Abstract
We explore the theoretical consequences of limiting selection to males for the evolution of imprinted genes. We find that the efficiency of male-limited selection depends on the pattern of imprinting at an imprinted locus. When selection is strong, the maternally expressed pattern of imprinting allows faster genetic change than the reciprocal, paternally expressed pattern. When selection is relatively weak, the pattern of imprinting that permits a greater rate of genetic response to selection depends on the frequency of the favored allele: the paternally expressed pattern permits faster genetic change than does the maternally expressed pattern at low frequencies of a favored allele; at higher frequencies of a favored allele, however, the maternally expressed pattern is again more conducive to a genetic response. To our knowledge, this is the first theoretical description of a difference between the two reciprocal patterns of imprinting. The selective efficiency bias we identify between the two patterns of imprinting has implications for natural and livestock populations, which we discuss.
A given mammalian genome contains dozens to hundreds of imprinted genes, with new empirical studies adjusting this estimate as they are published (Morison et al. 2005; http://igc.otago.ac.nz/home.html maintains a current database). An imprinted gene is one that is expressed at different levels when paternally derived vs. when maternally derived (Reik and Walter 2001). This unique feature, first described >20 years ago, makes their pattern of heredity non-Mendelian (McGrath and Solter 1984; Surani et al. 1984). Imprinted genes have been described convincingly in mammals and seed plants (reviewed recently by Burt and Trivers 2006, Chap. 4). We limit our consideration to mammals, although many of the same principles could in theory apply to seed plants. The difference in expression at an imprinted diploid locus takes the form of silence of one allele and exclusive expression from the other allele, which is achieved by a mechanism that involves reversible, germ-line-specific methylation of nuclear DNA (Pfeifer 2000; Reik and Walter 2001). Imprinted expression is often tissue specific. For example, the maternally derived allele may be the only allele expressed in placenta with both alleles expressed in all other tissues.
Imprinted genes have received attention in the livestock production literature because of their effects on growth (Ruvinsky 1999). The study of imprinted genetic variation in agricultural populations has moved forward on two fronts: linear model studies, which aim to measure the extent of imprinted variation compared to other sources of variation, and quantitative trait locus (QTL) studies, which aim to identify the specific imprinted genes or regions of the genome responsible for that variation.
First, the animal model, from which best linear unbiased predicted (BLUP) breeding values can be assessed, has been modified in two ways to permit the detection of imprinted genetic effects. Schaeffer et al. (1989) were the first to add a gametic effect (equivalent to “parent-specific” or “imprinted” effect) to the animal model (for a review of linear models and BLUP, see Lynch and Walsh 1998 and Mrode 2005). However, a widely used approach for estimating the genetic variances with restricted maximum likelihood (REML) cannot handle this mixture of animal and gametic levels (Essl and Voith 2002a). Tier and Solkner (1993) describe a model that cleverly assigns gametic effects as animal-level effects to sidestep this problem. This latter model has been used in a handful of studies that have found evidence for imprinted genetic effects on meat and production traits (De Vries et al. 1994; Engellandt and Tier 2002; Essl and Voith 2002b; Stella et al. 2003).
Second, the standard F2 design was modified by Knott et al. (1998) to allow for the detection of parent-specific QTL. More studies employing a QTL approach exist than those using linear models for imprinted effects. Further, these studies have not been limited solely to livestock species. Imprinted loci have now been detected in sheep, pig, and mouse for traits such as growth and other agriculturally relevant features (Cockett et al. 1996; De Koning et al. 2000; Mantey et al. 2005). Most notably, in two separate studies, an imprinted QTL for meat quality traits was found in a cross between pig breeds (Jeon et al. 1999; Nezer et al. 1999). The QTL was mapped to a region that contained Igf2—easily the best studied of all imprinted genes in mammals—and the single-nucleotide difference responsible for the effect was later demonstrated to lie within an intron of this gene (Van Laere et al. 2003), suggesting a regulatory role. The variant in this region accounted for as much as 20–30% of the variation in certain measures of meat quality. Further designs for detecting imprinted QTL in agricultural data sets have also been suggested and employed (De Koning et al. 2002; Cui et al. 2006, 2007).
