 6 and 7). For example, the appearance of extra sex combs in the metathoracic legs results from the ectopic expression of the homeotic Sex comb-reduced protein (Figure 6), while the transformation of wing into haltere (Figure 7, D–H) results from the ectopic expression of the UBX protein (Figure 7C).
Curiously, overexpression of dRYBP in the wing and leg imaginal discs does not result in the ectopic expression of UBX and SCR throughout the entire overexpression domain (Figure 6). This suggests that there is a partial inactivation of the PcG proteins in these regions, which could be due to transcriptional repression of PcG genes or to sequestration of PcG proteins. We investigated whether PC expression is decreased in en-GAL4>UAS-dRYBP wing imaginal discs and found no difference between the anterior and the posterior compartments (not shown). This indicates that the homeotic-like phenotypes are probably due to sequestration of the PcG proteins mediated by dRYBP overexpression. This possibility is supported by the observed modulation of the homeotic phenotypes by PcG and trxG mutations (Bejarano et al. 2005). The proposed sequestration of the PcG/trxG protein complexes is mediated through the carboxy terminus of the protein, as the ectopic expression of the UBX protein in the wing disc results from the overexpression of both the full-length dRYBP and the dRYBP-ΔZF proteins (Figure 7).
The phenotypes associated with the overexpression of some PcG/trxG proteins have been reported (Martin and Adler 1993; Peterson et al. 2004). Psc and Su(z)2 have been overexpressed by heat treatment of the transgenic flies containing hs-Psc and hs-Su(z)2 and neither one shows mutant phenotypes in any tissue (Martin and Adler 1993). Overexpression of the SPM domain of the Sex comb in midleg (SCM) protein produces homeotic-like phenotypes that can be modulated by PcG mutations (Peterson et al. 2004). Perhaps genes of PcG/trxG involved in recruitment or aggregation of PcG/trxG proteins produce, when overexpressed, sequestration of PcG/trxG proteins and, therefore, homeotic-like phenotypes.
Within the haltere, where homeotic-like phenotypes are also produced by the carboxy-terminal domain of the dRYBP protein, the effect of high-level dRYBP expression on UBX expression is distinct: UBX expression is repressed throughout the entire domain of dRYBP overexpression. As a result, the adult haltere shows a phenotypic transformation toward wing but without taking on the size and shape of a wing (Figure 7, M and R). Recently, it has been shown that the size of the haltere is determined by the combined levels of decapentaplegic (DPP) and UBX (de Navas et al. 2006). Perhaps high levels of dRYBP expression repress Dpp expression and this, together with the reduction of UBX expression, results in the small-haltere phenotype (Figure 7, M and R). Further characterization of this phenomenon may allow the separation of the mechanisms of size regulation and differentiation.
Here we have presented an examination of the phenotypes associated with inactivation of the dRYBP gene and an initial functional and biochemical characterization of the dRYBP protein. dRYBP was previously classified as a PcG gene on the basis of the protein molecular interactions and the phenotypes resulting from its overexpression (Garcia et al. 1999; Bejarano et al. 2005). However, the results from the present study indicate that dRYBP is a PcG- and trxG-interacting gene that participates in many biological processes, but it is not a “classical” PcG gene because dRYBP loss-of-function mutations do not show homeotic phenotypes. This demonstrates the danger of classification of a gene function based on overexpression phenotypes. Our results indicate that dRYBP is a PcG- and trxG-interacting gene that participates, together with PcG/trxG, in the mechanisms of cell-identity control. Furthermore, these results should provide the basis for a more complete description and understanding of the mechanisms in which dRYBP is involved, including the control of the nuclear divisions in the syncytial embryo and the regulation of cell differentiation.
Acknowledgments
We are very grateful to Fernando Diaz Benjumea, Keith Harshman, and Ernesto Sánchez-Herrero for discussions and critically reading the manuscript. We especially thank Fernando Diaz Benjumea for his help with the somatic dRYBP mutant clones and Thierry Cheutin and people of the laboratory of Giacomo Cavalli for their help with the dRYBP/PC/H3K27m3 colocalization experiments; Miguel A. Vidal for discussions; Judy Kassis, Richard Jones, and Isabel Guerrero for the PHO, PC, and SCE antibodies, respectively; Terry Orr-Weaver, Renato Paro, and the Bloomington Stock Center for fly stocks; Rocio Simón for help with some of the experiments; and the Madrid Drosophila community for reagents and discussions. This work was supported by grants from Dirección General de Investigación Científica y Técnica (BMC-2002-00524 and BFU-2005-02319) and the Fundación Investigación Médica Mutua Madrileña (FMM-2006) to A.B. and by an institutional grant to the Centro de Biología Molecular Consejo Superior de Investigaciones Cientificas–Universidad Autónoma de Madrid from Fundación Ramón Areces.