There is budding interest in the possibility that imprinted genes play a major role in agriculturally relevant traits (Ruvinsky 1999). However, there exists scant theoretical work to suggest where to begin looking for imprinted genetic variation and what shape it may take. This is why we are interested here in the population genetic features of imprinted genes under agricultural-like schemes of selection. One salient feature of many agricultural populations is the difference in selective strengths placed on males and females when particular “stud” males are used. Pearce and Spencer (1992) demonstrated that under most selective schemes, the population genetics of imprinted genes reduce to formal equivalence with Mendelian population genetics. The exception was when selection differed between males and females (extended in Anderson and Spencer 1999). In both of Spencer and colleagues' articles, their interests were not in comparing the two patterns of imprinting but mostly concerned with the comparison of imprinted loci with Mendelian loci. Here we pay special attention to the differences between imprinted loci that are paternally expressed and loci with the opposite pattern of imprinting (maternally expressed). Specifically, we wish to know how imprinted genes with the two patterns of imprinting evolve in response to selective breeding in agricultural settings. Knowing the theoretical underpinnings of the selective response in imprinted genes is essential, as imprinted genes have proven to be important in production traits and will likely be given more attention in the future as the techniques to detect their effects improve. Finally, since artificial selection and natural selection are at their core the same process, the work here also merits consideration by those interested in natural populations.
MODEL
We present a simple one-locus, two-allele model of evolution at an imprinted, autosomal locus, akin to Equation 1 of Anderson and Spencer (1999). This model is analyzed for both maternally expressed and paternally expressed imprinted loci to compare allele-frequency changes under both patterns of imprinting.
Consider an autosomal locus with two alleles, A and a, and four genotypes (AA, Aa, aA, and aa—maternally derived allele written first). The A allele is favored by selection, which acts on males only, and occurs in frequency pe in eggs and ps in sperm. Zygotes are formed by random union of eggs and sperm. Before selection, the genotype frequencies are the same in the two sexes. We refer to the frequencies of egg- and sperm-derived alleles in females and males as pef, psf, pem, and psm, accordingly.
Selection on males at a maternally expressed locus:
At a maternally expressed locus, the phenotype is determined by the egg-derived allele. Therefore, AA and Aa genotypes are phenotypically equivalent, as are aA and aa genotypes. AA and Aa males are given a fitness of 1 and the aA and aa males are given a fitness of w (0 ≤ w < 1). Since selection does not “see” the paternally derived allele at such a locus and because there is no correlation between maternally derived and paternally derived genes, the selection process, which is acting exclusively on males, modifies only pem from pre- to postselection males. The three other allele frequencies are unchanged by a round of selection. The prime (′) notation denotes a postselection allele frequency. These are the frequencies of the A allele in the current generation after selection has acted:
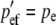 |
(1a) |
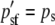 |
(1b) |
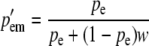 |
(1c) |
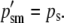 |
(1d) |
The corresponding changes at each of these four positions from zygotes to postselection adults within this generation are given by δp = p′ − p (we use δ for within-generation changes and Δ for between-generation changes):
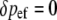 |
(2a) |
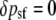 |
(2b) |