References
- Abrams, J. M., K. White, L. I. Fessler and H. Steller, 1993. Programmed cell death during Drosophila embryogenesis. Development 117 29–43. [DOI] [PubMed] [Google Scholar]
- Alam, S. L., J. Sun, M. Payne, B. D. Welch, B. K. Blake et al., 2004. Ubiquitin interactions of NZF zinc fingers. EMBO J. 23 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni, R., S. L. Alam, J. A. Wamstad, V. J. Bardwell, W. I. Sundquist et al., 2006. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 580 6233–6241. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bejarano, F., I. González, M. Vidal and A. Busturia, 2005. The Drosophila RYBP gene functions as a Polycomb-dependent transcriptional repressor. Mech. Dev. 122 1118–1129. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., A. S. Manoukian and N. Perrimon, 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44 635–654. [DOI] [PubMed] [Google Scholar]
- Breen, T. R., and I. M. Duncan, 1986. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 118 442–456. [DOI] [PubMed] [Google Scholar]
- Breen, T. R., and P. J. Harte, 1991. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech. Dev. 35 113–127. [DOI] [PubMed] [Google Scholar]
- Brock, H. W., and C. L. Fisher, 2005. Maintenance of gene expression patterns. Dev. Dyn. 232 633–655. [DOI] [PubMed] [Google Scholar]
- Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen and J. A. Kassis, 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1 1057–1064. [DOI] [PubMed] [Google Scholar]
- Buchenau, P., J. Hodgson, H. Strutt and D. J. Arndt-Jovin, 1998. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J. Cell Biol. 141 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia, A., and G. Morata, 1988. Ectopic expression of homeotic genes caused by the elimination of the Polycomb gene in Drosophila imaginal epidermis. Development 104 713–720. [DOI] [PubMed] [Google Scholar]
- Busturia, A., J. Casanova, E. Sanchez-Herrero, R. Gonzalez and G. Morata, 1989. Genetic structure of the abd-A gene of Drosophila. Development 107 575–583. [DOI] [PubMed] [Google Scholar]
- Calleja, M., E. Moreno, S. Pelaz and G. Morata, 1996. Visualization of gene expression in living adult Drosophila. Science 274 252–255. [DOI] [PubMed] [Google Scholar]
- Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage et al., 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 1039–1043. [DOI] [PubMed] [Google Scholar]
- Cao, R., Y. Tsukada and Y. Zhang, 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20 845–854. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., S. Sharma, D. J. Keelan and E. B. Lewis, 1990. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires, J. B., 1983. Daunomycin inhibits the B leads to Z transition in poly d(G-C). Nucleic Acids Res. 11 8485–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof et al., 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 185–196. [DOI] [PubMed] [Google Scholar]
- De Lorenzi, M., and M. Bienz, 1990. Expression of Abdominal-B homoeproteins in Drosophila embryos. Development 108 323–329. [DOI] [PubMed] [Google Scholar]
- de Napoles, M., J. E. Mermoud, R. Wakao, Y. A. Tang, M. Endoh et al., 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7 663–676. [DOI] [PubMed] [Google Scholar]
- de Navas, L. F., D. L. Garaulet and E. Sanchez-Herrero, 2006. The ultrabithorax Hox gene of Drosophila controls haltere size by regulating the Dpp pathway. Development 133 4495–4506. [DOI] [PubMed] [Google Scholar]
- Deshpande, A., P. Sicinski and P. W. Hinds, 2005. Cyclins and cdks in development and cancer: a perspective. Oncogene 24 2909–2915. [DOI] [PubMed] [Google Scholar]
- Dietzel, S., H. Niemann, B. Bruckner, C. Maurange and R. Paro, 1999. The nuclear distribution of Polycomb during Drosophila melanogaster development shown with a GFP fusion protein. Chromosoma 108 83–94. [DOI] [PubMed] [Google Scholar]
- Fang, J., T. Chen, B. Chadwick, E. Li and Y. Zhang, 2004. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 279 52812–52815. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco, D., I. Gutierrez-Garcia, D. M. Vallejo, J. Bolivar, F. J. Gutierrez-Avino et al., 2006. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439 430–436. [DOI] [PubMed] [Google Scholar]
- Ficz, G., R. Heintzmann and D. J. Arndt-Jovin, 2005. Polycomb group protein complexes exchange rapidly in living Drosophila. Development 132 3963–3976. [DOI] [PubMed] [Google Scholar]
- Foley, K. P., M. W. Leonard and J. D. Engel, 1993. Quantitation of RNA using the polymerase chain reaction. Trends Genet. 9 380–385. [DOI] [PubMed] [Google Scholar]
- Friedman, A., and N. Perrimon, 2007. Genetic screening for signal transduction in the era of network biology. Cell 128 225–231. [DOI] [PubMed] [Google Scholar]