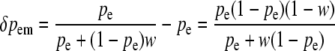 |
(2c) |
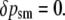 |
(2d) |
After selection, females produce eggs and males produce sperm in the frequencies pe″ and ps″. These frequencies are calculated by averaging (1a) and (1b) to give the next generation's egg-derived frequency and by averaging (1c) and (1d) to give the next generation's sperm-derived frequency. Their difference is due to selection on egg-derived alleles in males and gives rise to the difference in the frequency of the two heterozygotes in the next generation:
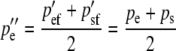 |
(3a) |
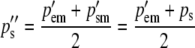 |
(3b) |
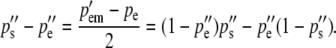 |
(3c) |
The changes in gametic frequencies from one generation to the next are simply
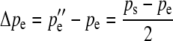 |
(4a) |
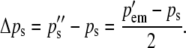 |
(4b) |
Selection on males at a paternally expressed locus:
With the reciprocal pattern of imprinting at the A locus, AA and aA males are phenotypically equivalent (fitness = 1) and aa and Aa males are phenotypically equivalent (fitness = w; 0 ≤ w < 1). Selection, in this case, acts solely on sperm-derived alleles in males:
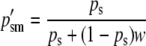 |
(5a) |
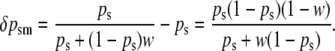 |
(5b) |
The allele frequencies among the gametes that produce the next generation of zygotes are calculated as before and the difference between them is due to selection on the sperm-derived alleles of males. This difference is again the cause of—and is numerically equal to—the difference in the frequencies of the reciprocal heterozygotes of the next generation:
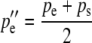 |
(6a) |
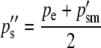 |
(6b) |
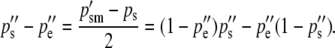 |
(6c) |
For this pattern of imprinting, the change in gametic frequencies across generations is given by
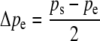 |
(7a) |
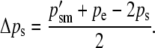 |
(7b) |
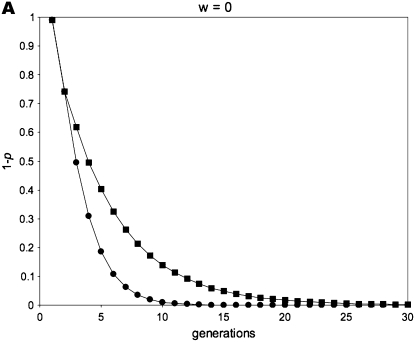
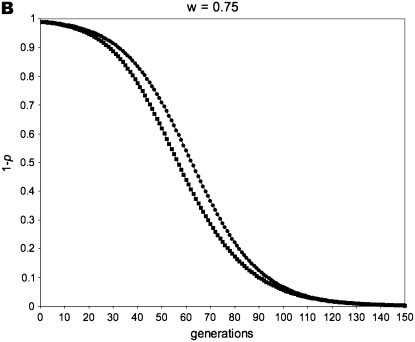
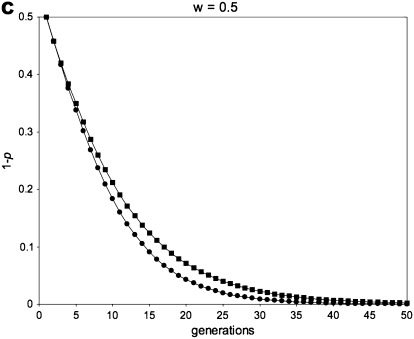
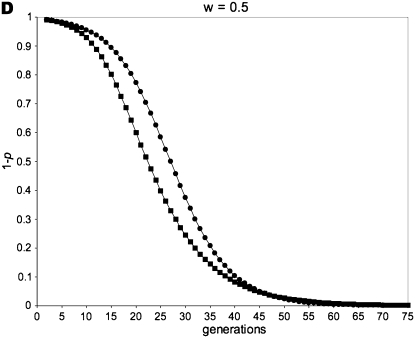
Figure 1 gives a simulation of all of the above recursion equations, averaging across sperm-derived and egg-derived frequencies to get a single value of p.
Figure 1.—
Allele-frequency dynamics under male-limited selection. (A) w = 0, po = 0.01; (B) w = 0 .75, po = 0.01; (C) w = 0.5, po = 0.5; (D) w = 0.5, po = 0.01. Each of the four graphs shows the decrease of the disfavored allele frequency, 1 − p, with fitness equal to w relative to that of the favored allele. We use the average of sperm-derived and egg-derived frequencies to give a single point for each generation at a maternally expressed locus (circles) and a paternally expressed locus (squares). A and C show instances in which maternally expressed loci evolve faster in response to selection: this occurs when selection is strong (w = 0 in A) and/or when the initial value of the favored allele frequency is high (p = 0.5 initially in C). B and D are examples in which a paternally expressed locus shows more change in the early part of the dynamic than a maternally expressed locus; this pattern reverses as the favored allele increases, however, with the reversal occurring at lower frequencies of p for smaller values of w. When selection is sufficiently weak, the difference between imprinting patterns is virtually negligible for both allele-frequency change and fixation time. When selection is strong, however, the difference between the imprinting patterns is more readily apparent.
Comparing the efficacy of the two forms of selection:
At a maternally expressed locus, the change in frequency due to selection on egg-derived alleles in males is given by
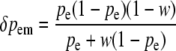 |
(see Equation 2c above).
At a paternally expressed locus, the change in frequency due to selection on sperm-derived alleles in males is given by
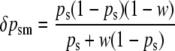 |
(see Equation 5b above).
Suppose that the genome contains an allele at a paternally expressed locus that increases the value of a quantitative trait and an allele at a maternally expressed locus with the same effect on the quantitative trait. Further suppose that allele frequencies, before selection, are the same at the two loci. Would one generation of artificial selection on the quantitative trait be more effective in changing the allele frequency at the paternally expressed locus or at the maternally expressed locus?:
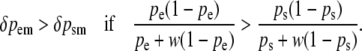 |
(8) |
Some rearrangement gives
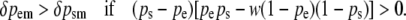 |
(9) |
One generation of male-limited selection ensures ps > pe. If this further restriction is accepted, then
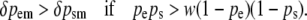 |
(10) |
This condition states that male-limited selection on a maternally expressed locus is more effective than male-limited selection on a paternally expressed locus when the frequency of the favored homozygote (AA) is greater than the frequency of the disfavored homozygote (aa) scaled by its relative fitness. For 0 < w < 1, this condition is satisfied when a is sufficiently rare but is not satisfied when A is sufficiently rare. Thus, male-limited selection is more effective at enriching rare favored alleles at paternally expressed loci but is more effective at purging rare disfavored alleles at maternally expressed loci. For the special case of w = 0, male-limited selection is more effective when acting on a maternally expressed locus at all gene frequencies.
DISCUSSION
First, we offer an intuitive explanation for the source of the bias in selection response that we have found between the two reciprocal patterns of imprinting. Imprinted genes differ from Mendelian genes in that reciprocal heterozygotes differ with respect to phenotype and fitness. Further, each heterozygote is lumped into the same fitness class as one of the homozygotes, creating inefficiencies in the removal of disfavored alleles and the promotion of favored alleles. As an illustration of this “efficiency gap” consider a maternally expressed locus. Among males, two genotypes (aA and aa) have lower fitness than the other two genotypes (Aa and AA). Selection is inefficient because favored A alleles are ignored in aA heterozygotes and disfavored a alleles are ignored in Aa heterozygotes. With male-limited selection, the aA heterozygote will always outnumber the Aa heterozygote (by a difference of ps − pe, from Equations 3c and 6c). When an allele is rare, it occurs only in heterozygotes. Thus, for maternally expressed loci, the average effect of the efficiency gap at low frequency of A is the loss of desired A alleles relative to an optimal system of selection. At high frequency, the a allele occurs only in heterozygotes and the result flips. Here, the paternally expressed pattern of imprinting has the average effect of inefficiently dragging along unwanted a alleles. The magnitude of w dictates the degree to which aA heterozygotes outnumber Aa heterozygotes, given by Equations 3c and 6c.
We have demonstrated for the first time, to our knowledge, an explicit consequence of the difference between the two reciprocal patterns of imprinting. We offer some discussion of the relevance of this consequence to livestock populations as well as to natural populations.
Consequences for livestock populations:
The most striking result of the models above is that when male-limited selection is strong, the response at maternally expressed loci is more efficient than the response at paternally expressed loci. Thus, if a breeder were aware that paternally expressed imprinted loci contribute to variation in the trait of interest, then consideration of alternatives to male-limited selection would be prudent. Female- rather than male-limited selection flips the result above: paternally expressed loci evolve more efficiently in response to female-limited selection than maternally expressed loci. However, selective deaths in females are undesirable as the number of breeding females is typically a limiting factor in production. Information from relatives may ameliorate some of the bias between maternally expressed and paternally expressed loci that male-limited selection introduces. What this information provides, essentially, is a window into the average effect of the silent allele. It is the difference between the average effect of the silent allele between paternally expressed and maternally expressed loci that causes the efficiency gap between the types of imprinting patterns. Paternally expressed loci are hiding an allele that certainly did not experience selection in the previous generation in a male; by contrast, maternally expressed loci have a 50% chance of having been seen by selection in the previous generation (that is, if they had been maternally derived in the sire of the focal individual). Thus, at maternally expressed loci, the allele that is hidden is more likely to be a favorable allele, relative to paternally expressed loci. Information from relatives helps recover some of the difference in what is known about the average effect of the silent allele at an imprinted locus.
Our models deal with single-generation changes. When selection is weak, it is not clear which type of imprinting pattern would lead to fixation of a rare A allele first. At low frequencies, paternally expressed imprinting is most efficient, while at higher frequencies (in fact, all above p > 0.5) the maternally expressed pattern is most efficient. An analytic solution to this problem would need to track four genotype frequencies with different fitnesses in males and females. Numerical simulations of our recursion equations find that the imprinting pattern that permits a faster fixation depends on the initial allele frequency (and depends on how one defines fixation in an infinite population). When favored alleles are initially rare, higher frequencies of favored alleles are reached faster at paternally expressed loci than at maternally expressed loci until the point in the dynamic at which the inefficiency of purging the disfavored alleles becomes limiting. This point is reached sooner for stronger selection. Some numerical examples are given in Figure 1.