- Garcia, E., C. Marcos-Gutierrez, M. del Mar Lorente, J. C. Moreno and M. Vidal, 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18 3404–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido, A., F. Cortes and M. Milan, 1994. Cell interactions in the control of size in Drosophila wings. Proc. Natl. Acad. Sci. USA 91 10222–10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti, M., and B. S. Baker, 1989. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 3 438–453. [DOI] [PubMed] [Google Scholar]
- Gearhart, M. D., C. M. Corcoran, J. A. Wamstad and V. J. Bardwell, 2006. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26 6880–6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glicksman, M. A., and D. L. Brower, 1988. Expression of the Sex combs reduced protein in Drosophila larvae. Dev. Biol. 127 113–118. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel, N., L. Fanti, T. Melgar, E. Garcia, S. Pimpinelli et al., 2004. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 121 449–462. [DOI] [PubMed] [Google Scholar]
- Grimaud, C., F. Bantignies, M. Pal-Bhadra, P. Ghana, U. Bhadra et al., 2006. a RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124 957–971. [DOI] [PubMed] [Google Scholar]
- Grimaud, C., N. Negre and G. Cavalli, 2006. b From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 14 363–375. [DOI] [PubMed] [Google Scholar]
- Jürgens, G., 1985. A group of genes controlling the spatial expression of the bithorax complex in drosophila. Nature 316 153–155. [Google Scholar]
- Kiernan, J. A., 2001. Classification and naming of dyes, stains and fluorochromes. Biotech. Histochem. 76 261–279. [DOI] [PubMed] [Google Scholar]
- Klymenko, T., B. Papp, W. Fischle, T. Kocher, M. Schelder et al., 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich, J. A., and C. F. Lehner, 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 12 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodjabachian, L., M. Delaage, C. Maurel, R. Miassod, B. Jacq et al., 1998. Mutations in ccf, a novel Drosophila gene encoding a chromosomal factor, affect progression through mitosis and interact with Pc-G mutations. EMBO J. 17 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst and D. Reinberg, 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 451–461. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276 565–570. [DOI] [PubMed] [Google Scholar]
- Lindsley, D., and G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, New York.
- Macias, A., J. Casanova and G. Morata, 1990. Expression and regulation of the abd-A gene of Drosophila. Development 110 1197–1207. [DOI] [PubMed] [Google Scholar]
- Martin, E. C., and P. N. Adler, 1993. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development 117 641–655. [DOI] [PubMed] [Google Scholar]
- Martinez, A. M., and G. Cavalli, 2006. The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 5 1189–1197. [DOI] [PubMed] [Google Scholar]
- Martinez, A. M., S. Colomb, J. Dejardin, F. Bantignies and G. Cavalli, 2006. Polycomb group-dependent Cyclin A repression in Drosophila. Genes Dev. 20 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H. H., J. G. Shorter, J. Seemann, D. Pappin and G. Warren, 2000. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H. H., Y. Wang and G. Warren, 2002. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21 5645–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, M. K., 2004. The use of double-stranded RNA to knock down specific gene activity. Methods Mol. Biol. 260 129–144. [DOI] [PubMed] [Google Scholar]
- Motzny, C. K., and R. Holmgren, 1995. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech. Dev. 52 137–150. [DOI] [PubMed] [Google Scholar]
- Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta et al., 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111 197–208. [DOI] [PubMed] [Google Scholar]
- Nagel, A. C., D. Maier and A. Preiss, 2002. Green fluorescent protein as a convenient and versatile marker for studies on functional genomics in Drosophila. Dev. Genes Evol. 212 93–98. [DOI] [PubMed] [Google Scholar]
- Netter, S., M. Faucheux and L. Theodore, 2001. Developmental dynamics of a polyhomeotic-EGFP fusion in vivo. DNA Cell Biol. 20 483–492. [DOI] [PubMed] [Google Scholar]
- O'Dor, E., S. A. Beck and H. W. Brock, 2006. Polycomb group mutants exhibit mitotic defects in syncytial cell cycles of Drosophila embryos. Dev. Biol. 290 312–322. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., 1994. Developmental modification of the Drosophila cell cycle. Trends Genet. 10 321–327. [DOI] [PubMed] [Google Scholar]
- Otte, A. P., and T. H. Kwaks, 2003. Gene repression by Polycomb group protein complexes: A distinct complex for every occasion? Curr. Opin. Genet. Dev. 13 448–454. [DOI] [PubMed] [Google Scholar]
- Patel, N. H., E. Martin-Blanco, K. G. Coleman, S. J. Poole, M. C. Ellis et al., 1989. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58 955–968. [DOI] [PubMed] [Google Scholar]
- Pattatucci, A. M., and T. C. Kaufman, 1991. The homeotic gene Sex combs reduced of Drosophila melanogaster is differentially regulated in the embryonic and imaginal stages of development. Genetics 129 443–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, A. J., D. R. Mallin, N. J. Francis, C. S. Ketel, J. Stamm et al., 2004. Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, M. D., and A. Shearn, 1990. Mutations in polycombeotic, a Drosophila polycomb-group gene, cause a wide range of maternal and zygotic phenotypes. Genetics 125 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirity, M. K., J. Locker and N. Schreiber-Agus, 2005. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol. Cell. Biol. 25 7193–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, R., and M. P. Scott, 1990. Expression and function of the homoeotic genes Antennapedia and Sex combs reduced in the embryonic midgut of Drosophila. Development 109 289–303. [DOI] [PubMed] [Google Scholar]
- Ringrose, L., and R. Paro, 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38 413–443. [DOI] [PubMed] [Google Scholar]
- Sanchez, C., I. Sanchez, J. A. Demmers, P. Rodriguez, J. Strouboulis et al., 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell Proteomics 6 820–834. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero, E., 1991. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111 437–449. [DOI] [PubMed] [Google Scholar]
- Sanson, B., P. White and J. P. Vincent, 1996. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature 383 627–630. [DOI] [PubMed] [Google Scholar]
- Sato, T., and R. E. Denell, 1985. Homoeosis in Drosophila: anterior and posterior transformations of Polycomb lethal embryos. Dev. Biol. 110 53–64. [DOI] [PubMed] [Google Scholar]
- Sato, T., and R. E. Denell, 1987. Homoeosis in Drosophila: the lethal syndrome of the regulator of bithorax (or trithorax) locus and its interaction with other homoeotic loci. Genetics 116 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin, A. J., C. Shiels, J. Williamson, D. P. Satijn, A. P. Otte et al., 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, C., T. Yoshikawa, F. Matsuda-Suzuki, S. Delehouzee, M. Goto et al., 2002. YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J. Biol. Chem. 277 22484–22490. [DOI] [PubMed] [Google Scholar]
- Schlisio, S., T. Halperin, M. Vidal and J. R. Nevins, 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21 5775–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber, B., D. Chourrout, M. Vervoort, B. Leblanc and G. Cavalli, 2007. Genome regulation by polycomb and trithorax proteins. Cell 128 735–745. [DOI] [PubMed] [Google Scholar]
- Schwartz, Y. B., and V. Pirrotta, 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Shao, Z., F. Raible, R. Mollaaghababa, J. Guyon, C. Wu et al., 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 9 37–46. [DOI] [PubMed] [Google Scholar]
- Sparmann, A., and M. van Lohuizen, 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6 846–856. [DOI] [PubMed] [Google Scholar]
- Stelling, J., U. Sauer, Z. Szallasi, F. J. Doyle, 3rd and J. Doyle, 2004. Robustness of cellular functions. Cell 118 675–685. [DOI] [PubMed] [Google Scholar]
- Struhl, G., 1982. Genes controlling segmental specification in the Drosophila thorax. Proc. Natl. Acad. Sci. USA 79 7380–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram, U., B. Riggs and W. Sullivan, 2001. Cleavage and Gastrulation in Drosophila Embryos (Encyclopedia of Life Sciences). Nature Publishing Group, London.
- Trimarchi, J. M., B. Fairchild, J. Wen and J. A. Lees, 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek, M. E., S. W. Bruggeman and M. van Lohuizen, 2004. Stem cells and cancer; the polycomb connection. Cell 118 409–418. [DOI] [PubMed] [Google Scholar]
- van der Lugt, N. M., J. Domen, K. Linders, M. van Roon, E. Robanus-Maandag et al., 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8 757–769. [DOI] [PubMed] [Google Scholar]
- Wang, B., S. L. Alam, H. H. Meyer, M. Payne, T. L. Stemmler et al., 2003. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J. Biol. Chem. 278 20225–20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst et al., 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431 873–878. [DOI] [PubMed] [Google Scholar]
- White, R. A., and M. Wilcox, 1984. Protein products of the bithorax complex in Drosophila. Cell 39 163–171. [DOI] [PubMed] [Google Scholar]
- Wodarz, A., U. Hinz, M. Engelbert and E. Knust, 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82 67–76. [DOI] [PubMed] [Google Scholar]