One scenario that permits a clear prediction, regardless of selection strength, is when two breeds are hybridized with the intent of introgressing certain genetic traits from one breed into another. Hybridization will make all alleles begin at a frequency of p = 0.5. Thus, male-limited selection will reap more genetic gains if the gene of interest is maternally expressed than if it is paternally expressed because p = 0.5 is a high enough frequency such that for any strength of selection, the maternally expressed pattern of imprinted expression is more efficient. This follows from inequality (10), which can be used to show that the crossover between more efficiency at paternally expressed to maternally expressed imprinting patterns necessarily occurs when p ≤ 0.5.
Breeding programs that have not considered the possibility that imprinted loci contribute to traits of interest may have inefficiently tapped a store of imprinted genetic variation. A breeder who naively assigns fitness according to male breeding value (measured as the average value of his offspring) ignores any effects of maternally expressed imprinted genes. Under such a scheme, heritability will be higher on dams than on sires once paternally expressed imprinted variation is quenched. Conversely, if a breeder has naïvely assigned fitness according to phenotypes only, then such a breeder will be subject to the bias that we model above. Thus, this breeder is expected to have higher heritability for selected traits on sires than on dams. A combined approach, such as BLUP, which incorporates information from all relatives including the focal individual, will make up for some of the naïveté of the pure breeding value or pure phenotype breeding plans sketched above.
Consequences for natural populations:
Finally, in naturally selected populations, all favorable mutations arise in low frequency. Since selection is more efficient at driving change at paternally expressed loci when favorable alleles are rare, this may bias which types of alleles can invade a population under male-limited selection. Therefore, novel variants that are adaptive in males and unselected in females may more likely be lost by drift at maternally expressed loci early in their evolutionary dynamic. In this sense, maternally expressed loci may be disadvantaged in adaptation under male-limited selection.
Models with sex-specific fitness and parent-specific gene expression, such as the one presented here, present the possibility of unexpected evolutionary dynamics. Previous authors have shown that alleles at a modifier of imprinted expression can be selectively favored when a second trait-coding locus is subject to selection of varying direction or strength in the two sexes (Iwasa and Pomiankowski 2001, Day and Bonduriansky 2004, Spencer and Clark 2006, and Van Cleve and Feldman 2007 examine the X-linked version of this). Our model differs in several regards from these: ours is a one-locus model, imprinted expression is already established, and the imprinted pattern that is most conducive to genetic evolution depends on allele frequency. While all of these models address the complex situation in which imprinted expression meets a fitness scheme with two sexes, it is not clear how to translate between our model and these previous models.
Our model can be applied to natural and agricultural settings because selection is the same process whether it is a breeder or nature that imposes fitness differences. However, the effects of the bias demonstrated here are most likely to be seen in agricultural populations because these have the requisite degree of strong, sex-limited selection.
Acknowledgments
K. Donohue, J. Engelstädter, Z. Kaliszewska, T. Székely, and W. Tong all made helpful comments on early versions of this manuscript. We are also grateful to two anonymous reviewers for their constructive suggestions. M.M.P. was supported by a National Science Foundation Graduate Research Fellowship.
References
- Anderson, R. J. E., and H. G. Spencer, 1999. Population models of genomic imprinting. I. Differential viability in the sexes and the analogy with genetic dominance. Genetics 153 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, A., and R. L. Trivers, 2006. Genes in Conflict. Harvard University Press, Cambridge, MA.
- Cockett, N. E., S. P. Jackson, T. L. Shay, F. Farnir, S. Berghmans et al., 1996. Polar overdominance at the ovine callipyge locus. Science 273 236–238. [DOI] [PubMed] [Google Scholar]
- Cui, Y. H., 2007. A statistical framework for genome-wide scanning and testing of imprinted quantitative trait loci. J. Theor. Biol. 244 115–126. [DOI] [PubMed] [Google Scholar]
- Cui, Y. H., Q. Lu, J. M. Cheverud, R. C. Littell and R. L. Wu, 2006. Model for mapping imprinted quantitative trait loci in an inbred F-2 design. Genomics 87 543–551. [DOI] [PubMed] [Google Scholar]
- Day, T., and R. Bonduriansky, 2004. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 167 1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning, D. J., A. P. Rattink, B. Harlizius, J. A. M. van Arendonk, E. W. Brascamp et al., 2000. Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc. Natl. Acad. Sci. USA 97 7947–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning, D. J., H. Bovenhuis and J. A. M. van Arendonk, 2002. On the detection of imprinted quantitative trait loci in experimental crosses of outbred species. Genetics 161 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, A. G., R. Kerr, B. Tier, T. Long and T. H. E. Meuwissen, 1994. Gametic imprinting effects on rate and composition of pig growth. Theor. Appl. Genet. 88 1037–1042. [DOI] [PubMed] [Google Scholar]
- Engellandt, T., and B. Tier, 2002. Genetic variances due to imprinted genes in cattle. J. Anim. Breed. Genet. 119 154–165. [Google Scholar]
- Essl, A., and K. Voith, 2002. a Estimation of variance components due to imprinting effects with DFREML and VCE: results of a simulation study. J. Anim. Breed. Genet. 119 1–14. [Google Scholar]
- Essl, A., and K. Voith, 2002. b Genomic imprinting effects on dairy- and fitness-related traits in cattle. J. Anim. Breed. Genet. 119 182–189. [Google Scholar]
- Iwasa, Y., and A. Pomiankowski, 2001. The evolution of X-linked genomic imprinting. Genetics 158 1801–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J. T., O. Carlborg, A. Tornsten, E. Giuffra, V. Amarger et al., 1999. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat. Genet. 21 157–158. [DOI] [PubMed] [Google Scholar]
- Knott, S. A., L. Marklund, C. S. Haley, K. Andersson, W. Davies et al., 1998. Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics 149 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Press, Sunderland, MA.
- Mantey, C., G. A. Brockmann, E. Kalm and N. Reinsch, 2005. Mapping and exclusion mapping of genomic imprinting effects in mouse F-2 families. J. Hered. 96 329–338. [DOI] [PubMed] [Google Scholar]
- McGrath, J., and D. Solter, 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37 179–183. [DOI] [PubMed] [Google Scholar]
- Morison, I. M., J. P. Ramsay and H. G. Spencer, 2005. A census of mammalian imprinting. Trends Genet. 21 457–465. [DOI] [PubMed] [Google Scholar]
- Mrode, R. A., 2005. Linear Models for the Prediction of Animal Breeding Values. CABI, Cambridge, MA.
- Nezer, C., L. Moreau, B. Brouwers, W. Coppieters, J. Detilleux et al., 1999. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat. Genet. 21 155–156. [DOI] [PubMed] [Google Scholar]
- Pearce, G. P., and H. G. Spencer, 1992. Population genetic models of genomic imprinting. Genetics 130 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer, K., 2000. Mechanisms of genomic imprinting. Am. J. Hum. Genet. 67 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik, W., and J. Walter, 2001. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2 21–32. [DOI] [PubMed] [Google Scholar]
- Ruvinsky, A., 1999. Basics of gametic imprinting. J. Anim. Sci. 77 228–237. [DOI] [PubMed] [Google Scholar]
- Schaeffer, L. R., B. W. Kennedy and J. P. Gibson, 1989. The inverse of the gametic relationship matrix. J. Dairy Sci. 72 1266–1272. [Google Scholar]
- Spencer, H. G., and A. G. Clark, 2006. A chip off the old block: a model for the evolution of genomic imprinting via selection for parental similarity. Genetics 174 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella, A., K. J. Stalder, A. M. Saxton and P. J. Boettcher, 2003. Estimation of variances for gametic effects on litter size in Yorkshire and Landrace swine. J. Anim. Sci. 81 2171–2178. [DOI] [PubMed] [Google Scholar]
- Surani, M. A. H., S. C. Barton and M. L. Norris, 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308 548–550. [DOI] [PubMed] [Google Scholar]
- Tier, B., and J. Solkner, 1993. Analyzing gametic variation with an animal-model. Theor. Appl. Genet. 85 868–872. [DOI] [PubMed] [Google Scholar]
- Van Cleve, J., and M. W. Feldman, 2007. Sex-specific viability, sex linkage and dominance in genomic imprinting. Genetics 176 1101–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere, A. S., M. Nguyen, M. Braunschweig, C. Nezer, C. Collette et al., 2003. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425 832–836. [DOI] [PubMed] [Google Scholar]